3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(11):2480-2487. doi:10.7150/jca.29489 This issue Cite
Research Paper
SNAI2 3'untranslated region promotes the invasion of ovarian cancer cells by inducing MARCKS expression
1. Obstetrics and Gynecology Hospital, Fudan University, Shanghai 200011, China
2. Department of Obstetrics and Gynecology of Shanghai Medical College, Fudan University, Shanghai 200032
3. Shanghai Key Laboratory of Female Reproductive Endocrine Related Diseases, Shanghai 200011, China
Received 2018-8-26; Accepted 2019-5-2; Published 2019-6-2
Abstract
Objective: Recently, accumulating evidence has indicated that the 3' untranslated regions (3'UTRs) of protein coding genes play critical roles in the progression of various cancers, including ovarian cancer. This study is aimed to identify the potential role of SNAI2-3'UTR in ovarain cancer progression.
Study Design: First, we tried to explore the clinical significance of SNAI2 in ovarian cancer using TCGA and GSE26712 dataset. Then, gain-of-function studies were performed to establish the role of SNAI2-3'UTR in invasion and migration of ovarian cancer cells. Finally, efforts were made to identify the downstream targets of SNAI2-3'UTR.
Results: Our data indicated that the expression of SNAI2 was significantly correlated with FIGO stage (P=0.015) and lymphatic invasion status (P=0.004), whereas not with age(P>0.05) and histological grade(P>0.05). Patients with higher SNAI2 expression had a shorter overall survival (OS) in both TCGA dataset (P=0.039, HR=1.54(1.02-2.33)) and GSE26712 dataset (P=0.0017, HR=1.77(1.24-2.54)). Functional studies revealed that SNAI2-3'UTR promoted the invasion of both OVCA433 and SKOV-3 cells without significantly affecting their migratory abilities. MARCKS, which was also involved in the invasion of ovarian cancer cells, was identified as a potential downstream target of SNAI2-3'UTR. SNAI2-3'UTR may function as a ceRNA to upregulate MARCKS expression in ovarian cancer.
Conclusion: In conclusion, our study demonstrated that SNAI2-3'UTR cloud promote the invasion of ovarian cancer cells by upregulating MARCKS expression, which proposed a new mechanism by which SNAI2 contributed to progression of ovarian cancer.
Keywords: Ovarian cancer, SNAI2, MARCKS, 3' untranslated regions, Invasion
1. Introduction
Ovarian cancer is the most lethal gynecological cancer and the majority of the patients are not diagnosed until advanced stages. Over the past decades, little improvement in overall survival has been achieved despite advances in chemotherapeutic agents. The standard treatment for epithelial ovarian cancer is aggressive debulking surgery followed by a platinum and taxane combination chemotherapy, and debulking status is a crucial prognostic factor for patients with ovarian cancer(1). However, due to the aggressive metastatic nature of ovarian cancer, surgery rarely leads to complete tumor resection. Thus, a better understanding of the mechanisms involved in ovarian cancer invasion and metastasis is of great importance to overcome this malignancy.
SNAI2 is a key epithelial-to-mesenchymal trasition (EMT) associated transcription factor. Previous studies have indicated that repression of EMT is a complex process controlled by multi-layer regulations, including but not limited to EMT-inducing transcription factors(EMT-TFs), non-coding RNAs, alternative splicing, and translational and post-translational control (which affect protein stabilization and localization). Among them, the network built around EMT-TFs is most extensively studied. SNAI2 is one of these key EMT-TFs and previous studies have indicated that repression of SNAI2 by RNA interference, which could reduce both SNAI2 mRNA and SNAI2 protein, greatly reverses the EMT process and thereby decreases metastatic ability of various cancer cells(2-6). Since the competing endogenous RNA(ceRNA) hypothesis indicates that the 3'untranslated regions (3'UTR) of protein coding genes can function as natural miRNA sponges(7-14), functionally freeing other transcripts targeted by that panel of miRNAs, it is possible that the SNAI2-3'UTR could function independently of the protein in cancer progression. That is to say SNAI2-3'UTR may regulate the progression process of ovarian cancer by functioning as a ceRNA.
In this study, we try to establish the roles of SNAI2-3'UTR in ovarian cancer and to identify its downstream targets, which may add a layer of regulation to the progression process controlled by SNAI2.
2. Materials and Methods
2.1 TCGA datasets
We downloaded the level 3 Affymetrix HG-U133A gene expression data for SNAI2 and MARCKS from 208 patients with serous ovarian cancer in TCGA dataset to analyze the association of SNAI2 expression with the clinicopathological features, and to determine the correlation between SNAI2 expression and MARCKS expression.
2.2 GEO datasets
GSE26712 dataset is used to explore the relationship between SNAI2 expression and the clinical outcome in 185 patients with serous ovarian cancer, and to determine the correlation between SNAI2 expression and MARCKS expression.
2.3 Cell lines
Human ovarian cancer cell lines OVCA433 and SKOV-3 were purchased from the cell bank of Chinese Academy of Science (Shanghai) and cultured in RPMI1640 medium plus 10% fetal bovine serum with penicillin/streptomycin. 293T cell line was originally obtained from the cell bank of Chinese Academy of Science (Shanghai) and cultured in DMEM plus 10% fetal bovine serum with penicillin/streptomycin.
2.4 Constructs
The full length of SNAI2-3'UTR (Gene ID: 6591; NCBI Reference Sequence: NM_003068.4) was cloned into pCDH vector (No.CD510B-1, System Biosciences) using restriction enzymes NheI-HF and NotI-HF (New England Biolabs). And the full length of SNAI2-3'UTR was also cloned into pcDNA3.1 vector (Invitrogen). Additionally, a fragment of MARCKS-3'UTR (1124bp, from+43 to +1166; Gene ID: 6591; NCBI Reference Sequence: NM_003068.4) was cloned into psiCHECKTM-2 vector (Promega) using restriction enzymes XhoI and NotI (New England Biolabs). The reconstructed vector was named psi-MARCKS-3'UTR. Primers for cloning were shown in Table S1 (supplementary data). PLKO-Scramble-shRNA was purchased from Addgene (Addgene #1864). PLKO-MARCKS-shRNA plasmids are constructed according to PLKO.1 protocol. The sequences of MARCKS shRNAs are obtained from The RNAi Consortium (TRC, MISSION® TRC shRNA library, Sigma) and shown in Table S1 (supplementary data).
2.5 Lentiviral infection
For producing lentiviral particles, 293T cells were seeded in 3.5-cm dishes at a density of 4×105 per dish and transfected with pCDH-Vector/pCDH-SNAI2-3'UTR plasmids or PLKO-Scramble-shRNA/PLKO-MARCKS-shRNA, as well as packing plasmids psPAX2 (0.75µg) (Addgene#12260) and pMD2.G (0.25µg) (Addgene #12259) using Attractene Transfection Reagent (QIAGEN). Forty-eight hours post transfection, virus-containing medium (2ml) was harvested, filtered, mixed with 2ml of freshsly prepared medium, supplemented with 8µg/ml polybrene (Sigma) and added to 3×105 OVCA433 cells or SKOV-3 cells seeded in a 3.5-cm dish the day before. Puromycin (2µg/ml) was supplemented 48h after infection. The cells were selected for 2 days and then used for various assays.
2.6 Real-time RT-PCR
Briefly, reverse transcription reactions were carried out using RevertAidTM First Strand cDNA Synthesis Kit (Fermentas). Real-Time PCR was performed using SYBR® Premix Ex Taq™ II (Perfect Real Time) Kit (TaKaRa, Dalian, China) in ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Quantitative analysis was performed using Comparative CT method. The relative expression of each gene was normalized to the expression of GAPDH. Primers used in present study were shown in Table S1 (supplementary data).
2.7 Western Blot
Whole cell extracts were prepared in chilled RIPA lysis buffer (Beyotime, China). 630 µg of lysates protein were separated by SDS-PAGE using a 10% polyacrylamide gel and transferred to 0.45µm PVDF membrane (Millipore). Membranes were blocked with 5% non-fat milk in PBS containing 0.05% Tween-20, blotted with MARCKS antibody (1:1000, CST) or Actin antibody (1:3000, beyotime) overnight, followed by goat anti-rabbit antibody or goat anti-mouse antibody. The images were scanned by LAS4000 device.
2.8 Real-time measurement of cell migration and invasion
RTCA CIM-Plate was used to explore the migration and invasion of ovarian cancer cells. Migration and invasion assays were performed according to protocols supplied by the manufacturers. For invasion assay, the inserts were coated with BD Matrigel (B.D. Biosciences). Briefly, 4×104 Scramble-shRNA- and MARCKS-shRNA-transfected OVCA433 cells or pCDH-vector- and pCDH-SNAI2-3'UTR-transfected OVCA433 cells or SKOV-3 cells were added into the RTCA CIM-Plate and monitored by xCELLigence (Roche) for 24 hours respectively.
2.9 Dual luciferase reporter assay
For the dual luciferase reporter assays, 1×105 OVCA433 cells were seeded in 24-well culture plate and cotransfected with 25ng psi-MARCKS-3'UTR, and an increasing dose of pcDNA3.1-Vector or pcDNA3.1-SNAI2-3'UTR, using Attractene Transfection Reagent (QIAGEN). Twenty-four hours later, luciferase activities were measured by dual luciferase reporter assay kit (Promega).
2.10 Statistical analysis
All data were analyzed with SPSS statistic software (SPSS 16.0). Student's t test (two-tailed) was used to compare two groups. Chi-square test was used to evaluate the differences between groups for categorical variables. The Log-rank test was used to determine the relationship between SNAI2 expression and clinical outcomes. The Kaplan-Meier method was used to generate survival curves. Pearson correlation analysis was performed to determine the coexpression status between SNAI2 and MARCKS. P-value<0.05 was considered statistically significant.
3. Results
3.1 The relationship between SNAI2 expression and the clinicopathological parameters in ovarian cancer patients
Initially, we sought to explore the relationship between SNAI2 expression and the clinicopathological features for 208 patients with serous ovarian cancer using data from the TCGA data portal. The median expression value is used as the cutoff point. The results revealed that increased expression of SNAI2 was significantly correlated with FIGO stage (P=0.015, χ2 test) and lymphatic invasion status (P=0.004, χ2 test), whereas not with age and histological grade (Table 1). Namely, the expression of SNAI2 was higher in patients classified as FIGO Stage III/IV than that classified as FIGO Stage I/II (Fig. 1A), and was higher in patients with lymphatic invasion than that without lymphatic invasion (Fig. 1B). Furthermore, Log Rank (Mantel-Cox) analysis revealed that increased expression of SNAI2 was associated with shorter overall survival (OS) (P=0.039, HR=1.54 (1.02-2.33); Fig. 1C). This result was confirmed in GSE26712 dataset (P=0.0017, HR=1.77 (1.24-2.54); Fig. 1D).
The relationship between SNAI2 expression and clinicopathological parameters in 208 primary ovarian cancer from TCGA datasets
| Variables | No. of patients | SNAI2 expression | P value | |
|---|---|---|---|---|
| Low, n(%) | High, n(%) | |||
| Age (years) | 0.325 | |||
| ≤60 | 121 | 57(47.1%) | 64(52.9%) | |
| >60 | 87 | 47(54.0%) | 40(46.0%) | |
| Histologic grade | 0.149 | |||
| G1/G2 | 27 | 17(63.0%) | 10(37.0%) | |
| G3 | 181 | 87(48.1%) | 94(51.9%) | |
| FIGO stage | 0.015 | |||
| I-II | 23 | 17(73.9%) | 6(26.1%) | |
| III-IV | 185 | 87(47.0%) | 98(53.0%) | |
| Lymphatic invasion | 0.004 | |||
| Yes | 130 | 55(42.3%) | 75(57.7%) | |
| No | 78 | 49(62.8%) | 29(37.2%) | |
3.2 SNAI2-3'UTR promoted invasion of ovarian cancer cells
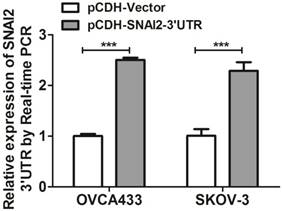
Next, we tried to establish the role of SNAI2-3'UTR in invasion and migration of ovarian cancer cells. We used lentivirus to stably overexpress SNAI2-3'UTR in OVCA433 and SKOV-3 cells, respectively (Fig. 2). Our data indicated that overexpression of SNAI2-3'UTR increased the invasive ability of both OVCA433 and SKOV-3 cells (Fig. 3A & 3B) without significantly affecting their migratory property (Fig. 3C & 3D).
3.3 MARCKS was a potential downstream targets of SNAI2-3'UTR
To identify the potential targets of SNAI2-3'UTR, we examined the differential expression of several invasion-associated genes between control group and SNAI2-3'UTR overexpression group. It was found that overexpression of SNAI2-3'UTR significantly upregulated the mRNA level of MARCKS and TWIST1 in OVCA433 cells (Fig. 4A), and the mRNA level of MARCKS in SKOV-3 cells (Fig. 4B). Next, we focused on MARCKS because a significant alteration of MARCKS abundance was observed after SNAI2-3'UTR overexpression in both OVCA433 and SKOV-3 cells. Western blot analysis further confirmed the positive regulation of SNAI2-3'UTR on MARCKS protein level (Fig. 4C & 4D).
The differential expression of SNAI2 in different groups. (A) The expression of SNAI2 was higher in patients classified as FIGO Stage III/IV than that classified as FIGO Stage I/II; (B) The expression of SNAI2 was higher in patients with lymphatic invasion than that without lymphatic invasion; (C, D) Log Rank (Mantel-Cox) analysis revealed that increased expression of SNAI2 was associated with shorter overall survival in both the TCGA dataset (C) and the GEO dataset (D). ''Low'' and “High'' were classified according to the SNAI2 expression level. The median expression value for SNAI2 was used as the cutoff point.
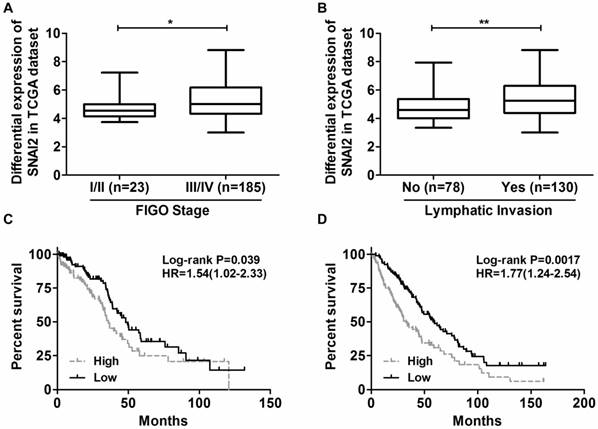
Overexpression of SNAI2-3'UTR in OVCA433 and SKOV-3 cells validated by Real-time PCR.
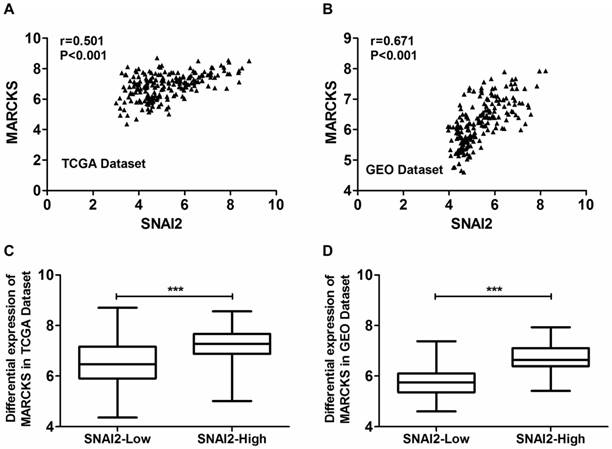
Moreover, the expression of MARCKS was significantly positively correlated with the expression of SNAI2 in both TCGA dataset (Fig. 5A) and GSE26712 dataset (Fig. 5B). Moreover, MARCKS showed differential expression when samples are subdivided according to SNAI2 expression levels in both TCGA dataset (Fig. 5C) and GSE26712 dataset (Fig. 5D).
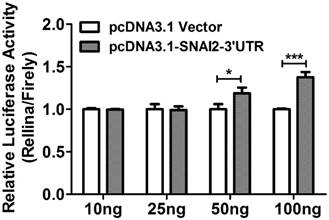
Bioinformatics analysis based on TargetScan 6.2 revealed that SNAI2 and MARCKS shared multiple miRNA binding sites (MREs), and these MREs were mainly located on the former 1000bp region of MARCKS-3'UTR. To ascertain whether the observed regulatory effect of SNAI2-3'UTR on the expression of MARCKS is dependent upon its interaction with MARCKS-3'UTR, we constructed a luciferase reporter construct fused to a fragment of MARCKS-3'UTR (1124bp, from +43 to +1166). Interestingly, overexpression of SNAI2-3'UTR led to a significant increase of the luciferase activity in a dose dependent manner (Fig.6).
3.4 Attenuated expression of MARCKS impaired the invasion and migration of OVCA433 cells
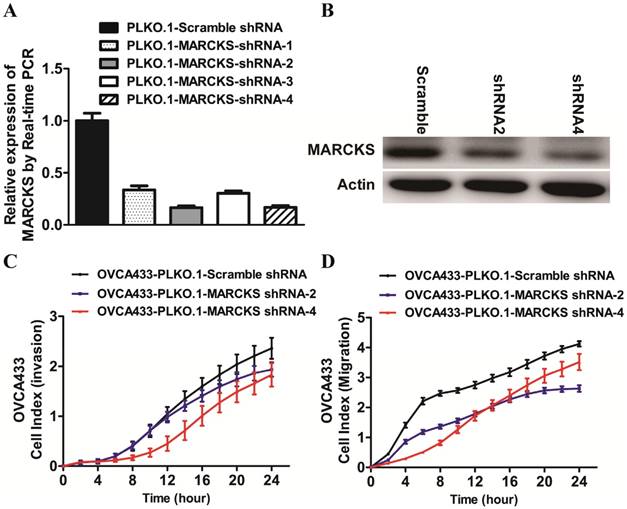
To determine whether deregulated expression of MARCKS contributed to the progression of ovarian cancer, we performed loss-of-function studies in OVCA433 cells. Four lentiviral vectors encoding MARCKS-shRNA1, MARCKS-shRNA2, MARCKS-shRNA3, and MARCKS-shRNA4 were constructed and found to downregulate MARCKS mRNA level by 66.4%, 83.4%, 69.5%, and 83.0% in OVCA433 cells respectively (Fig. 6A). Western blot analysis further confirmed the inhibitory effect of MARCKS-shRNA2 and MARCKS-shRNA4 on MARCKS expression (Fig. 6B). So, we chose MARCKS-shRNA2 and MARCKS-shRNA4 for subsequent studies. Interestingly, it was found that the invasion and migration of OVCA433 cells was significantly suppressed by MARCKS downregulation (Fig. 6C & 6D).
The effect of SNAI2-3'UTR on migration and invasion of ovarian cancer cells monitored by xCELLigence. (A, B, C, D) Overexpression of SNAI2-3'UTR significantly induced invasion in OVCA433 (A) and SKOV-3 (B) cells, without significantly affecting their migratory ability (C, D).
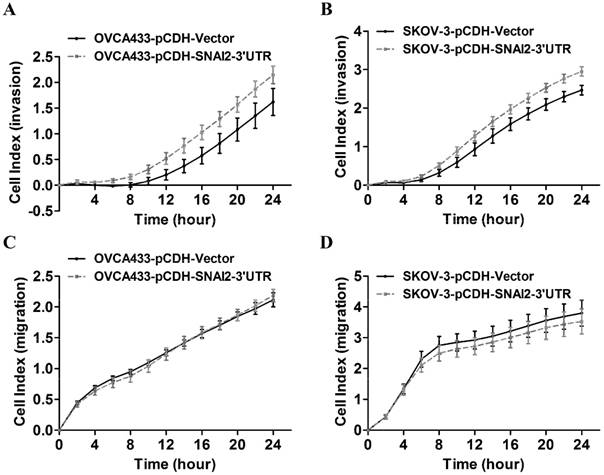
Screen and identification of the potential downstream targets of SNAI2-3'UTR. (A, B) The regulation of SNAI2 3'UTR on the mRNA level of invasion-associated genes in OVCA433 (A) and SKOV-3 (B) cells; (C, D) SNAI2 3'UTR overexpression significantly induced the protein level of MARCKS in OVCA433 (C) and SKOV-3 (D) cells;
Coexpression analysis of SNAI2 and MARCKS in primary ovarian cancer tissues from the TCGA dataset and GEO dataset. (A, B) The expressionf of SNAI2 was significantly positively correlated with the expression of MARCKS in both TCGA (A) and GEO (B) datasets. (C, D) MARCKS was differentially expressed between “SNAI2-low” group and “SNAI2-high” group in both TCGA (C) and GEO (D) datasets.
The relative luciferase activity in OVCA433 cells co-transfected with psi-MARCKS-3'UTR, and an increasing dose of pcDNA3.1-Vector or pcDNA3.1-SNAI2-3'UTR.
4. Discussion
The findings presented herein have allowed us to reach some important conclusions. First, elevated expression of SNAI2-3'UTR was associated with shorter OS in patients with serous ovarian cancer. Second, SNAI2-3'UTR could promote the invasion of ovarian cancer cells at least partly by upregulating MARCKS expression. These findings propose a new mechanism by which SNAI2 promotes the progression of ovarian cancer.
Traditionally, the functional characterization of protein-coding genes has mainly focused on the role of their coding sequences. However, the untranslated regions of protein-coding genes have received undue attention. In our previous review, we have made a summary that the reprogramming of 3'UTRs by various means is a common phenomenon in cancer and disturbances of 3'UTRs significantly increase the risk of cancer susceptibility by acting both in cis and in trans(15). In this study, we demonstrated that SNAI2-3'UTR could promote the invasion of ovarian cancer cells at least partly by upregulating MARCKS expression.
Previously, artificial miRNA decoys termed “miRNA sponges” were employed to create loss-of-function phenotypes for specific miRNAs in various disease models(16, 17). It is tempting to speculate that the nature may also create such natural miRNA sponges to sequence-specifically sequester miRNAs. Indeed, evidence supporting this hypothesis has emerged and a theory termed ceRNA hypotheis was born(7, 18). The ceRNA hypothesis indicates that specific RNAs, including but not limited to mRNAs, transcribed pseudogenes, circular RNAs, and other lncRNAs, can function as natural miRNA sponges, functionally freeing other transcripts targeted by that panel of miRNAs and thereby leading to phenotypic changes(7). In this study, we demonstrated that SNAI2-3'UTR could upregulate MARCKS expression by interacting with MARCKS-3'UTR. Correlation analysis further revealed that SNAI2 and MARCKS were coexpressed in clinical samples from GEO dataset. Subsequent loss-of-function studies demonstrated that MARCKS was involved in the invasion of ovarian cancer cells. These findings supported the notion that SNAI2-3'UTR may function as a ceRNA to regulate MARCKS expression.
The effect of MARCKS inhibition on the EMT process in OVCA433 cells. (A)The mRNA level of MARCKS was significantly inhibited in cells transfected with MARCKS-shRNA-1/-shRNA-2/-shRNA-3/shRNA-4 compared with cells transfected with scramble shRNA. (B) The protein level of MARCKS was significantly decreased in cells transfected with MARCKS-shRNA-2/-shRNA-4 compared with cells transfected with scramble shRNA. (C, D) Downregulation of MARCKS expression significantly repressed invasion (C) and migration (D) of OVCA433 cells monitored by RTCA xCELLigence.
However, it should be noted that the noncoding function of SNAI2-3'UTR in promoting invasion may go beyond its ability to modulate the levels of MARCKS only. Some other invasion associated genes may be also subjected to the regulation by SNAI2-3'UTR. For example, TWIST1 was another potential target of SNAI2-3'UTR as indicated by our data. In addition to the 3'UTR, the binding sites for miRNAs may be also located in other regions of the mRNA transcript, including the coding regions and 5'UTR(19, 20). Thus, the entire transcript of SNAI2 mRNA could have decoy activity. The challenge ahead is to identify the whole network of SNAI2 associated ceRNAs. Furthermore, the 3'UTRs could function as decoys for not only miRNAs but also for other trans-acting factors(21). For instance, overexpression of HMGA1 pseudogene (HMGA1-p) 3'UTR led to the destabilization of HMGA1 mRNA transcript possibly by competing for negative trans factors(21). Thus, it remains to be seen whether SNAI2-3'UTR has such functions. Efforts toward this would help us fully understand the noncoding function of SNAI2.
In conclusion, our study demonstrated that SNAI2-3'UTR could function as a ceRNA to regulate MARCKS expression, which proposed a new mechanism by which SNAI2 induced EMT in ovarian cancer.
Supplementary Material
Supplementary table.
Acknowledgements
This study is sponsored by grants from Shanghai Sailing Program (No.16YF1401100), Natural Science Foundation of China (No.81802596), and Natural Science Foundation of Shanghai (No.17ZR1403500).
Author Contribution
Conception and design: Jun Li, Xin Lu.
Collection and assembly of data: All authors.
Data analysis and interpretation: All authors.
Manuscript writing: Jun Li, Xin Lu.
Final approval of manuscript: All authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. NAT REV CLIN ONCOL. 2013;10:211-24
2. Olmeda D, Montes A, Moreno-Bueno G. et al. Snai1 and Snai2 collaborate on tumor growth and metastasis properties of mouse skin carcinoma cell lines. ONCOGENE. 2008;27:4690-701
3. Emadi BM, Soheili ZS, Essmann F. et al. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol. 2010;31:297-307
4. Wang C, Liu X, Huang H. et al. Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. INT J CANCER. 2012;130:2249-58
5. Uygur B, Wu WS. SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. MOL CANCER. 2011;10:139
6. Toiyama Y, Yasuda H, Saigusa S. et al. Increased expression of Slug and Vimentin as novel predictive biomarkers for lymph node metastasis and poor prognosis in colorectal cancer. CARCINOGENESIS. 2013;34:2548-57
7. Salmena L, Poliseno L, Tay Y. et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? CELL. 2011;146:353-8
8. Poliseno L, Salmena L, Zhang J. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. NATURE. 2010;465:1033-8
9. Karreth FA, Tay Y, Perna D. et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. CELL. 2011;147:382-95
10. Tay Y, Kats L, Salmena L. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. CELL. 2011;147:344-57
11. Cesana M, Cacchiarelli D, Legnini I. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. CELL. 2011;147:358-69
12. Jeyapalan Z, Deng Z, Shatseva T. et al. Expression of CD44 3'-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. NUCLEIC ACIDS RES. 2011;39:3026-41
13. Kumar MS, Armenteros-Monterroso E, East P. et al. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. NATURE. 2014;505:212-7
14. Fang L, Du WW, Yang X. et al. Versican 3'-untranslated region (3'-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013;27:907-19
15. Li J, Lu X. The emerging roles of 3' untranslated regions in cancer. CANCER LETT. 2013;337:22-5
16. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. NAT METHODS. 2007;4:721-6
17. Franco-Zorrilla JM, Valli A, Todesco M. et al. Target mimicry provides a new mechanism for regulation of microRNA activity. NAT GENET. 2007;39:1033-7
18. Li J, Lu X. The emerging roles of 3' untranslated regions in cancer. CANCER LETT. 2013;337:22-5
19. Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667-72
20. Tay Y, Zhang J, Thomson AM. et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. NATURE. 2008;455:1124-8
21. Chiefari E, Iiritano S, Paonessa F. et al. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. NAT COMMUN. 2010;1:40
Author contact
![]() Corresponding author: Prof. Xin Lu, Obstetrics and Gynecology Hospital, Fudan University, No.419, Fangxie Road, Shanghai, 200011, China, 86; Tel: 86-21-33189900; E-mail: xin_luedu.cn
Corresponding author: Prof. Xin Lu, Obstetrics and Gynecology Hospital, Fudan University, No.419, Fangxie Road, Shanghai, 200011, China, 86; Tel: 86-21-33189900; E-mail: xin_luedu.cn

 Global reach, higher impact
Global reach, higher impact