Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(12):2670-2678. doi:10.7150/jca.32072 This issue Cite
Research Paper
SRPX2 and RAB31 are effective prognostic biomarkers in pancreatic cancer
1. Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, Shanghai 20032, China.
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
3. Shanghai Pancreatic Cancer Institute, Shanghai 200032, China.
4. Pancreatic Cancer Institute, Fudan University, Shanghai 200032, China.
Hao Li and Shi-Rong Zhang contributed equally to this work.
Received 2018-12-7; Accepted 2019-5-1; Published 2019-6-2
Abstract
Introduction: SRPX2 and RAB31 play important roles in tumorigenesis and metastasis; however, their prognostic value in pancreatic cancer remains unclear. This study aimed to investigate the potential interactions and effects of SRPX2 and RAB31 on the diagnosis and prognosis of pancreatic cancer.
Methods: The expression of SRPX2 and RAB31 in pancreatic tumor tissues and cells was evaluated through database mining of the Oncomine, Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases, and validated the results through immunohistochemistry (IHC) and Western blot in our clinical database. Protein-protein interactions were explored by immunofluorescence and Co-immunoprecipitation (Co-IP). Two hundred tissue microarray specimens from patients (79 training and 121 validation), who underwent curative pancreatectomy for pancreatic ductal adenocarcinoma (PDAC) were used. Additionally, the association between the SRPX2 and RAB31 and prognosis of PDAC patients after surgery was analyzed.
Results: The expression of SRPX2 and RAB31 was highly increased in pancreatic cancer, and there was a significant positive correlation between these two proteins. Co-IP showed the direct interaction between SRPX2 and RAB31. Kaplan-Meier analysis showed that positive expression of SRPX2 and RAB31 was associated with reduced disease-free survival (DFS) and overall survival (OS) of PDAC patients in the training set and the validation sets. Furthermore, multivariate analysis indicated that the 8th edition TNM stage and combination of SRPX2 and RAB31 were independent prognostic factors that associated with OS and DFS in the training, and the validation sets, respectively.
Conclusions: The combination of SRPX2 and RAB31 can be important markers for the prognosis of pancreatic cancer.
Keywords: Pancreatic ductal adenocarcinoma, SRPX2, RAB31, Prognosis
Introduction
Pancreatic cancer is one of the leading causes of cancer mortality in developed countries, resulting in a substantial global burden, and pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer [1]. The incidence of PDAC is projected to increase dramatically, becoming the second leading cause of cancer-related deaths before 2030 [2]. An investigation of 678,842 patients with invasive cancer in China between 2003 and 2013 indicated that 5-year relative survival has not improved in the past decade, being only 7.3% [3]. Therefore, prognostic factors are indispensable for providing personalized treatment to PDAC patients with an individualized balance between treatment efficacy and side effects. Some progress has been made in identifying prognostic molecules for PDAC. For example, SPARC overexpression is correlated with poor prognosis [4], elevated CA19-9 serum levels are associated with poor prognosis [5], and miRNAs have emerged as potential prognostic biomarkers for PDAC. However, due to the poor sensitivity and specificity of currently available markers, the identification of new biomarkers with increased specificity is urgently needed.
Sushi repeat-containing protein, X-linked 2 (SRPX2) is a secreted extracellular matrix protein that has recently emerged as a multifunctional molecule involved in seizure disorders, angiogenesis, and cellular adhesion [6]. Recently, SRPX2 expression was demonstrated to be upregulated in colorectal cancer [7], and gastric cancer [8]. In these malignancies, SRPX2 is commonly considered an oncogene that promotes the proliferation, migration, and invasion of cancer cells. However, the clinical significance and biological role of SRPX2 in pancreatic cancer remains largely unknown.
RAB31, a member of the Ras superfamily, has been reported to play a role in tumor development and progression, and dysregulation of Rab small GTPases leads to various diseases, including cancers [9]. Therefore, many studies have also reported that RAB31 is widely involved in the molecular pathways underlying cancer progression [10]. For instance, Grismayer et al reported that high expression of RAB31 is frequently observed in breast cancer tissues and is associated with uncontrolled cell proliferation [11]. Additionally, RAB31 protein has also been reported to be correlated with the overall survival of patients with liver cancer [12]. However, no study has reported whether there is a significant association between RAB31 expression and the progression and prognosis of pancreatic cancer.
The present study sought to examine the expression of SRPX2 and RAB31 in tumor tissues and cells and to investigate the possible correlation and interaction between SRPX2 and RAB31 expression in pancreatic cancer. Additionally, we analyzed the associations between clinicopathological variables, including the expression of SRPX2 and RAB31, and clinical outcomes of postsurgical PDAC patients. Finally, SRPX2 and RAB31 were shown to be potential prognostic biomarkers for pancreatic cancer.
Materials and Methods
Patients and Specimens
79 clinical specimens as training set and 121 validation set were obtained from patients who underwent R0 resection for PDAC at Shanghai Cancer Center (Shanghai, China) from January 2012 to December 2013 and from January 2014 to December 2015. All specimens were independently assessed by two experienced pathologists according to the 8th edition of the American Joint Committee on Cancer (AJCC) TNM staging system. All patients in this study were resectable PDAC without distant metastasis, and none of the patients received preoperative anticancer treatment.
Overall survival (OS) was calculated as the interval between surgery and death or the date of last follow up. Disease-free survival (DFS) is defined as the interval between surgery and recurrence or the last date of follow up. All patients were followed through June 2018. Research Ethics Committees at the associated pancreatic centers approved the use of human tissues, and informed consent was obtained from all patients according to the committees' regulations.
Cell lines
Human pancreatic cancer cell lines (MIA, PaCa2) was obtained from the American Type Culture Collection (Manassas, VA, USA) and were cultured in DMEM with 10% fetal bovine serum (Gibco) supplemented with 100 μ g/ml streptomycin and 100 U/ml penicillin, and were maintained in humidified atmosphere of 5% CO2 at 37 °C.
ONCOMINE analysis
Oncomine is a cancer microarray database and web-based data mining platform by which people accelerate new and significant discoveries using genome-wide expression analyses. Expression and co-expression of distinct SRPX2 and RAB31 at the mRNA level in cancers was detected through analysis of the ONCOMINE database (www.Oncomine.org). In this study, Student's t-test was used to generate a p-value for comparison between cancer specimens and normal control datasets. Fold change was defined as 2 and P-value was set as 0.05.
GEO and TCGA data sources
Public gene expression profiles of GSE15471 were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo), a public functional genomic data repository. GSE15471 contains 39 pairs of tissues (PDAC tissues and abnormal tissues). Additionally, the RNA-sequencing (RNA-seq) dataset of SRPX2 and RAB31 were downloaded from TCGA (http://portal.gdc.cancer.gov/) for co-expression analysis. Fold change was defined as 2 and p-value was set as 0.05.
CCLE analysis
CCLE is a popular online encyclopedia containing a compilation of gene expression, chromosomal copy number, and massively parallel sequencing data, which facilitates identification of genetic, lineage, and predictors of drug sensitivity. Expression of SRPX2 and RAB31 at the mRNA level in pancreatic cancer cells was analyzed using the CCLE database (https://portals.broadinstitute.org/ccle/home).
Immunofluorescence staining assay
PDAC tumors were dissected and fixed in 2% PAF overnight at 4℃, dehydrated, and embedded in paraffin. Sections (5 µm thickness) were then incubated with anti-SRPX2 antibody (1:400, Abcam, Shanghai, China) or anti-RAB31 antibody (1:100, Abcam, Shanghai, China). Finally, images were acquired using immunofluorescence microscopy.
Immunohistochemistry (IHC) and Western Blot Analysis
IHC and western blot were performed as previously described [13].
Co-immunoprecipitation (Co-IP)
Cell lysates were immunoprecipitated with rabbit anti-SRPX2, anti-RAB31 antibody, and IgG to detect the interactions of endogenous proteins. The precipitates were separated by SDS-PAGE and analyzed by immunoblotting.
Statistical analysis
All statistical analyses were performed using SPSS v22.0 software (IBM SPSS, Chicago, IL). Differences between two groups were assessed using Student t-test. The association between SRPX2 and RAB31 expression in pancreatic cancer tissues was analyzed using Spearman's correlation. OS and DFS were analyzed by Kaplan-Meier survival curves with 95% confidence intervals (CIs), and the differences between subgroups were compared by log-rank test. Univariate and multivariate Cox regression analyses were used to identify independent prognostic factors. P< 0.05 was the criterion for variable deletion to perform backward stepwise selection. All tests were 2-sided, and P < 0.05 was considered statistically significant.
Results
Overexpression of SRPX2 and RAB31 are identified in pancreatic cancer
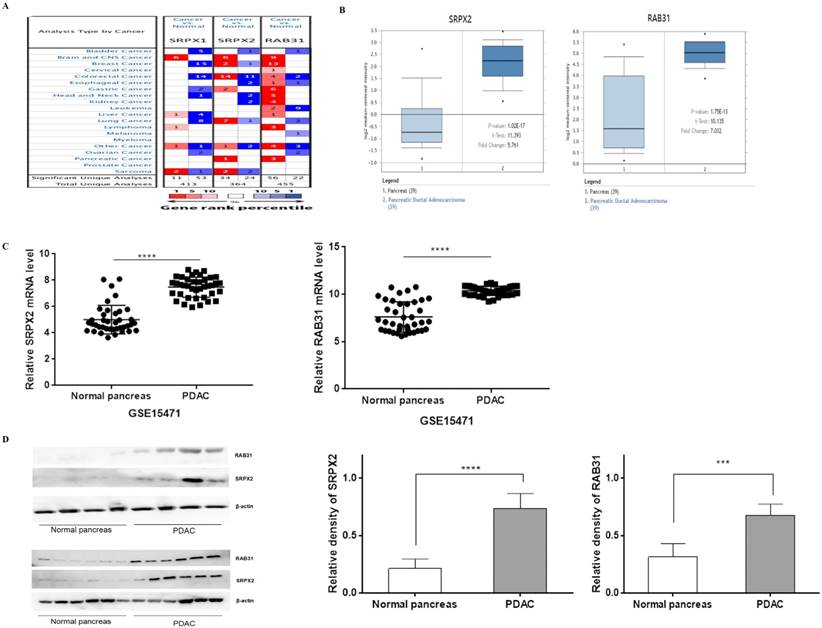
To explore the potential roles of SRPXs (SRPX1 and SRPX2) in pancreatic cancer, we analyzed the expression of these genes by data mining and validated by our data. First, we investigated the mRNA levels of SRPXs in human cancers using the Oncomine database. The results revealed that SRPX2 mRNA expression was significantly higher in pancreatic tumors than in normal tissues across a wide variety of datasets in different cancer types, whereas no significant difference was found in SRPX1 mRNA expression (Figure 1A). CCLE analysis was performed to evaluate the expression of these genes in human cancer cell lines. The results were consistent with those of the Oncomine analysis, demonstrating that SRPX2 and RAB31 are highly expressed in pancreatic cancer cell lines (Supplementary Figure 1). Oncomine analysis demonstrated that the SRPX2 and RAB31 transcripts were increased by 5.761-fold and 7.032-fold, respectively, in PDAC samples compared with normal tissues from the Oncomine database (Figure 1B). Additionally, from GSE15471 data (39 PDAC tissues vs. 39 normal tissues), GEO analysis revealed that both SRPX2 and RAB31 were significantly upregulated in PDAC tissues compared with normal tissues (Figure 1C). Then, this result was validated by Western blot in another 10 pairs of specimens (Figure 1D). These results demonstrate that SRPX2 and RAB31 are significantly upregulated in pancreatic cancer.
The positive correlation between SRPX2 and RAB31 expression in pancreatic cancer
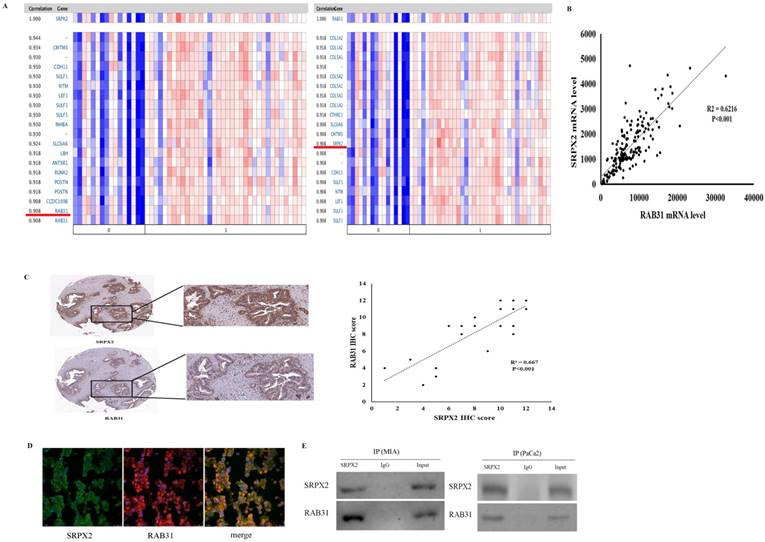
Based on these data that revealed high expression of both SRPX2 and RAB31 in pancreatic cancer, co-expression analysis was performed to further study the relationships in pancreatic cancer. Oncomine co-expression analysis revealed that SRPX2 expression is significantly correlated with RAB31 in pancreatic cancer (r = 0.908) and that RAB31 expression was significantly correlated with SRPX2 in pancreatic cancer (r = 0.908) (Figure 2A). TCGA database also showed a strong positive correlation (P < 0.001, r = 0.788) exists between SRPX2 and RAB31 (Figure 2B). Then, the SRPX2 and RAB31 proteins were stained on 20 PDAC specimens by immunohistochemistry. The results showed that in the same tumor tissues from PDAC patients, the expression of SRPX2 was positively correlated with RAB31 expression (Figure 2C). To further analysis of relationship, SRPX2 and RAB31 expression and subcellular localization in PDAC tissues were assessed by immunofluorescence staining. This assay showed that SRPX2 and RAB31 staining intensity was prominently increased in both the cytoplasm and cell membrane in tumor tissues (Figure 2D). Additionally, when we merged these two slices with SRPX2 and RAB31 staining, they merged very well, indicating that these two proteins are co-expressed (Figure 2D). In co-IP analysis, input and immunoglobulin G (IgG) served as positive and negative controls, respectively. The reciprocal co-IP analysis revealed that endogenous SRPX2 interacted directly with RAB31 in MIA cells (Figure 2E). These results demonstrate that SRPX2 and RAB31 are positive correlation and direct interaction in PDAC.
SRPX2 and RAB31 are upregulated in pancreatic cancer. (A) mRNA expression of genes in cancer tissue versus normal matched tissue. Up and down regulated expression of target genes shown in red and blue, respectively. Color transparency shifted in the top 1% and top 10% in both up and down regulated gene expression. The number in each square denotes the number of analyses that satisfy the threshold. (B) Representative SRPX1 and RAB31 mRNA expression in pancreatic tumor tissues versus normal samples from the Oncomine database. The p-value was set as 0.01 and fold change was defined as 2. (C) Representative SRPX1 and RAB31 mRNA expression in pancreatic tumor tissues versus normal samples from the GEO database. (D) The expression level of SRPX1 and RAB31 was significantly upregulated in pancreatic cancer tissue compared to that in the adjacent noncancerous tissue (WB: P < 0.001). ****P < 0.0001, ***P < 0.001, *P < 0.05.
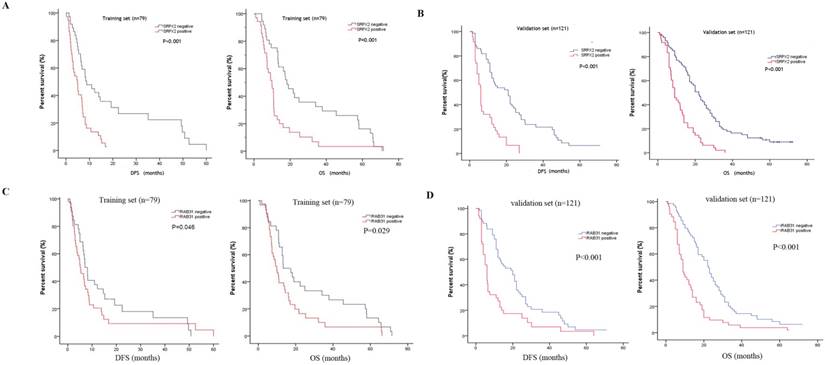
SRPX2 and RAB31 predict a poor prognosis for PDAC patients
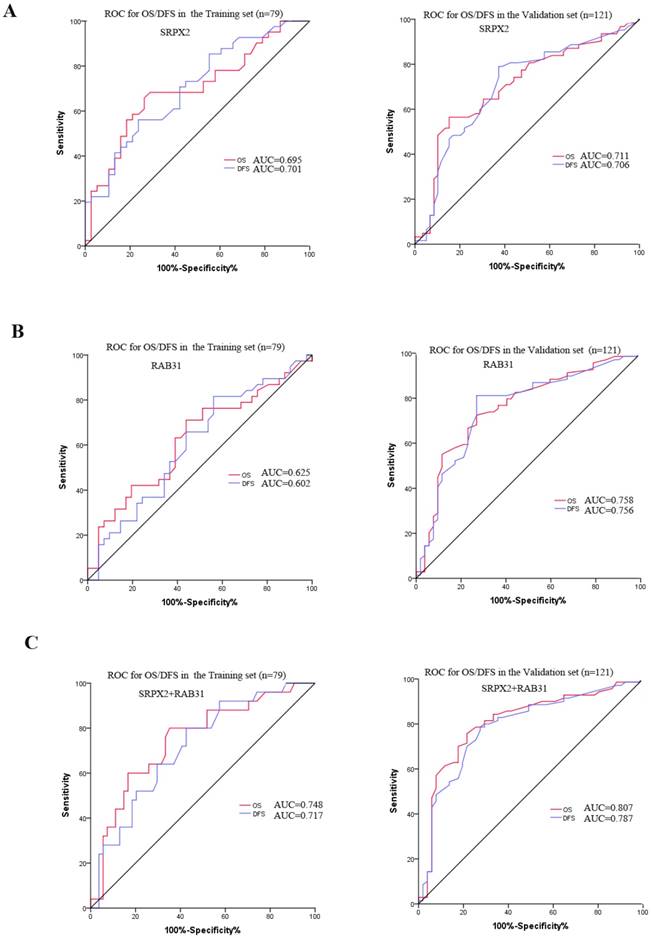
The expression of SRPX2 and RAB31 in training set and validation set was examined by immunohistochemistry. The ratio of SRPX2 high expression in training set and validation set were 53.2% and 60.3%, respectively. There was no heterogeneity in expression between the two sets (P = 0.316). The ratio of RAB31 high expression in training set and validation set were 44.3% and 56.2%, respectively. There was no heterogeneity in expression between the two sets (P = 0.100) (Supplementary table 1). The prognostic effects of SRPX2 and RAB31 for the disease-free survival (DFS) and overall survival (OS) of PDAC patients were evaluated by Kaplan-Meier survival analysis and the log-rank test. These findings revealed that increased expression of SRPX2 is associated with shorter DFS and OS than patients with low SRPX2 expression in the training set (P < 0.001, and P = 0.001, respectively) and the validation set (P < 0.001, and P < 0.001, respectively) (Figure 3A,B). Similarly, the patients with high expression of RAB31 have poor DFS and OS than patients with low SRPX2 expression in the training set (P = 0.046, 0.029 respectively) and the validation set (P < 0.001, and P < 0.001, respectively) (Figure 3C,D). A receiver operating characteristic (ROC) curve analysis were performed to evaluate the prognosis value of SRPX2 and RAB31 and the results showed that the AUC for SRPX2 expression associated with OS was 0.695 and 0.711 (DFS was 0.701 and 0.706) in the training set and validation set, respectively (Figure 4A). The Area Under the Curve (AUC) for RAB31 expression associated with OS was 0.625 and 0.758 (DFS was 0.602 and 0.756) in the training set and validation set, respectively (Figure 4B).The AUC for SRPX2+RAB31 expression associated with OS was 0.748 and 0.807 (DFS was 0.717 and 0.787) in the training set and validation set, respectively (Figure 4C). In summary, these results demonstrated that combination of SRPX2 and RAB31 had better predictive ability for PDAC patients.
The combination of SRPX2 and RAB31 is an independent prognostic factor for PDAC patients
To determine the association between SRPX2 and RAB31 expression and clinicopathological characteristics with respect to PDAC prognosis, these variables were subjected to Cox regression analysis. By multivariate Cox regression analysis, 8th edition T classification, 8th edition N classification, SRPX2, and RAB31 were excluded to avoid collinearity (tolerance > 0.1). Multivariate Cox regression analysis showed that 8th edition TNM stage (HR = 3.847, CI 2.946-4.352, P < 0.001; HR =2.938, CI 1.835-4.027, P < 0.001) and SRPX2+RAB31 (HR = 2.869, CI 1.938-3.304, P < 0.001; HR = 3.929, CI 2.908-5.456, P < 0.001) were independent prognostic factors that were associated with DFS in the training sets and the validation sets, respectively. Additionally, the results also showed that 8th edition TNM stage (HR = 3.960, CI 3.015-4.245, P < 0.001; HR = 2.551, CI 1.426-3.856, P < 0.001) and SRPX2+RAB31 (HR = 3.125, CI 2.679-3.864, P < 0.001; HR = 3.436, CI 2.081-6.143, P < 0.001) were independent prognostic factors that were associated with DFS in the training sets and the validation sets, respectively. Therefore, 8th edition TNM stage and SRPX2+RAB31 were considered independent prognostic factors for PDAC (Table 1, 2).
The positive correlation between SRPX2 and RAB31 expression in pancreatic cancer. (A) Correlation between SRPX2 with RAB31 expression in pancreatic cancer tissues analyzed in the Oncomine database (shown in red frame). (B) Correlation between SRPX2 with RAB31 expression in pancreatic cancer tissues analyzed by TCGA database. (C) Expression of SRPX2 and RAB31 in the same slice of PDAC tissue analyzed by immunohistochemistry staining. Magnification=200x. (D) Immunofluorescence staining of SRPX2 and SAB31 protein in PDAC tissues. (E) Co-IP assay was used to examine the association between SRPX2 and RAB31. Input and IgG served as positive and negative controls, respectively.
Kaplan-Meier estimates of OS and DFS according to the expression of SRPX2 and RAB31 in patients with resectable PDAC. (A) SRPX2 in Training set, OS (P < 0.001), DFS (P = 0.001); (B) SRPX2 in Validation set, OS (P < 0.001), DFS (P < 0.001). (C) RAB31 in Training set, OS (P =0.046), DFS (P = 0.029); (D) RAB31 in Validation set, OS (P< 0.001), DFS (P< 0.001).
Univariate and multivariate analysis of DFS and OS in the training sets of patients with PDAC
| Variables | DFS | OS | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | p value | Hazard ratio (95% CI) | P value | |
| Univariate analysis | ||||
| Age (>62 vs. ≤62) | 1.009 (0.981-1.039) | 0.529 | 1.015 (0.986-1.044) | 0.320 |
| Gender (male vs. female) | 1.315 (0.835-1.967) | 0.146 | 1.529 (0.941-2.486) | 0.086 |
| Tumor location (head/body vs. tail) | 1.687 (1.214-2.145) | 0.169 | 1.732 (1.458-2.232) | 0.130 |
| Neural invasion (yes vs. no) | 1.372 (0.751-1.957) | 0.352 | 1.316 (0.625-1.825) | 0.098 |
| Microvascular invasion (yes vs. no) | 1.845 (1.420-2.318) | 0.041 | 1.976 (1.385-2.252) | 0.036 |
| Differentiation (Poorly vs. well+moderately) | 2.136 (2.014-2.675) | 0.085 | 2.548 (2.364-2.973) | 0.167 |
| 8th edition TNM stage ( I/ II/ III) | 2.742 (1.852-3.685) | <0.001 | 2.264 (1.982-3.579) | <0.001 |
| 8th edition T classification (T1+T2 vs. T3) | 2.928 (2.146-3.352) | <0.001 | 2.743 (2.294-3.952) | <0.001 |
| 8th edition N classification (N0/N1/N2) | 2.862 (1.989-3.157) | <0.001 | 2.548 (2.046-2.869) | <0.001 |
| CA19-9 (U/mL, >37 vs. ≤37) | 1.854 (0.873-2.425) | 0.032 | 1.466 (1.023-1.989) | 0.069 |
| SRPX2 (positive vs. negative) | 2.982 (1.854-3.215) | 0.003 | 2.367 (1.454-3.854) | 0.002 |
| RAB31 (positive vs. negative) | 1.326 (0.817-2.152) | 0.042 | 1.747 (1.073-2.845) | 0.025 |
| SRPX2+RAB31 (positive vs. negative) | 2.467 (2.193-3.244) | 0.001 | 2.923 (1.333-3.708) | <0.001 |
| Multivariate analysis | ||||
| 8th edition TNM stage ( I/ II/ III) | 3.847 (2.946-4.352) | <0.001 | 3.960 (3.015-4.245) | <0.001 |
| SRPX2+RAB31 (positive vs negative) | 2.869 (1.938-3.304) | <0.001 | 3.125 (2.679-3.864) | <0.001 |
P < 0.05 was considered statistically significant
Univariate and multivariate analysis of DFS and OS in the validation sets of patients with PDAC
| Variables | DFS | OS | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| Univariate analysis | ||||
| Age (>62 vs. ≤62) | 1.034 (0.682-1.491) | 0.922 | 1.008 (0.757-1.514) | 0.621 |
| Gender (male vs. female) | 0.917 (0.434-1.264) | 0.085 | 1.019 (0.716-1.884) | 0.135 |
| Tumor location (head/body vs. tail) | 1.072 (0.714-1.455) | 0.863 | 0.933 (0.487-1.534) | 0.757 |
| Neural invasion (yes vs. no) | 1.152 (0.853-1.865) | 0.742 | 1.014 (0.423-1.138) | 0.608 |
| Microvascular invasion (yes vs. no) | 1.675 (1.132-2.524) | 0.022 | 1.769 (1.058-2.974) | 0.011 |
| Differentiation (Poorly vs. well+moderately) | 2.033 (1.854-4.358) | 0.025 | 2.141 (1.573-3.864) | 0.031 |
| 8th edition TNM stage ( I/ II/ III) | 3.013 (2.172-4.056) | <0.001 | 2.863 (1.368-5.477) | <0.001 |
| CA19-9 (U/mL, >37 vs. ≤37) | 1.324 (0.638-2.265) | 0.029 | 1.784 (1.008-2.339) | 0.179 |
| SRPX2 (positive vs. negative) | 3.127 (1.484-5.326) | <0.001 | 1.667 (1.067-2.746) | 0.021 |
| RAB31 (positive vs. negative) | 2.434 (1.228-4.017) | <0.001 | 1.991 (1.164-3.065) | 0.002 |
| SRPX2+RAB31 (positive vs. negative) | 3.983 (2.268-6.157) | <0.001 | 4.123 (2.667-7.403) | <0.001 |
| Multivariate analysis | ||||
| 8th edition TNM stage ( I/ II/ III) | 2.938 (1.835-4.027) | <0.001 | 2.551 (1.426-3.856) | <0.001 |
| SRPX2 (positive vs. negative) | 2.037 (1.146-4.235) | 0.004 | 1.739 (1.004-3.316) | 0.037 |
| SRPX2+RAB31 (positive vs. negative) | 3.929 (2.908-5.456) | <0.001 | 3.436 (2.081-6.143) | <0.001 |
P < 0.05 was considered statistically significant
ROC curve for the prediction of OS and DFS in patients with resectable PDAC. (A) ROC curve of SRPX2 in Training set and Validation set. (B) ROC curve of RAB31 in Training set and Validation set. (C) ROC curve of combination of SRPX2 and RAB31 in Training set and Validation set. AUC = area under the curve.
Discussion
As pancreatic cancer is unfortunately typically detected during the late stages, fewer than 20% of patients are eligible for curative resections [14]. Identifying sensitive and specific markers for improving diagnosis and accurately evaluating prognosis remain urgent issues that need to be solved. In this study, we explored proteins associated with the diagnosis and prognosis of pancreatic cancer. First, the results from the Oncomine analysis and GEO databases revealed that there was a significant difference in SRPX2, RAB31 expression between tumor tissues and normal tissues in pancreatic cancer. Second, we found that SRPX2 directly interact with RAB31 in PDAC. Furthermore, cox regression analysis indicated that the combination of SRPX2 and RAB31 can be considered an independent predictive factor for the clinical outcomes of PDAC patients.
SRPX2 has been identified as a novel secreted chondroitin sulfate proteoglycan (GSPG) and found to play roles in a series of biological processes in tumor cells, including cellular motility and cellular attachment [6]. In several previous studies, researchers have reported that the expression of SRPX2 is upregulated in a variety of malignant tumor tissues [15]. Therefore, we analyzed the Oncomine and GEO databases to further evaluate the expression of SRPX2 in pancreatic cancer. The results showed that SRPX2 expression was increased by 5.761-fold in pancreatic cancer tissues compared with normal tissues. Additionally, CCLE analysis and immunofluorescence assays revealed that the expression of SRPX2 was increased in pancreatic tumor cells. Hence, as has been reported in other cancer studies, the expression of SRPX2 is significantly upregulated in pancreatic tumor tissues, implying that SRPX2 may be essential for the tumorigenesis and progression of pancreatic cancer. Additionally, the potential signaling pathways underlying SRPX2's involvement in the proliferation, migration, adhesion, and invasion of tumor cells have been persistently explored. One of the most widely recognized is the ability of SRPX2 overexpression to promote cell migration and invasion through FAK signaling in gastric cancer and hepatocellular carcinoma [16, 17]. Additionally, Zemmour et al demonstrated that SRPX2 directly interacts with urokinase plasminogen activator receptor (uPAR) [18]. uPAR is well known for its role in various tumor processes, so the SRPX2/uPAR interaction might provide one possible molecular mechanism for the function of SRPX2 in cancer.
Next, co-expression analysis was performed and indicated that there is a significantly positive correlation between the expression of SRPX2 and RAB31 in pancreatic tumor tissues. In the differential expression analysis, it was demonstrated that RAB31 is highly expressed not only in pancreatic tumor tissues but also in pancreatic cancer cells. Undoubtedly, RAB31 has long been considered a vital tumor promoter in other types of cancers [10]. Furthermore, several recent studies have demonstrated possible potential molecular pathways for the roles in cancer, one of which is that RAB31 induces cancer cell proliferation and migration through the ERK1/2 and PI3k/AKT pathways [19]. Additionally, increasing evidence indicates that the expression of RAB31 is correlated with uPAR-del4/5, and uPAR may be regulated by the Rab protein-driven pathway [20]. The combination of the involved molecular pathways of SRPX2 and RAB31 in cancer suggests that these two proteins share a common signaling pathway, which may be responsible for the positive correlation between their expression in pancreatic cancer.
The present study firstly reports the correlation and clinical significance of SRPX2 and RAB31 in PDAC patients, but it has some limitations. First, this study is retrospective, we will evaluate the prognosis value of SRPX2 and RAB31 in the prospective study. Second, although we found the positive correlation and direct interaction between SRPX2 and RAB31 in PDAC, we did not provide the mechanism between SRPX2 and RAB31 to affect the prognosis of patients with PDAC.
Conclusion
This study indicated that expression of SRPX2 and RAB31 was significantly increased in PDAC. Additionally, SRPX2 directly interact with RAB31 in PDAC. Furthermore, survival analysis showed that increased expression of SRPX2 and RAB31 was significantly associated with poorer prognosis for PDAC patients. Cox regression analysis indicated that the combination of SRPX2 and RAB31 can be considered an independent predictive factor for the clinical outcomes of PDAC patients. Thus, SRPX2 and RAB31 can be promising prognostic factors in pancreatic cancer.
Supplementary Material
Supplementary figure and table.
Acknowledgements
This work was supported by grants from the National Science Foundation for Distinguished Young Scholars of China (81625016), the National Natural Science Foundation of China (81472670, 81871941, 81872366, 81802380, 81702341, and 81802675), the Outstanding Academic Leader Program of the “Technological Innovation Action Plan” in Shanghai Science and Technology Commission (18XD1401200), and the Young Talented Specialist Training Program of Shanghai.
The funding agencies had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-386
2. Rahib L, Smith BD, Aizenberg R. et al. Projecting cancer incidence and deaths to 2030:the unexpected burden of thyroid,liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921
3. Zeng H, Chen W, Zheng R. et al. Changing cancer survival in China during 2003-15:a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:E555-E567
4. Infante JR, Matsubayashi H, Sato N. et al. Peritumoral fibroblast SPARC expression and patient outcome with resectable pancreaticadenocarcinoma. J Clin Oncol. 2007;25:319-325
5. Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma:An evidence based appraisal. J Gastrointest Oncol. 2012;3:105-119
6. Sia GM, Clem RL, Huganir RL. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science. 2013;342:987-991
7. Royer B, Soares DC, Barlow PN. et al. Molecular evolution of the human SRPX2 gene that causes brain disorders of the Rolandic and Sylvian speech areas. BMC Genet. 2007;8:72
8. Tang H, Zhao J, Zhang L. et al. SRPX2 Enhances the Epithelial-Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Cells. Cell Mol Neurobiol. 2016;36:1067-1076
9. Cheng KW, Lahad JP, Gray JW. et al. Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 2005;65:2516-2519
10. Chua CE, Tang BL. The role of the small GTPase Rab31 in cancer. J Cell Mol Med. 2015;19:1-10
11. Grismayer B, Sölch S, Seubert B. et al. Rab31 expression levels modulate tumor-relevant characteristics of breast cancer cells. Mol Cancer. 2012;11:62
12. Sui Y, Zheng X, Zhao D. Rab31 promoted hepatocellular carcinoma (HCC) progression via inhibition of cell apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumour Biol. 2015;36:8661-8670
13. Li H, Zhou ZQ, Yang ZR. et al. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66:136-151
14. Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039-1049
15. Pan Y, Zhang Y, Chen L. et al. The Critical Role of Rab31 in Cell Proliferation and Apoptosis in Cancer Progression. Mol Neurobiol. 2016;53:4431-4437
16. Tanaka K, Arao T, Maegawa M. et al. SRPX2 is overexpressed in gastric cancer and promotes cellular migration and adhesion. Int J Cancer. 2009;124:1072-1080
17. Lin X, Chang W, Wang Y. et al. SRPX2, an independent prognostic marker, promotes cell migration and invasion in hepatocellular carcinoma. Biomed Pharmacother. 2017;93:398-405
18. Royer-Zemmour B, Ponsole-Lenfant M, Gara H. et al. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum Mol Genet. 2008;17:3617-3630
19. Sui Y, Zheng X, Zhao D. Rab31 promoted hepatocellular carcinoma (HCC) progression via inhibition of cellapoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumour Biol. 2015;36:8661-8670
20. Kotzsch M, Dorn J, Doetzer K. et al. mRNA expression levels of the biological factors uPAR, uPAR-del4/5, and rab31, displaying prognostic value in breast cancer, are not clinically relevant in advanced ovarian cancer. Biol Chem. 2011;392:1047-1051
Author contact
![]() Corresponding authors: Xian-Jun Yu, MD, PhD, FACS, Liang Liu, MD, PhD, and Chun-Tao Wu, MD. Shanghai Pancreatic Cancer Institute; Pancreatic Cancer Institute, Fudan University; Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center; 270 Dong An Road, Shanghai 200032, China; Tel/Fax: +86-21-6403-1446; Email: yuxianjunorg (X.J.Yu); liuliangorg (L.Liu); or wuchuntaoorg (C.T.Wu).
Corresponding authors: Xian-Jun Yu, MD, PhD, FACS, Liang Liu, MD, PhD, and Chun-Tao Wu, MD. Shanghai Pancreatic Cancer Institute; Pancreatic Cancer Institute, Fudan University; Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center; 270 Dong An Road, Shanghai 200032, China; Tel/Fax: +86-21-6403-1446; Email: yuxianjunorg (X.J.Yu); liuliangorg (L.Liu); or wuchuntaoorg (C.T.Wu).

 Global reach, higher impact
Global reach, higher impact