3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(12):2783-2787. doi:10.7150/jca.31450 This issue Cite
Research Paper
The characteristics of HPV integration in cervical intraepithelial cells
1. Department of Obstetrics and Gynecology, Chinese PLA General Hospital, Beijing, P. R. China
2. MyGenostics Inc., Beijing, P. R. China
3. ME Genomics Inc., Shenzhen, P. R. China
4. Collaborative Innovation Center for Birth Defect Research and Transformation of Shandong Province, Jining Medical University, Jining, Shandong 272067, China
Received 2018-11-12; Accepted 2019-4-25; Published 2019-6-2
Abstract
Cervical cancer is one of the most common malignant tumors in gynecology. Deploying HIVID, a cost-effective technique to detect HPV integration sites, our team had studied the characteristics of HPV integrations in cervical exfoliated cells. Our results indicated that both the sample proportion and the number of HPV integrations gradually increased following the development of cervical lesion. Meanwhile, our data also revealed that there were recurrent genes integrated by HPV in cervical exfoliated cells. Collectively, the HPV integration breakpoints were highly enriched in the intron and promoter regions. Intriguingly, the gene pathway analysis indicated that the HPV-integrated genes were strongly inclined to pathways of metabolism of xenobiotics by cytochrome P450, chemical carcinogenesis and steroid hormone biosynthesis. In conclusion, this study unveiled the HPV integration patterns and the associated recurrent genes in cervical epithelial exfoliated cells. Altogether, our data suggested that the HPV integrations in cervical exfoliated cells might have vital clinical significance, and probably also diagnostic and/or prognostic values in future clinical applications.
Keywords: HPV integration, Cervical exfoliated cells, Cervical cancer, Characteristics
1. Introduction
Cervical cancer is one of the most common malignant tumors in gynecology, with an occurring incidence only second to that of breast cancer among all female malignant tumors. In recent years, due to the development of cervical exfoliative cytology, the rate of cervical cancer had been decreased by 70%-80%, as well as the corresponding mortality [1]. It is believed that the sensitivity and specificity of cervical cancer and precancerous lesion diagnosis by cytology were around 47%-62% and 60%-90%, respectively [2]. Suggesting further improvement of the current diagnostic mean is urgently demanded to help identify the high-risk group.
More recently, HPV screening had been shown better at predicting cervical cancer than cytology as confirmed by an increasing number of studies. It is acknowledged that HPV infection could be cleared by the host immune system in most of women. Nevertheless, HPV infection in approximately 1-2% of the affected might persist and eventually leading to cervical intraepithelial neoplasia or cervical cancer [3]. The high-risk HPV had a clear association with the development of carcinogenesis. During the persist infection of the high-risk HPV, HPV integration events could often be detected, which were well known to cause genome instability, abnormal gene expression in cells [4-6]. In addition, the viral E2 region is often being abrogated upon the HPV integration, resulting in uncontrolled viral replication [7]. An elevating number of research data had now suggested that the DNA integration of high-risk HPV might be pre-requisite and/or driven force of HPV-induced carcinogenesis, the maintenance of malignant phenotype, and the development of cervical cancer [8]. As followed by the ongoing research, the association of HPV integration have been established with the CIN level of the cervix, which could potentially being used as a marker for the cervical cancer risk assessment for those who suffered HPV infection [9]. Bearing in mind that most of the current research on HPV integration had focused and established on the pathological tissues. In clinical practice, cervical exfoliated cells might be collected relatively at ease compared to pathological tissues, providing a distinct advantage to be applied to generate HPV integration databases, and hence assessing the risks of HPV integrations. To find out the regularity of HPV integration in cervical exfoliated cells and also the correlation of HPV integration characteristics of exfoliated cells and different degrees of cervical lesions, we deployed the cost-effective HIVID detection technology. Altogether, this study had provided partial theoretical basis on whether HPV integration events of cervical exfoliated cells could have potential to be regarded as an effective clinical indicator.
2. Materials and Method
A total of 230 HPV positive samples were obtained from the General Hospital of the People's Liberation Army (PLA). All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and the later amendments or comparable ethical standards. The study had been approved by the Ethics Review Committee in the General Hospital of the PLA. HPV integration sites were detected using HIVID method [10]. Functional annotation analysis was performed using DAVID based on Gene Ontology and KEGG pathway databases [11, 12]. The categories of KEGG Pathways served as background databases. The breakpoints were annotated through the latest ANNOVAR in hg19 coordinates [13].
It is now documented that HPV integration could affect gene expression in following aspects. 1. Influence flanking genes via cis-activation; 2. mimicking the effects of cis-acting enhancers to influence target genes over long distances [14, 15] (up to 1 Mb for upstream enhancers and 850 kb for downstream enhancers). In this study, genes located within a distance of <500kb away from breakpoints were regarded as the affected genes, and being assessed in the HPV-positive samples [16].
3. Results
3.1 Analysis of Integration Ratio and Breakpoint Number
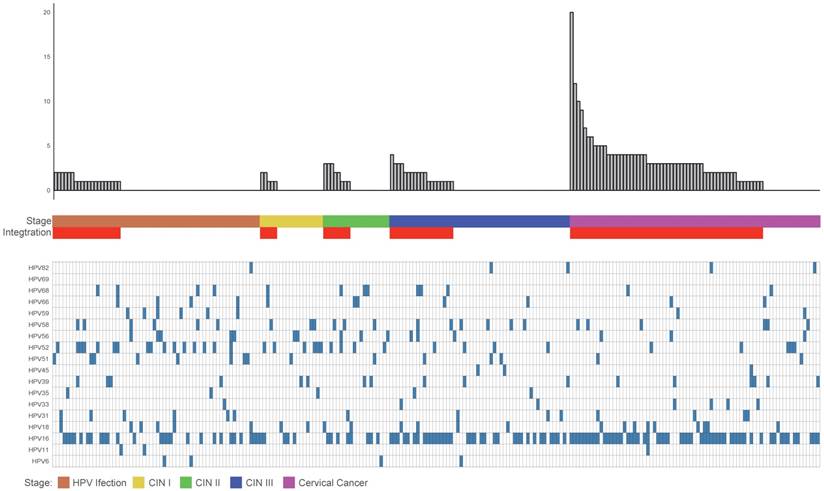
The data analysis of these clinical samples suggested that the proportion of HPV integrated samples and the average number of breakpoints in these samples positively correlated with the stage of disease progression. The integration ratios of HPV infection, CINI, CINII, CINIII, and cervical cancer were 32%, 26%, 40%, 35%, and 77%, respectively. The average breakpoint numbers of HPV infection, CINI, CINII, CINIII and cervical cancer were 1.3, 1.4, 2, 1.8 and 3.7 respectively. As in the samples with more advanced stages of the disease, the diversity of the HPV displayed a gradual decease (Figure 1).
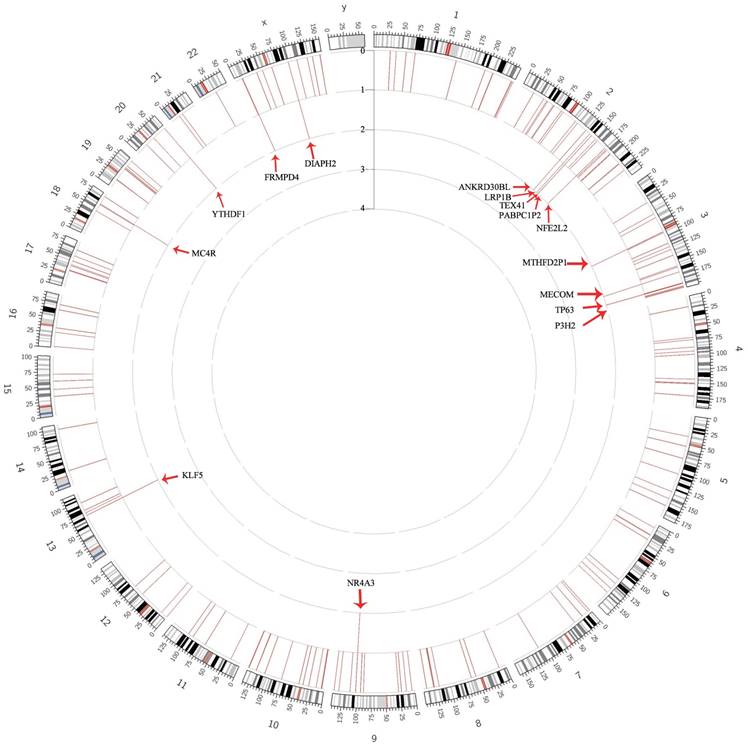
3.2 The Recurrent Genes of HPV Integration in the DNA of Cervical Epithelial Samples
As documented in the frequency analysis, most genes only appeared once in the samples, despite a number of recurrent genes (Figure 2). Overall, 15 recurrent genes (frequency = 2) were filtered out from the HPV positive cervical epithelial samples (Table S1, Table S2). Pathway analysis revealed that the disrupted genes were enriched in the pathway of Metabolism of xenobiotics by cytochrome P450, Chemical carcinogenesis, Steroid hormone biosynthesis and so on (Table S3).
3.3 Distribution of Genetic Elements
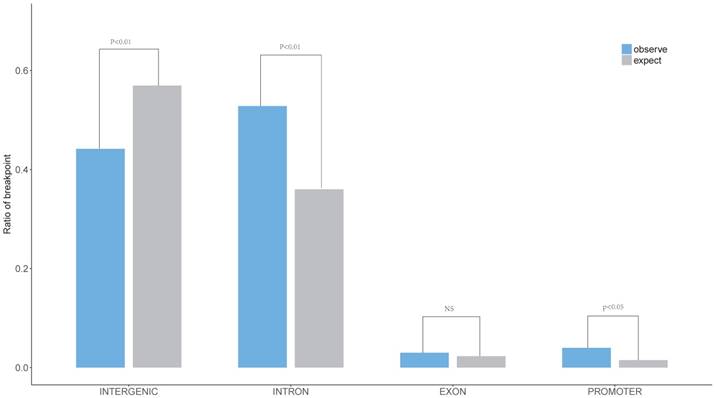
The characteristics of HPV breakpoints distribution had been surveyed. The results revealed that HPV breakpoints were prone to the INTRON (P < 0.01, Chi-squared test, Figure 3) and PROMOTER (P < 0.05, Chi-squared test, Figure 3) regions, but not the INTERGENIC (P < 0.01, Chi-squared test, Figure 3) region.
4. Discussion
The cervical epithelial exfoliated cells had been widely used for early screening of HPV, due to its obvious advantage of being non-invasive, which had greatly reduced the incidence of cervical cancer. In spite of that the screening accuracy is yet to be improved further. The development of new generation of HPV screening (enrich HPV reads and localise HPV integration sites) had provided technical basis allowing the risk assessment of cervical epithelial lesions. Having provided in the previous studies, persistent HPV infection is the causal cause of cervical epithelial lesions, and the occurrence of HPV integrations often indicating a greater risk of epithelial lesions [17, 18]. Herein, our results suggested that the proportion of HPV integrated samples and the average number of breakpoints in the samples positively correlated with the stage of disease progression. Further implying that certain categories of HPV integration events could be good indicators of the progressive stage of developing cervical cancer. In fact, detecting HPV integration of cervical epithelial cells has gradually been practiced in clinical diagnosis and treatment. Generally speaking, for patients positive for HPV high-risk infection and HPV integration, even with negative TCT, the patients will be advised to carry out colposcopy examination, hence to prevent false diagnosis. For patients positive for HPV high-risk infection, but negative for HPV integration and TCT, the patients will be required to carry out short-term observation. In circumstances where patients are positive for HPV high-risk infection and integration, and biopsy of CINI, LEEP or cervical conization is usually recommended. For cervical cancer and HPV integration positive patients, it is recommended to expand the scope of surgery and increase the number of post-operative examinations. Furthermore, HPV integration hotspots and the recurrent genes had also been filtered out. However, it is still unclear whether the recurrent genes could be useful in terms of helping with prognosis. More interestingly, the breakpoints are significantly enriched in the INTRON and PROMOTER regions. Therefore, it might be directly related to the disruption and alteration of gene function [19, 20]. Gene pathway analysis results indicated that the HPV-integrated genes were significantly inclined to the metabolic and steroid hormone bio-synthesis pathways, and it is speculated that certain abnormalities in some of the metabolic and bio-synthesis pathways may be affected contributing to the progression of cervical cancer. In addition, HPV integrations had been evident to affect the cancer-associated pathways and modify target gene expression in the host cells, which could further promote carcinogenesis [21, 22]. Moreover, we have begun to conduct follow-up studies on patients with HPV integration for future research. It will be necessary to further determine the correlation of between disease progression and HPV integration. Meanwhile, it will provide efficient means to evaluate disease progression.
In this study, deploying a genome-wide scanning method for HPV integrations had enabled us to determine the close correlation between viral integration and clinical disease progression. Our data had identified the inclination of HPV integration and the recurrent HPV-integrated genes in cervical epithelial exfoliated cells which might be useful in establishing a more sensitive and accurate future clinical application, helping with the diagnosis of HPV positive cervical cancer.
Clinical annotation of 230 samples. All panels are aligned with vertical tracks representing 230 individuals. The data are sorted by the number of breakpoints, HPV infection stage, CIN stage, Cancer and HPV type.
Gene frequency of HPV integration throughout the human genomes in 230 samples. Gene frequency of breakpoints across the human genome in 230 samples. Each bar represents the sample frequency of HPV integration in the human genome (hg19). Histogram axis units represent number of samples. Some loci with a high frequency of integration are marked.
Supplementary Material
Supplementary tables.
Acknowledgements
The study was funded by Cancer foundation of China (N2013016), Technology Development Project of Medical and Health Science in Shandong Province (no. 2017WS516), Research Fund for Lin He' s Academician Workstation of New Medicine and Clinical Translation in Jining Medical University (JYHL2018MS05) and Supporting Fund for Teachers Research of Jining Medical University (JYFC2018KJ034).
Authors' Contributions
WYL, YQY and WPL conceived and wrote the paper. ST, PPW and YKZ analyzed the data.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study had been approved by the Chinese PLA General Hospital. All subjects signed written informed consent forms.
Distribution of breakpoints in gene element region. The expected (assuming uniform, random distribution, grey) and the observed (actual numbers, blue) ratio of HPV breakpoints in Promoter, Exon, Intron, Intergenic region are shown. P values were calculated by Chi-square test.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lowy DR, Schiller JT. Reducing HPV-associated cancer globally. Cancer prevention research. 2012;5:18-23
2. Nanda K, McCrory DC, Myers ER, Bastian LA, Hasselblad V, Hickey JD. et al. Accuracy of the Papanicolaou test in screening for and follow-up of cervical cytologic abnormalities: a systematic review. Annals of internal medicine. 2000;132:810-9
3. Villa LL, Bernard HU, Kast M, Hildesheim A, Amestoy G, Franco EL. Past, present, and future of HPV research: highlights from the 19th International Papillomavirus Conference-HPV2001. Virus research. 2002;89:163-73
4. Groves IJ, Coleman N. Human papillomavirus genome integration in squamous carcinogenesis: what have next-generation sequencing studies taught us? The Journal of pathology. 2018;245:9-18
5. Warburton A, Redmond CJ, Dooley KE, Fu H, Gillison ML, Akagi K. et al. HPV integration hijacks and multimerizes a cellular enhancer to generate a viral-cellular super-enhancer that drives high viral oncogene expression. PLoS genetics. 2018;14:e1007179
6. Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B. et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome research. 2014;24:185-99
7. Xu B, Chotewutmontri S, Wolf S, Klos U, Schmitz M, Durst M. et al. Multiplex Identification of Human Papillomavirus 16 DNA Integration Sites in Cervical Carcinomas. PloS one. 2013;8:e66693
8. Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P. et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer research. 2008;68:307-13
9. Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer research. 2004;64:3878-84
10. Li W, Zeng X, Lee NP, Liu X, Chen S, Guo B. et al. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics. 2013;102:338-44
11. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1-13
12. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44-57
13. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164
14. Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Human molecular genetics. 2003;12:1725-35
15. Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, Gounari F. et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902-11
16. Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X. et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nature genetics. 2015;47:158-63
17. Li W, Qi Y, Cui X, Huo Q, Zhu L, Zhang A. et al. Characteristic of HPV Integration in the Genome and Transcriptome of Cervical Cancer Tissues. BioMed research international. 2018;2018:6242173
18. Whitham HK, Hawes SE, Chu H, Oakes JM, Lifson AR, Kiviat NB. et al. A Comparison of the Natural History of HPV Infection and Cervical Abnormalities among HIV-Positive and HIV-Negative Women in Senegal, Africa. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2017;26:886-94
19. Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ. et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371-5
20. Holmes A, Lameiras S, Jeannot E, Marie Y, Castera L, Sastre-Garau X. et al. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ genomic medicine. 2016;1:16004
21. Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, McGovern R. et al. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233-42
22. Cancer Genome Atlas Research N, Albert Einstein College of M, Analytical Biological S, Barretos Cancer H, Baylor College of M, Beckman Research Institute of City of H. et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378-84
Author contact
![]() Corresponding authors: Yuanqing Yao, Department of Obstetrics and Gynecology, Chinese PLA General Hospital, Beijing, P. R. China, E-mail: yqyaocom, or Weiyang Li, Collaborative Innovation Center for Birth Defect Research and Transformation of Shandong Province, Jining Medical University, Jining, Shandong 272067, China. E-mail: 163.lwycom
Corresponding authors: Yuanqing Yao, Department of Obstetrics and Gynecology, Chinese PLA General Hospital, Beijing, P. R. China, E-mail: yqyaocom, or Weiyang Li, Collaborative Innovation Center for Birth Defect Research and Transformation of Shandong Province, Jining Medical University, Jining, Shandong 272067, China. E-mail: 163.lwycom

 Global reach, higher impact
Global reach, higher impact