Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(13):2935-2941. doi:10.7150/jca.31134 This issue Cite
Research Paper
Correlations between angiopoietin-2 gene polymorphisms and lung cancer progression in a Chinese Han population
1. Department of Thoracic Surgery, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China
2. School of Medicine, China Medical University, Taichung, Taiwan
3. Chinese Medicine Research Center, China Medical University, Taichung, Taiwan
4. Department of Biotechnology, College of Health Science, Asia University, Taichung, Taiwan
5. Department of Sports Medicine, College of Health Care, China Medical University, Taichung, Taiwan
6. Department of Orthopaedic Surgery, China Medical University Hospital, Taichung, Taiwan
7. Department of Biomedical Sciences Laboratory, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China
8. Department of Pathology, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China
# These authors have contributed equally to this work
Received 2018-10-31; Accepted 2019-4-22; Published 2019-6-2
Abstract
Lung cancer is the most common malignancy in China and is associated with a poor survival rate amongst Han Chinese. The high mortality is largely attributed to late-stage diagnosis, when treatment is largely ineffective. Identification of genetic variants could potentially assist with earlier diagnosis and thus more effective treatment. The development and progression of lung cancer is stimulated by angiopoietin-2 (Ang2), a ligand for Tie2, an endothelial tyrosine kinase. Patients with lung cancer with higher serum Ang2 levels have significantly poorer survival than patients with lower serum Ang2 levels. We explored the effects of Ang2 single nucleotide polymorphisms (SNPs) on lung cancer susceptibility. We used lung cancer tissue and serum samples to measure Ang2 expression in a Chinese Han population. Five Ang2 SNPs (rs2442598, rs734701, rs1823375, 11137037, and rs12674822) were analyzed using TaqMan SNP genotyping in 695 patients with lung cancer and 900 cancer-free controls. Carriers of the variant GT allele of rs12674822 had a higher risk of lung cancer than wild-type (GG) carriers, while the presence of the CC genotype at rs11137037 was associated with higher clinical stage disease compared with having the AA genotype. Our study is the first to document a correlation between Ang2 polymorphisms and lung cancer development and progression in people of Chinese Han ethnicity.
Keywords: Angiopoietin-2, lung cancer, polymorphism
Introduction
As a result of worldwide trends in aging populations, increasing air pollution, smoking rates, environmental carcinogens and a genetic predisposition, lung cancer has become the most common malignancy worldwide, affecting around 13% of all cancer cases globally [1]. In 2012, worldwide age-standardized mortality rates for males and females with lung cancer were 30.0/100,000 and 11.1/100,000, respectively, reflecting poor 5-year relative survival rates [2]. In 2014, a total of 782,000 new lung cancer cases and 626,000 new lung cancer deaths were documented in China [3]. The specific genetic risks underlying lung cancer development and progression are still unclear. Genetic variants and epigenetic changes have been documented to influence the development and progression of non-small cell lung cancer [4]. Thus, specific information about genetic aberrations could potentially assist with risk prediction and earlier diagnosis of lung cancer in individual patients.
Angiopoietin-2 (Ang2) is a 75-kDa protein expressed in endothelium that is localized to human chromosomes 8p23.1 and first defined at sites of vascular remodeling [5]. Ang2 acts as an autocrine factor in supporting endothelial cell homeostasis and counteracts Ang1 to sensitize endothelium [6]. Genetic factors and single nucleotide polymorphisms (SNPs) genotyping are both pivotal to explore the risk and prognosis of tumorigenesis [7-9]. SNPs in the Ang2 intron are known to be associated with breast cancer, lung injury, or acute respiratory distress syndrome [10-12]. A recent study reported an association between the Ang2 variants, rs2442598, and the risk of psoriasis vulgaris [13]. These studies suggest that it is worth exploring a possible correlation between Ang2 gene polymorphisms and lung cancer diagnosis. Our case-control study therefore explored possible associations between five SNPs in the Ang2 gene and lung cancer.
Materials and Methods
Patients and blood samples
We collected blood specimens from 695 patients (cases) diagnosed with lung cancer at Dongyang People's Hospital between 2014 and 2018. Nine hundred healthy participants without any history of cancer served as controls. All participants provided written informed consent. The study was approved by the Ethics Committee of Dongyang People's Hospital Ethics Committee and Institutional Review Board (2016-YB003). Medical records were reviewed for the clinicopathological characteristics. A standardized questionnaire and electronic medical records were used to acquire detailed clinical data on age, sex, smoking history and alcohol consumption. The pathological stages of lung cancer in all patients were determined by their medical records and the Revised International System for Staging Lung Cancer. Whole blood samples (3 mL) were collected from all study participants and stored at -80°C for subsequent DNA extraction.
Selection of Ang-2 polymorphisms
Five Ang-2 SNPs (rs2442598, rs734701, rs1823375, 11137037, and rs12674822) were selected from the intron of Ang-2; all SNPs had minor allele frequencies of greater than 5%. Most Ang-2 SNPs were known to be associated with lung injury or breast cancer [11, 14]. The sequence of five Ang2 SNP probes were described as follows: rs2442598, TATGTGTGCGAGGACAGTGTGTGTT[A/T]ATTTTGTCCTCTTCTTGATGGTTGA; rs734701, TGTGATATTGTGGAAAGACCTGGTA[T/C]TCAAGTAATTTGTTATTCTATTCTC; rs1823375, GTGACTTCTCTTAGGGAGCACACTT[C/G]CCTTCACCTGCCCTGACCACATGGA; rs11137037, CCCACCATCCCCCATTGCATGCCCT[A/C]AGCAAAGATACTCGTTTTGTGTTTC; rs12674822, GCAATCACTTGTCTGGCCCAACCCT[G/T]TATATTATTTGAGGCCCAGAAAAGG.
Genomic DNA extraction and genotyping by real-time PCR
Total genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA blood mini kit (Qiagen, CA, USA), according to the manufacturer's instructions [15]. Extracted DNA was stored at -20°C and prepared for genotyping by polymerase chain reaction (PCR). DNA was dissolved in TE buffer (10 mM Tris pH 7.8, 1 mM EDTA) and stored at -20°C until quantitative PCR analysis. Five Ang-2 SNP probes were purchased from Thermo Fisher Scientific Inc. (USA), and allelic discrimination analysis of Ang-2 SNPs was assessed using a QuantStudio™ 5 Real-Time PCR system (Applied Biosystems, CA, USA), according to the manufacturer's instructions. Data were further analyzed with QuantStudio™ Design & Analysis Software (Applied Biosystems) [16]. Genotyping PCR was carried out in a total volume of 10 μL containing 20-70 ng genomic DNA, 1 U Taqman Genotyping Master Mix (Applied Biosystems) and 0.25 μL probes. The protocol included an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Cell culture
The human lung cancer cell line A549 was purchased from Jennio Biotech Co., Ltd. (Guangzhou, China). F12, fetal bovine serum (FBS) and all other cell culture reagents were purchased from Thermo Fisher Scientific (MA, USA). A549 cells were cultured in F12 supplemented with 10% FBS (Invitrogen, USA) and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin). Cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C as previous description [17]. We changed the culture medium twice a week and passaged the cells when they reached 80% confluence.
Western blot analysis
We used 11 lung cancer tissues to lyse and resolve through sodium dodecyl sulfate-polyacrylamide gel electrophoresis [18]. The target protein was immunoblotted to Immobilon polyvinyl difluoride membranes (Millipore, Billerica, MA, USA). The blots were blocked with 4% non-fat milk for 1 h at room temperature and then probed with rabbit anti-human antibodies against β-actin, and with mouse anti-human antibodies against Ang2 (#ab155106, Abcam, CA, USA) for 1 h at room temperature. After 3 washes, the blots were incubated with goat anti-rabbit or goat anti-mouse peroxidase-conjugated secondary antibody for 1 h at room temperature. The blots with horseradish peroxidase-labeled substrate were analyzed using a Molecular Imager ChemiDocTM XRS+ chemiluminescence detection system (Bio-Rad, California, USA).
Enzyme-linked immunosorbent assay
527 serum from the lung cancer patients and healthy controls was measured using an Ang2 ELISA kit, according to the manufacturer's instructions [19]. The kit was purchased from R&D Systems.
Statistical analysis
Differences between the two groups were considered significant if p values were less than 0.05. Hardy-Weinberg equilibrium (HWE) was assessed using Chi-square goodness-of-fit tests for bi-allelic markers. Since the data was independent and normal distribution, Fisher's exact test was used to compare differences in demographic characteristics between healthy controls and patients with lung cancer. Odds ratios (ORs) combined with 95% confidence intervals (CIs) calculated associations between genotype frequencies and the risk of lung cancer. To further exclude the impact of confounding variables, adjusted odds ratios (AORs) with 95% CIs were estimated by multiple logistic regression models that controlled for age.
A Cox proportional hazard model was used to evaluate the effects of Ang2 genotyping results on the probability of recurrence or overall survival after adjusting for age in multivariate analysis relative to recurrence or survival time. When the follow-up period was included in survival or recurrence analysis, the patients were enrolled for overall survival analysis including the 5-year survival rate or probability of recurrence between primary surgery and death or recurrence or the end of the study (August, 2018) using the Kaplan-Meier model, as well as multivariate and univariate Cox regression models. All data were analyzed using Statistical Analytic System software (v. 9.1, 2005; SAS Institute, Cary, NC, USA).
Results
In the present study, we enrolled 695 patients with lung cancer and 900 cancer-free healthy participants; all were of Chinese Han ethnicity. Between-group differences in general demographic characteristics are listed in Table 1. The mean age was 43.35 ± 18.01 years for the lung cancer cohort and 59.72 ± 10.81 years for the controls (p<0.001). There were no between-group differences for gender distribution (p=0.891), although significantly higher proportions of lung cancer patients compared with controls were either current or historical cigarette smokers, or consumed alcohol (both p<0.001). According to the American Joint Committee on Cancer (AJCC) tumor/node/metastasis (TNM) classification and staging system, we identified 512 patients (73.7%) with clinical stage I and 110 patients (26.3%) with clinical stage II, III, or IV disease. The patients were categorized by type of lung cancer, i.e., non-small cell lung cancer (NSCLC) (97.2%), or small cell lung cancer (SCLC) (2.8%), or by histology, i.e., adenocarcinoma (80.3%), squamous cell carcinoma (14.7%), and other carcinomas (5.0%) (Table 1).
Demographic and clinical characteristics of healthy cancer-free controls and patients with lung cancer.
| Variable | Controls (N=900) | Patients (N=695) | p value |
|---|---|---|---|
| Age (yrs) | Mean ± S.D. | Mean ± S.D. | |
| 43.35 ± 18.01 | 59.72 ± 10.81 | p<0.001 | |
| Gender | N (%) | N (%) | |
| Female | 470 (52.2) | 358 (51.5) | |
| Male | 430 (47.8) | 337 (48.5) | p=0.891 |
| Historical or current cigarette smokers | |||
| No | 800 (88.9) | 446 (64.2) | |
| Yes | 100 (11.1) | 249 (35.8) | p<0.001 |
| Alcohol consumption | |||
| No | 783 (87.0) | 472 (67.9) | |
| Yes | 117 (13.0) | 223 (32.1) | p<0.001 |
| Clinical stage | |||
| I | 512 (73.7) | ||
| II, III, IV | 183 (26.3) | ||
| Tumor size | |||
| <T2 | 476 (68.5) | ||
| ≧T2 | 219 (31.5) | ||
| Lymph node status | |||
| N0 | 552 (79.4) | ||
| N1, N2, N3 | 143 (20.6) | ||
| Metastasis | |||
| M0 | 623 (89.6) | ||
| M1 | 72 (10.4) | ||
| Types of lung cancer | |||
| NSCLC | 674 (97.2) | ||
| SCLC | 21 (2.8) | ||
| Histology | |||
| Adenocarcinoma | 541 (77.8) | ||
| Squamous cell carcinoma | 99 (14.2) | ||
| Other carcinoma | 55 (7.9) |
The Fisher's exact test was used to compare independent data between controls and patients with lung cancer. * p value <0.05 was considered statistically significant. NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer.
First, we genotyped the patients with lung cancer (cases) and healthy participants (controls), to evaluate whether the five Ang2 SNPs (rs2442598, rs734701, rs1823375, 11137037, and rs12674822) were associated with the risk of lung cancer. The results are illustrated in Table 2. Alleles with the highest distribution frequency at Ang2 rs2442598, rs734701, rs1823375, 11137037, and rs12674822 in both cases and controls were heterozygous AT, heterozygous TC, homozygous CC, homozygous AA, and heterozygous GT, respectively (Table 2). Individuals carrying the GT heterozygote at rs12674822 had a 1.281-fold (95% CI: 1.016-1.617, p<0.05) higher risk of lung cancer compared with individuals carrying the wild-type GG polymorphic allele. Individuals with polymorphisms at rs2442598, rs734701, rs1823375, and rs11137037 showed no significant associations with lung cancer risk relative to the risk in individuals with the wild-type genotype (Table 2).
Distribution frequencies of Ang2 genotypes in healthy cancer-free controls and patients with lung cancer.
| Variable | Controls N=900 (%) | Patients N=695 (%) | OR (95% CI) | p value |
|---|---|---|---|---|
| rs2442598 | ||||
| AA | 252 (28.0) | 188 (27.1) | 1.00 (reference) | |
| AT | 464 (51.6) | 337 (48.5) | 0.974 (0.769-1.232) | p=0.823 |
| TT | 184 (20.4) | 170 (24.5) | 1.238 (0.935-1.641) | p=0.136 |
| AT+TT | 648 (72.0) | 507 (73.0) | 1.049 (0.840-1.309) | p=0.674 |
| rs734701 | ||||
| TT | 305 (33.9) | 226 (32.5) | 1.00 (reference) | |
| TC | 403 (44.8) | 351 (50.5) | 1.175 (0.940-1.470) | p=0.157 |
| CC | 192 (21.3) | 118 (17.0) | 0.829 (0.623-1.105) | p=0.201 |
| TC+CC | 595 (66.1) | 469 (67.5) | 1.064 (0.862-1.313) | p=0.565 |
| rs1823375 | ||||
| CC | 451 (50.1) | 342 (49.3) | 1.00 (reference) | |
| CG | 372 (41.3) | 282 (40.6) | 1.000 (0.811-1.232) | p=0.998 |
| GG | 77 (8.6) | 71 (10.2) | 1.216 (0.855-1.729) | p=0.276 |
| CG+GG | 449 (49.9) | 353 (50.8) | 1.037 (0.851-1.264) | p=0.721 |
| rs11137037 | ||||
| AA | 417 (46.3) | 344 (49.5) | 1.00 (reference) | |
| AC | 331 (36.8) | 221 (31.8) | 0.809 (0.648-1.011) | p=0.062 |
| CC | 152 (16.9) | 130 (18.7) | 1.037 (0.788-1.364) | p=0.796 |
| AC+CC | 483 (53.7) | 351 (50.5) | 0.881 (0.723-1.074) | p=0.210 |
| rs12674822 | ||||
| GG | 283 (31.4) | 191 (27.5) | 1.00 (reference) | |
| GT | 407 (45.2) | 352 (50.6) | 1.281 (1.016-1.617)* | p=0.036 |
| TT | 210 (23.3) | 152 (21.9) | 1.072 (0.812-1.416) | p=0.622 |
| GT+TT | 617 (68.6) | 504 (72.5) | 1.210 (0.973-1.505) | p=0.086 |
Odds ratios and their confidence intervals were estimated by logistic regression models. * p value <0.05 was considered statistically significant. OR, odds ratio; CI, confidence interval; Ang2, angiopoietin-2.
Next, Ang2 genotypes in patients with lung cancer were evaluated to clarify the role of Ang2 polymorphisms in TNM stage, primary tumor size, lymph node metastasis, and distant metastasis. For 475 patients with lung cancer, a significant correlation between rs11137037 variants (GG vs AG) and clinical stage (OR: 1.584, 95% CI: 1.021-2.456, p<0.05) was observed (Table 3). However, no significant differences were observed between Ang2 rs12674822 genotypes and clinicopathological status (the data are not shown). Distribution frequencies for clinical status in the NSCLC and SCLC cohorts are shown in Table 4. Most of the patients were in the NSCLC category. Significant correlations were observed between types of lung cancer (NSCLC vs SCLC) and clinical stage (OR: 29.543, 95% CI: 6.809-128.181, p<0.001), tumor size (OR: 22.515, 95% CI: 5.196-97.568, p = 0.001), lymph node metastasis (OR: 42.137, 95% CI: 9.688-183.263, p<0.001) and distant metastasis (OR: 11.054, 95% CI: 4.513-27.077, p<0.001), respectively (Table 4). For 527 patients with NSCLC, a significant correlation was observed between rs12674822 variants (GG vs GT) and clinical stage (OR: 1.555, 95% CI: 1.011-2.391, p<0.05; AOR: 1.589, 95% CI: 1.029-2.455, p<0.05, respectively) (Table 5). However, there were no significant differences observed between histology subgroups and polymorphisms of the Ang2 genotypes (the data are not shown).
Odds ratios and their confidence intervals for the clinical status and Ang2 rs11137037 genotypic frequencies of 474 lung cancer patients.
| Variable | AA N=344 (%) | CC N=130 (%) | OR (95% CI) | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I | 260 (75.6) | 86 (66.2) | 1.00 | |
| Stage II/III/IV | 84 (24.4) | 44 (33.8) | 1.584 (1.021-2.456)* | p=0.039 |
| Tumor size | ||||
| <T2 | 239 (69.5) | 79 (60.8) | 1.00 | |
| ≧T2 | 105 (30.5) | 51 (39.2) | 1.469 (0.965-2.237) | p=0.072 |
| Lymph node metastasis | ||||
| No | 279 (81.1) | 95 (67.9) | 1.00 | |
| Yes | 65 (18.9) | 35 (32.1) | 1.581 (0.986-2.536) | p=0.056 |
| Distant metastasis | ||||
| No | 310 (90.1) | 112 (86.2) | 1.00 | |
| Yes | 34 (9.9) | 18 (13.8) | 1.465 (0.796-2.699) | p=0.218 |
Odds ratios with their confidence intervals were estimated by logistic regression models. * p value <0.05 was considered statistically significant. OR, odds ratio; CI, confidence interval; Ang2, angiopoietin-2.
Clinical details of patients with NSCLC or SCLC.
| Variable | NSCLC N=674 (%) | SCLC N=21 (%) | OR (95% CI) | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I | 510 (75.7) | 2 (9.5) | 1.00 | |
| Stage II/III/IV | 164 (24.3) | 19 (90.5) | 29.543 (6.809-128.181)* | p<0.001 |
| Tumor size | ||||
| <T2 | 474 (70.3) | 2 (9.5) | 1.00 | |
| ≧T2 | 200 (29.7) | 19 (90.5) | 22.515 (5.196-97.568)* | p=0.001 |
| Lymph node metastasis | ||||
| No | 550 (81.6) | 2 (9.5) | 1.00 | |
| Yes | 124 (18.4) | 19 (90.5) | 42.137 (9.688-183.263)* | p<0.001 |
| Distant metastasis | ||||
| No | 613 (90.9) | 10 (47.6) | 1.00 | |
| Yes | 61 (9.1) | 11 (52.4) | 11.054 (4.513-27.077)* | p<0.001 |
Odds ratios with their confidence intervals were estimated by logistic regression models.
* p value <0.05 as statistically significant. OR, odds ratio; CI, confidence interval; NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer.
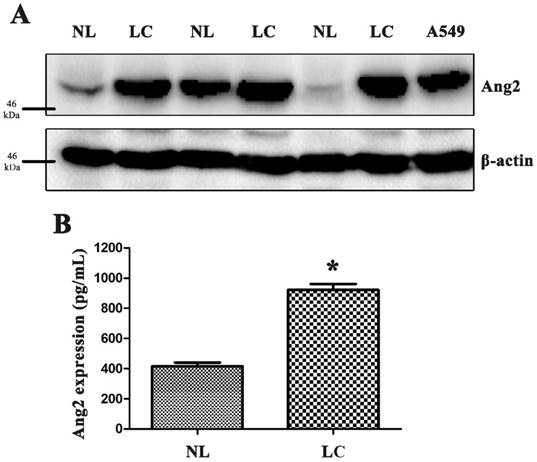
We measured Ang2 protein expression in primary tissues and blood samples from our patients as well as lung cancer cell line, A549 (Fig. 1A and B). We used the Kaplan-Meier survival curve to analyze correlations of Ang2 polymorphisms with overall survival and progression-free survival (supplementary Fig. S1). Cox proportional hazards analysis revealed that patients carrying AC+CC at rs11137037 had longer five-year overall survival (p=0.049; hazard ratio: 0.645, 95% CI: 0.417-0.998), while those carrying GT+TT at rs12674822 had longer progression-free survival (p=0.027; hazard ratio: 0.437, 95% CI: 0.213-0.896), compared with those carrying wild-type variants (the data are not shown).
High expression of Ang2 in lung cancer tissue and serum samples. (A) Human lung cancer and normal lung tissue samples and lung cancer cell line A549 were used to purify proteins, and Ang2 expression was measured by Western blot. (B) Blood serum samples were collected from patients and controls and analyzed by enzyme-linked immunosorbent assay (ELISA). The results are expressed as the mean ± standard error of the mean (S.E.M.) of three separate experiments.
Associations of Ang2 rs12674822 genotypic frequencies with laboratory status in the NSCLC cohort.
| Variable | GG N=182 (%) | GT N=345 (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I | 145 (79.7) | 247 (71.6) | 1.00 | 1.00 |
| Stage II/III/IV | 37 (21.8) | 98 (28.4) | 1.555 (1.011-2.391)* | 1.589 (1.029-2.455)* |
| Tumor size | ||||
| <T2 | 133 (73.1) | 233 (67.5) | 1.00 | 1.00 |
| ≧T2 | 49 (26.9) | 112 (32.5) | 1.305 (0.877-1.942) | 1.332 (0.890-1.994) |
| Lymph node metastasis | ||||
| No | 154 (84.6) | 269 (78.0) | 1.00 | 1.00 |
| Yes | 28 (15.4) | 76 (22.0) | 1.554 (0.965-2.502) | 1.583 (0.980-2.559) |
| Distant metastasis | ||||
| No | 166 (91.2) | 312 (90.4) | 1.00 | 1.00 |
| Yes | 16 (8.8) | 33 (9.6) | 1.097 (0.587-2.052) | 1.111 (0.592-2.083) |
Odds ratios with their confidence intervals were estimated by logistic regression models. Ang2, angiopoietin-2; NSCLC, non-small cell lung cancer; OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio. * p value <0.05 as statistically significant.
Discussion
It is recognized that several SNP genes affect an individual's susceptibility to lung cancer [20-22]. Correlations between Ang2 gene polymorphisms and lung cancer have rarely been documented and most of the evidence is associated with lung injury syndromes [11, 23]. It is essential that we know more about genetic polymorphisms, in order to help us to explore novel therapeutic strategies for lung cancer [24, 25]. To the best of our knowledge, our study provides the first clinical evidence on associations between Ang2 polymorphisms (rs2442598, rs734701, rs1823375, 11137037, and rs12674822) and lung cancer susceptibility, as well as on interactions between these polymorphisms and clinical characteristics in a Chinese Han population. In this study, we increased our overall numbers of participants by incorporating the patients and controls included in our previous study [25]. Our results show that Ang2 gene polymorphisms only slightly increased the susceptibility for lung cancer. This might be because most of our study participants were nonsmokers (i.e., controls and patients), and because the ratios of cigarette smokers/nonsmokers among the controls (11.1:88.9) and patients (35.8:64.2) were relatively normal, as were the proportions of those who consumed alcohol. Although our results demonstrated that smoking and alcohol consumption were significant risk factors for lung cancer (Table 1; both p<0.001), the significance of this association did not persist in Ang2 SNPs genotyping analyses that controlled for smoking and alcohol consumption (these data are not shown).
Ang2 involvement has been observed in lung cancer metastasis [26, 27]. Further investigations have revealed that Ang2 is not only pro-angiogenic but also pro-inflammatory factors in cancer development [28, 29]. We therefore sought to determine if our data could emulate this finding. We initially measured Ang2 expression in lung cancer tissue and serum samples, but the low number of lung cancer samples meant that we could not analyze the immunostaining results for Ang2. Moreover, although the 5-year survival rates among our patients were significantly associated with clinical status (these data are not shown), this significant association disappeared in an Ang2 SNP genotyping analysis (Fig. S1). Owing to this unusual protective survival correlation between Ang2 polymorphism rs11137037 and clinical stage, it might be associated with early clinical stage and well survive of these lung cancer patients. In a previous study, two Ang2 SNPs (rs1868554 and rs2442598) were significantly associated with acute lung injury [11]. However, our results are the first to demonstrate that Ang2 gene polymorphisms at rs12674822 but not at rs2442598 are significantly related to the risk of developing lung cancer. We also found that patients with the CC genotype at rs11137037 were 1.58 times more likely to develop stage II, III or IV disease. The correlation between Ang2 polymorphism and the lung cancer cells required to be further discussed in the future.
Although blockade of the programmed death-1 receptor ligand 1 pathway is a powerful therapeutic strategy for NSCLC [30], this disease still has a dismal prognosis [31, 32], so genetic information that can be useful for lung cancer prognosis or prediction is essential. Our results reveal a significant correlation between NSCLC and clinical status in 695 Chinese Han patients; in 527 patients with NSCLC, those with Ang2 polymorphisms carrying the GT genotype at rs12674822 were 1.56-fold more likely to develop stage II, III or IV disease. Others Ang2 polymorphisms were not associated with a certain type of lung cancer since the populations were too small to analyze. On the other hand, a reconstructed linkage disequilibrium plot of the five Ang2 SNPs failed to reveal any significant difference between the clinical stages of the disease (supplementary Fig. S2).
In summary, our results demonstrate an association between one Ang2 gene variant and risk of lung cancer, with a significant association between Ang2 SNP rs12674822 and stage II, III, or IV disease in a Chinese Han population. This study is the first to report any such correlation between Ang2 polymorphisms and risk of lung cancer. Our evidence indicates that Ang2 could be developed as a genetic prognostic marker for lung cancer prognosis.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by grants from China's National Natural Science Foundation (No. 81702117) and China Medical University Hospital (DMR-108-073).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
3. Chen W, Sun K, Zheng R. et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1-12
4. Zhu J, Zeng Y, Xu C. et al. Expression profile analysis of microRNAs and downregulated miR-486-5p and miR-30a-5p in non-small cell lung cancer. Oncol Rep. 2015;34:1779-86
5. Maisonpierre PC, Suri C, Jones PF. et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55-60
6. Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512-23
7. Tung MC, Wen YC, Wang SS. et al. Dopamine receptor D2 genetic variations is associated with the risk and clinicopathological variables of urothelial cell carcinoma in a Taiwanese population. Int J Med Sci. 2018;15:1187-1193
8. Lee CY, Ng SC, Hsiao YH. et al. Impact of the Receptor for Advanced Glycation End Products Genetic Polymorphisms on the Progression in Uterine Cervical Cancer. J Cancer. 2018;9:3886-3893
9. Yang WH, Wang SJ, Chang YS. et al. Association of Resistin Gene Polymorphisms with Oral Squamous Cell Carcinoma Progression and Development. Biomed Res Int. 2018;2018:9531315
10. Makhoul I, Todorova VK, Siegel ER. et al. Germline Genetic Variants in TEK, ANGPT1, ANGPT2, MMP9, FGF2 and VEGFA Are Associated with Pathologic Complete Response to Bevacizumab in Breast Cancer Patients. PLoS One. 2017;12:e0168550
11. Meyer NJ, Li M, Feng R. et al. ANGPT2 genetic variant is associated with trauma-associated acute lung injury and altered plasma angiopoietin-2 isoform ratio. Am J Respir Crit Care Med. 2011;183:1344-53
12. Su L, Zhai R, Sheu CC. et al. Genetic variants in the angiopoietin-2 gene are associated with increased risk of ARDS. Intensive Care Med. 2009;35:1024-30
13. He L, Dang L, Zhou J. et al. Association of angiopoietin-1, angiopoietin-2 and caspase-5 polymorphisms with psoriasis vulgaris. Clin Exp Dermatol. 2015;40:556-63
14. Miaskowski C, Dodd M, Paul SM. et al. Lymphatic and angiogenic candidate genes predict the development of secondary lymphedema following breast cancer surgery. PLoS One. 2013;8:e60164
15. Wang L, Tang CH, Lu T. et al. Resistin polymorphisms are associated with rheumatoid arthritis susceptibility in Chinese Han subjects. Medicine (Baltimore). 2018;97:e0177
16. Wang CQ, Tang CH, Wang Y. et al. FSCN1 gene polymorphisms: biomarkers for the development and progression of breast cancer. Sci Rep. 2017;7:15887
17. Su CM, Hsu CJ, Tsai CH. et al. Resistin Promotes Angiogenesis in Endothelial Progenitor Cells Through Inhibition of MicroRNA206: Potential Implications for Rheumatoid Arthritis. Stem Cells. 2015;33:2243-55
18. Su CM, Hu SL, Sun Y. et al. Myostatin induces tumor necrosis factor-alpha expression in rheumatoid arthritis synovial fibroblasts through the PI3K-Akt signaling pathway. J Cell Physiol. 2018
19. Su CM, Tang CH, Chi MJ. et al. Resistin facilitates VEGF-C-associated lymphangiogenesis by inhibiting miR-186 in human chondrosarcoma cells. Biochem Pharmacol. 2018;154:234-242
20. Liu C, Cui H, Gu D. et al. Genetic polymorphisms and lung cancer risk: Evidence from meta-analyses and genome-wide association studies. Lung Cancer. 2017;113:18-29
21. Hu W, Liu PY, Yang YC. et al. Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects. Int J Med Sci. 2017;14:1197-1202
22. Huang CY, Hsieh MJ, Wu WJ. et al. Association of endothelial nitric oxide synthase (eNOS) polymorphisms with EGFR-mutated lung adenocarcinoma in Taiwan. J Cancer. 2018;9:2518-2524
23. Wang H, Cade BE, Chen H. et al. Variants in angiopoietin-2 (ANGPT2) contribute to variation in nocturnal oxyhaemoglobin saturation level. Hum Mol Genet. 2016;25:5244-5253
24. Ishikawa T. Genetic polymorphism in the NRF2 gene as a prognosis marker for cancer chemotherapy. Front Genet. 2014;5:383
25. Hu WW, Tang CH, Sun Y. et al. Correlation between resistin gene polymorphism and clinical aspects of lung cancer. Medicine (Baltimore). 2017;96:e9485
26. Park JS, Kim IK, Han S. et al. Normalization of Tumor Vessels by Tie2 Activation and Ang2 Inhibition Enhances Drug Delivery and Produces a Favorable Tumor Microenvironment. Cancer Cell. 2016;30:953-967
27. Coelho AL, Gomes MP, Catarino RJ. et al. CSF-1 and Ang-2 serum levels - prognostic and diagnostic partners in non-small cell lung cancer. ESMO Open. 2018;3:e000349
28. Scholz A, Plate KH, Reiss Y. Angiopoietin-2: a multifaceted cytokine that functions in both angiogenesis and inflammation. Ann N Y Acad Sci. 2015;1347:45-51
29. Wu FT, Man S, Xu P. et al. Efficacy of Cotargeting Angiopoietin-2 and the VEGF Pathway in the Adjuvant Postsurgical Setting for Early Breast, Colorectal, and Renal Cancers. Cancer Res. 2016;76:6988-7000
30. Rolfo C, Caglevic C, Santarpia M. et al. Immunotherapy in NSCLC: A Promising and Revolutionary Weapon. Adv Exp Med Biol. 2017;995:97-125
31. Blanco-Prieto S, De Chiara L, Rodriguez-Girondo M. et al. Highly Sensitive Marker Panel for Guidance in Lung Cancer Rapid Diagnostic Units. Sci Rep. 2017;7:41151
32. Ramalingam S, Dinan MA, Crawford J. Survival comparison in patients with Stage IV lung cancer in Academic versus Community Centers in the United States. J Thorac Oncol. 2018
Author contact
![]() Corresponding author: Chen-Ming Su, PhD, Department of Biomedical Sciences Laboratory, Affiliated Dongyang Hospital of Wenzhou Medical University, No.60 Wuning west Rd., Dongyang, Zhejiang, China 322100. Tel: (86) 579-86857846; E-mail: ericsucmcom; proof814com
Corresponding author: Chen-Ming Su, PhD, Department of Biomedical Sciences Laboratory, Affiliated Dongyang Hospital of Wenzhou Medical University, No.60 Wuning west Rd., Dongyang, Zhejiang, China 322100. Tel: (86) 579-86857846; E-mail: ericsucmcom; proof814com

 Global reach, higher impact
Global reach, higher impact