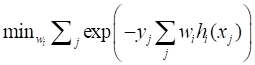Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(15):3323-3332. doi:10.7150/jca.29693 This issue Cite
Research Paper
Molecular Decision Tree Algorithms Predict Individual Recurrence Pattern for Locally Advanced Nasopharyngeal Carcinoma
1. Department of Radiation Oncology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou 510655, China
2. School of Computer Science and Engineering, South China University of Technology, Guangzhou 510641, China
3. Department of Rhinology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China
4. Department of Pathology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou 510060, China
5. Department of Nasopharyngeal Carcinoma, State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, Guangzhou 510060, China
6. Department of Medical Oncology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou 510060, China
7. Department of Medical Radiology, State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, China
*These authors contributed equally to this work.
Received 2018-9-4; Accepted 2019-4-25; Published 2019-6-2
Abstract
Background: Recurrence remains one of the key reasons of relapse after the radical radiation for locally advanced nasopharyngeal carcinoma (NPC). Here, the multiple molecular and clinical variables integrated decision tree algorithms were designed to predict individual recurrence patterns (with VS without recurrence) for locally advanced NPC.
Methods: A total of 136 locally advanced NPC patients retrieved from a randomized controlled phase III trial, were included. For each patient, the expression levels of 33 clinicopathological biomarkers in tumor specimen, 3 Epstein-Barr virus related serological antibody titer and 5 clinicopathological variables, were detected and collected to construct the decision tree algorithm. The expression level of 33 clinicopathological biomarkers in tumor specimen was evaluated by immunohistochemistry staining.
Results: Three algorithm classifiers, augmented by the adaptive boosting algorithm for variable selection and classification, were designed to predict individual recurrence pattern. The classifiers were trained in the training subset and further tested using a 10-fold cross-validation scheme in the validation subset. In total, 13 molecules expression level in tumor specimen, including AKT1, Aurora-A, Bax, Bcl-2, N-Cadherin, CENP-H, HIF-1α, LMP-1, C-Met, MMP-2, MMP-9, Pontin and Stathmin, and N stage were selected to construct three 10-fold cross-validation decision tree classifiers. These classifiers showed high predictive sensitivity (87.2-93.3%), specificity (69.0-100.0%), and overall accuracy (84.5-95.2%) to predict recurrence pattern individually. Multivariate analyses confirmed the decision tree classifier was an independent prognostic factor to predict individual recurrence (algorithm 1: hazard ration (HR) 0.07, 95% confidence interval (CI) 0.03-0.16, P < 0.01; algorithm 2: HR 0.13, 95% CI 0.04-0.44, P < 0.01; algorithm 3: HR 0.13, 95% CI 0.03-0.68, P = 0.02).
Conclusion: Multiple molecular and clinicopathological variables integrated decision tree algorithms may individually predict the recurrence pattern for locally advanced NPC. This decision tree algorism provides a potential tool to select patients with high recurrence risk for intensive follow-up, and to diagnose recurrence at an earlier stage for salvage treatment in the NPC endemic region.
Keywords: nasopharyngeal carcinoma, decision tree algorithms, classifiers, recurrence pattern
Introduction
Nasopharyngeal carcinoma (NPC), an Epstein-Barr virus (EBV) associated malignancy, has the highest incidence in endemic Southern China [1-3]. Although concurrent radiochemotherapy partially improves progression free survival, the disease recurrence remains the major cause of cancer mortality for locally advanced NPC [4]. For local recurrences, re-irradiation always causes a series of severe late toxicity, such as radiation encephalopathy, carotid blowout, cervical skin and muscle fibrosis [5-7]. The treatment efficacy was however dissatisfied that the 3-year overall survival was always less than 50.0%, and coupled with low quality of life [5,6]. By contrast, for early stage residual or recurrence (stage I and II), either the treatment of endoscopic nasopharyngectomy or interstitial intensity-modulated brachytherapy, the 3-year disease free survival may reach to 85.7-97.4%, with acceptable early and late complications [8,9]. Therefore, identifying the high recurrence risk individuals at or prior to their earlier recurrence would greatly benefit patients from timely salvage treatment for NPC.
Aberrant expression of tumor related biomarkers have been reported to be potential predictor of tumor recurrence [10,11]. However, the single prognostic tumor biomarker or Tumor-Node-Metastasis prognostication system was also found to have the limitation of low sensitivity and specificity to predict tumor recurrence individually [12]. By contrast, multi-molecules or variables integrated data mining methods provided a novel way to personally predict cancer patients' outcome. In breast cancer, a 21-gene recurrence algorithm approach showed high predictive power to categorize the individual patient as low risk and high risk recurrence (6.8% vs. 30.5%) at the 10-year follow-up [13,14]. Using the LASSO Cox regression model and miRNA microarrays, the six-miRNA-based classifier was able to classify patients between those at high risk of disease progression and those at low risk of disease progression in stage II colon cancer [15]. Similarly, a gene-expression-based radiation-Sensitivity index and the linear quadratic model integrated approach allowed the individualisation of radiotherapy dose to tumor radiosensitive [16]. These studies revealed that patient-specific molecular signature might be a precision way to predict cancer patient outcome.
Here, we sought to identify the 5-year recurrence pattern (with VS without recurrence) individually by designing 10-fold cross-validation decision tree algorithm classifiers for locally advanced NPC. The expression level of 33 tumor related biomarkers and 3 EBV-related serological biomarkers, coupled with 5 clinicopathological variables, were integrated into the classical decision tree learning algorisms. The trained decision tree model was further validated in separated subgroups to test its efficacy in predicting the individual recurrence pattern for locally advanced NPC.
Materials and methods
Study design
The study population of 136 locally advanced NPC patients was originated from a prospective randomized controlled phase III trial as previously described [17]. The start date of the enrolled was August 2002, and the latest date of each patient being followed up was May 2010. Clinical stage was defined according to the NPC staging system of China [18]. The patients were treated with induction chemotherapy plus concurrent adjuvant chemoradiotherapy (IC/CCRT) or induction chemotherapy plus radiotherapy (IC/RT). In IC/RT subset, patients received two cycles of floxuridine plus carboplatin (floxuridine 750 mg/m2, d1-5; carboplatin AUC = 6) chemotherapy and underwent radiotherapy one-week thereafter. In the IC/CCRT subgroup, one week after completion of two cycles of floxuridine plus carboplatin (floxuridine 750 mg/m2, d1-5; carboplatin AUC = 6) chemotherapy, patients received radiotherapy and concurrent carboplatin (AUC = 6) chemotherapy on day 7, 28 and 49, respectively. Here, 72 IC/RT patients and 64 IC/CCRT cases were included. In this study, patients received two-dimensional (2D) radiotherapy based on traditional Co60 γ-ray or linear accelerator 6-8 MV photon. The radiation fields were determined by the invasion field of the tumor and local regional cervical lymph node. The radiotherapy fields arrangement was divided into 2 parts. At the first course, two lateral opposing faciocervical portals were administered to 36-40 Gy irradiation. At the second course, facio-cervical splitting portals course was received. The accumulated radiation to the primary tumor was 68-72 Gy. For the neck region, 50 to 70 Gy was obtained according to the extent of the lymph node invasiveness. For lymph node negative and positive invaded necks, 50 Gy and 60-70 Gy radiation would respectively be given.
This study was approved by the institutional review board at the Sixth Affiliated Hospital of Sun Yat-sen University, and written informed consent was obtained from all participants.
Immunohistochemical (IHC) staining measurement and EBV-related serological antibodies assay
Tissue microassays and IHC staining were performed to detect the tumor biomarkers expression level under the protocol as previously reported [19]. In total, 33 tumor biomarkers were selected and subjected to IHC staining: cyclin D1, 14-3-3σ, Aurora-A, CENP-H, Stathmin, P21, CDC2, P27, ERK, p-ERK, Ki-67, E-Cadherin, β-Catenin, N-Cadherin, Snail, Twist, C-Met, nm23-H1, HIF-1α, COX2, MMP-2, MMP-9, TIMP-2, CD31, CD34, Bax, Bcl-2, Survivin, AKT 1, Pontin, Beclin 1, EZH2 and LMP 1. The detailed antibodies and dilution information are summarized in Table S1. The serological titer of EBV related antibodies, EA-IgA, VCA-IgA and anti-enzyme rate (AER) of EBV DNase-specific neutralizing antibody, were tested by ELISA assay [20]. The microvessel densities were evaluated by counting CD31-positive and CD34-positive capillaries (vascular endothelial cell markers) in the three most vascularised areas (''hotspots''). Prior to IHC staining, each primary antibody was tested according to the manufacturer's datasheet with recommended positive control. Moreover, a non-immune serum immunoglobulin at the 1:200 dilutions was also used to replace the primary antibody as a negative control.
We semi-quantitatively assessed each tumor biomarker expression level by measuring the staining intensity and extent as previously reported [21]. Briefly, the staining intensity was graded as follows: negative (score 0), bordering (score 1), weak (score 2), moderate (score 3) and strong (score 4). The staining extent was ranked into four parts according to the percentage of positive staining cells in the field: 0-25% (score 1), 26-50% (score 2), 51-75% (score 3) and 76-100% (score 4). The overall score was obtained by multiplying the staining intensity and extent. The scores were assessed by two independent pathologists blinded to the clinical follow-up. Any discrepancies between these two pathologists were referred to a third pathological expert.
Selection of variable for decision tree algorithm
The univariate analysis is limited in prognostic variable selection, since it ignores the combinational potentials among individual factors which may provide joint beneficiaries. Earlier studies had shown that favourable classification accuracy could be obtained by a sophisticated feature subset selection (FSS) approach [22,23]. Here, we conducted a FSS method to select a pool of informative biomarkers, which were called feature herein, that were able to dichotomize the individuals into high and low risk to recurrence. To achieve the task of FSS, a hybrid filter-wrapper algorithm was utilized [24]. Specifically, we firstly ranked the importance of feature by LH-RELIEF model to select the top 20 variables [25]. These selected 20 variables were then scrutinized by FSS via wrapping of classical classification model to remove the redundant variables, which were believed to rarely contribute to the classification. Further, the classification model of random tree was used to generate decision rules. The resampling technique, also known as Adaptive Boosting (AdaBoost), was subsequently employed to enhance the classification performances [26]. To alleviate the computation cost in wrapping, a genetic algorithm was used to seek an informative feature subset. Once the candidate subset was obtained, it was further scrutinized to remove the redundant features by adding or removing of a particular variable to quantify its loss energy. Finally, a compact yet highly informative feature subset was obtained by preserving of the variables that were highly relevant to the classification process. The main advantage of this hybrid approach lies in that it keeps a great part of wrapper advantages while reducing the computation cost greatly. The detailed description of the FSS model is shown in the Supplementary Methods.
Recurrence pattern classification by decision tree algorithm
In order to validate the predictive power of the identified biomarkers pool, simulation experiments were conducted on the enrolled cases by using the AdaBoost algorithm with decision tree serving as the weak classifier. The strong point of the AdaBoost algorithm was that each case of the training set acted in a different role for discrimination at different training stages. Those cases that were incorrectly classified in the previous rounds would be given more attention. Therefore, the weak learner was forced to focus on the more informative examples of the training set. The AdaBoost algorithm was implemented by inducing decision trees using the gain ratio criterion for feature selection. The algorithm to generate the ensemble was identical to the idea proposed by Freund and Schapire [27]. The algorithm can be viewed as stage-wise for minimizing a particular error function. In the i-th iteration, the learning algorithm is invoked to minimize the weighted error on the training set by returning a weak classifier hi. Then, the weighted error hi is computed to update the weights on the training samples. The weight-updating scheme places more weight on training examples that were misclassified by hi and less weight on examples that were correctly classified. Therefore, AdaBoost constructs progressively more difficult learning problems in subsequent iterations. The final classifier is obtained by a weighted vote of each individual classifier via minimizing a margin error function.
Here, the weight wi for the i-th classifier is decided by its accuracy in the weighted training set.
Participant characteristics and association with recurrence free survival
| Characteristic (n = 136) | No. (%) | Univariate, HR (95% CI) | P Value |
|---|---|---|---|
| Age, year (> 43 vs ≤ 43) | 47.8 vs 52.2 | 1.01 (0.58 to 1.77) | 0.97 |
| Gender (Male vs Female) | 78.7 vs 21.3 | 1.05 (0.54 to 2.05) | 0.89 |
| T stage (T1-T2 vs T3-T4) | 17.6 vs 82.4 | 0.64 (0.27 to 1.52) | 0.31 |
| N stage (N0-N1 vs N2-N3) | 44.9 vs 55.1 | 0.79 (0.45 to 1.41) | 0.43 |
| Overall stage (III vs IVa) | 54.4 vs 45.6 | 0.93 (0.53 to 1.63) | 0.80 |
| Treatment (IC/RT vs IC/CCRT) | 52.9 vs 47.1 | 1.10 (0.62 to 1.94) | 0.75 |
| 14-3-3σa (> 7.0 vs ≤ 7.0) | 54.2 vs 45.8 | 1.53 (0.81 to 2.89) | 0.19 |
| AKT1a (> 5.0 vs ≤ 5.0) | 47.5 vs 52.5 | 1.44 (0.79 to 2.62) | 0.23 |
| Aurora-Aa (≤ 7.0 vs > 7.0) | 46.7 vs 53.3 | 0.36 (0.20 to 0.66) | < 0.01 |
| Baxa (≤ 3.5 vs > 3.5) | 48.1 vs 51.9 | 0.75 (0.42 to 1.31) | 0.30 |
| Bcl-2a (> 3.5 vs ≤ 3.5) | 52.9 vs 47.1 | 0.78 (0.44 to 1.38) | 0.40 |
| Beclin 1a (≤ 8.0 vs > 8.0) | 70.1 vs 29.9 | 0.59 (0.33 to 1.06) | 0.07 |
| β-catenina (> 5.0 vs ≤ 5.0) | 26.7 vs 73.3 | 0.79 (0.42 to 1.46) | 0.45 |
| CDC2a (≤ 5.0 vs > 5.0) | 51.5 vs 48.5 | 0.71 (0.40 to 1.25) | 0.24 |
| CD31b (≤ 379.1 vs > 379.1) | 48.9 vs 51.1 | 0.71 (0.34 to 1.48) | 0.36 |
| CD34b (≤ 380.5 vs > 380.5) | 47.7 vs 52.3 | 0.64 (0.31 to 1.33) | 0.23 |
| CENP-Ha (≤ 5.0 vs > 5.0) | 43.7 vs 56.3 | 0.73 (0.42 to 1.28) | 0.28 |
| C-Meta (≤ 5.0 vs > 5.0) | 45.5 vs 54.5 | 0.89 (0.51 to 1.56) | 0.68 |
| COX2a (≤ 5.0 vs > 5.0) | 55.6 vs 44.4 | 0.83 (0.47 to 1.47) | 0.53 |
| Cyclin D1a (≤ 3.5 vs > 3.5) | 48.5 vs 51.5 | 0.64 (0.36 to 1.13) | 0.12 |
| E-Cadherina (> 3.5 vs ≤ 3.5) | 50.4 vs 49.6 | 1.10 (0.62 to 1.92) | 0.75 |
| ERKa (≤ 5.0 vs > 5.0) | 43.4vs 56.6 | 0.81 (0.46 to 1.43) | 0.47 |
| EZH2a (≤ 8.5 vs > 8.5) | 50.7 vs 49.3 | 0.82 (0.47 to 1.44) | 0.49 |
| HIF-1αa (≤ 5.0 vs > 5.0) | 45.2 vs 54.8 | 0.61 (0.34 to 1.07) | 0.08 |
| Ki-67a (≤ 5.0 vs > 5.0) | 46.7 vs 53.3 | 0.78 (0.44 to 1.36) | 0.37 |
| LMP1a (> 5.0 vs ≤ 5.0) | 42.1 vs 57.9 | 0.94 (0.51 to 1.73) | 0.84 |
| MMP-2a (≤ 7.0 vs > 7.0) | 46.7 vs 53.3 | 0.64 (0.37 to 1.13) | 0.13 |
| MMP-9a (> 1.5 vs ≤ 1.5) | 48.5 vs 51.5 | 1.60 (0.83 to 3.06) | 0.16 |
| N-Cadherina (≤ 5.0 vs > 5.0) | 44.0 vs 56.0 | 0.69 (0.39 to 1.21) | 0.19 |
| nm23-H1a (≤ 5.0 vs > 5.0) | 55.9 vs 44.1 | 0.62 (0.34 to 1.12) | 0.11 |
| P21a (≤ 3.5 vs > 3.5) | 48.9 vs 51.1 | 0.77 (0.44 to 1.35) | 0.35 |
| P27a (≤ 7.0 vs > 7.0) | 42.6 vs 57.4 | 0.49 (0.28 to 0.85) | 0.01 |
| p-ERKa (> 2.5 vs ≤ 2.5) | 48.0 vs 52.0 | 1.12 (0.58 to 2.16) | 0.73 |
| Pontina (> 3.5 vs ≤ 3.5) | 50.8 vs 49.2 | 0.86 (0.48 to 1.54) | 0.62 |
| Snaila (> 3.5 vs ≤ 3.5) | 57.8 vs 42.2 | 1.14 (0.65 to 2.00) | 0.65 |
| Stathmina (≤ 7.0 vs > 7.0) | 44.1 vs 55.9 | 0.69 (0.40 to 1.22) | 0.20 |
| Survivina (> 2.5 vs ≤ 2.5) | 50.0 vs 50.0 | 1.10 (0.63 to 1.92) | 0.75 |
| TIMP-2a (> 7.0 vs ≤ 7.0) | 50.0 vs 50.0 | 1.07 (0.61 to 1.88) | 0.81 |
| Twista (> 2.5 vs ≤ 2.5) | 54.9 vs 45.1 | 0.96 (0.55 to 1.70) | 0.90 |
| EA-IgAc (≤ 1:40 vs > 1:40) | 31.8 vs 68.2 | 0.89 (0.49 to 1.63) | 0.71 |
| VCA-IgAc (≤ 1:160 vs > 1:160) | 59.1 vs 40.9 | 0.86 (0.48 to 1.56) | 0.63 |
| AERc (≤ 64.5% vs > 64.5%) | 49.2 vs 50.8 | 0.63 (0.35 to 1.13) | 0.12 |
Statistical methods
The recurrence free survival (RFS) was defined as the time of diagnosis to the date of local and regional recurrence or the date of death or when censused at the latest date. After the completion of treatment, each patient was followed up at 3-month intervals during the first 3 years and at 6-month intervals thereafter. The univariate and multivariate proportional hazards model were employed to estimate the hazard ratio (HR) and 95% confidence interval (CI). The RFS was calculated by Kaplan-Meier analysis and log-rank tests. A two-tailed P < 0.05 was considered statistically significant. The statistical analysis was performed utilizing SPSS v.17.0.
Results
Study population
In our previously reported randomized controlled phase III clinical trial, we showed that IC/CCRT subgroup had a comparable recurrence probability with IC/RT subset for locally advanced NPC [17]. Here, we included 64 IC/CCRT and 72 IC/RT patients from this trial (Table 1). The molecular and clinicopathological features of these two subgroups are shown in Table 2 and Table 3. The median RFS was respectively 65.0 and 64.0 months for IC/CCRT and IC/RT subgroups. The 3-year and 5-year RFS ratios were 73.9% and 66.1% for the IC/CCRT subgroup, and 70.1% and 62.8%, for the IC/RT subgroup, respectively (all with P value > 0.05). Thus, the two subgroups had very similar clinicopathological features, and therefore were suitable for further training and validation by using the decision tree algorithm.
Construction of decision tree algorithm to individually predict recurrence pattern
In this study, the hybrid filter-wrapper algorithm was employed to select a subset of potential prognostic variables [24]. The proposed FSS process was conducted on 64 IC/CCRT, 72 IC/RT, and overall patients. Furthermore, the prognostic power of these identified biomarker panel was validated using the AdaBoost algorithm with decision tree serving as the weak classifier [27]. In particular, a rigorous 10-fold cross-validation scheme was used to quantify the prognostic performance of the trained model. In the 10-fold cross-validation scheme, patients were randomly divided into 10 equal sized subgroups wherein nine subsets were used as training set to construct the classification algorithm while the tenth one was utilized as validation set to test the predictive performance. This 10-fold cross-validation would be cycled 10 times to guarantee each subgroup was used as a validation set one time. The averaged performance was used to quantify the overall accuracy of the constructed recurrence prediction decision tree algorithm.
Prediction of individual recurrence pattern on overall patients by decision tree algorithm
Nine of the 33 molecular variables (Table 1), including Aurora-A, AKT1, Bcl-2, LMP-1, MMP-9, MMP-2, Stathmin, Pontin and C-Met, were selected to the recurrence prediction biomarker pool. By using this 9-variable pool, a recurrence decision tree model was built and then tested by 10-fold cross-validation scheme on training subset of the overall patients. To validate the performance of this recurrence prediction model, six quantitative measurements, including positive predictive value (PPV), negative predictive value (NPV), sensitivity, specificity, area under curve (AUC), and overall accuracy were calculated.
Participant characteristics and association with recurrence free survival in IC/CCRT subgroup
| Characteristic | No. (%)a | Univariate, HR (95% CI) | P Value |
|---|---|---|---|
| Age, year (> 43 vs ≤ 43) | 57.8 vs 42.2 | 0.92 (0.44 to 1.93) | 0.82 |
| Gender (Male vs Female) | 81.3 vs 19.7 | 1.63 (0.48 to 5.54) | 0.43 |
| T stage (T1-T2 vs T3-T4) | 25.0 vs 75.0 | 1.01 (0.31 to 3.35) | 0.98 |
| N stage (N0-N1 vs N2-N3) | 48.4 vs 51.6 | 1.25 (0.58 to 2.70) | 0.58 |
| Overall stage (III vs IVa) | 60.9 vs 39.1 | 1.07 (0.51 to 2.25) | 0.86 |
| 14-3-3σb (> 7.0 vs ≤ 7.0) | 52.9 vs 47.1 | 0.60 (0.26 to 1.42) | 0.25 |
| AKT1b (> 5.0 vs ≤ 5.0) | 43.1 vs 56.9 | 0.96 (0.43 to 2.14) | 0.92 |
| Aurora-Ab (≤ 7.0 vs > 7.0) | 60.3 vs 39.7 | 3.56 (1.50 to 8.42) | < 0.01 |
| Baxb (≤ 3.5 vs > 3.5) | 54.8 vs 45.2 | 1.71 (0.80 to 3.64) | 0.17 |
| Bcl-2b (> 3.5 vs ≤ 3.5) | 48.4 vs 51.6 | 1.50 (0.70 to 3.23) | 0.30 |
| Beclin 1b (≤ 8.0 vs > 8.0) | 71.9 vs 28.1 | 1.97 (0.90 to 4.30) | 0.08 |
| β-cateninb (> 5.0 vs ≤ 5.0) | 76.6 vs 23.4 | 1.11 (0.49 to 2.54) | 0.80 |
| CDC2b (≤ 5.0 vs > 5.0) | 50.0 vs 50.0 | 1.97 (0.91 to 4.28) | 0.09 |
| CD31c (≤ 379.1 vs > 379.1) | 46.3 vs 53.7 | 1.05 (0.38 to 2.89) | 0.93 |
| CD34c (≤ 380.5 vs > 380.5) | 43.9 vs 56.1 | 1.45 (0.54 to 3.86) | 0.46 |
| CENP-Hb (≤ 5.0 vs > 5.0) | 63.0 vs 37.0 | 1.359 (0.65 to 2.86) | 0.42 |
| C-Metb (≤ 5.0 vs > 5.0) | 52.4 vs 47.6 | 0.63 (0.28 to 1.39) | 0.25 |
| COX2b (≤ 5.0 vs > 5.0) | 42.9 vs 57.1 | 1.05 (0.50 to 2.20) | 0.91 |
| Cyclin D1b (≤ 3.5 vs > 3.5) | 53.1 vs 46.9 | 1.739 (0.81 to 3.72) | 0.15 |
| E-Cadherinb (> 3.5 vs ≤ 3.5) | 54.7 vs 45.3 | 1.24 (0.59 to 2.60) | 0.57 |
| ERKb (≤ 5.0 vs > 5.0) | 59.4 vs 40.6 | 0.94 (0.45 to 1.98) | 0.88 |
| EZH2b (≤ 8.5 vs > 8.5) | 46.9 vs 53.1 | 1.31 (0.62 to 2.76) | 0.47 |
| HIF-1αb (≤ 5.0 vs > 5.0) | 54.0 vs 46.0 | 1.23 (0.59 to 2.59) | 0.58 |
| Ki-67b (≤ 5.0 vs > 5.0) | 54.7 vs 45.3 | 1.13 (0.54 to 2.38) | 0.74 |
| LMP1b (> 5.0 vs ≤ 5.0) | 68.5 vs 31.5 | 1.00 (0.44 to 2.27) | 0.99 |
| MMP-2b (≤ 7.0 vs > 7.0) | 57.1 vs 42.9 | 1.50 (0.71 to 3.17) | 0.29 |
| MMP-9b (> 1.5 vs ≤ 1.5) | 47.9 vs 52.1 | 0.38 (0.15 to 1.00) | 0.05 |
| N-Cadherinb (≤ 5.0 vs > 5.0) | 54.7 vs 45.3 | 1.92 (0.90 to 4.11) | 0.09 |
| nm23-H1b (≤ 5.0 vs > 5.0) | 53.8 vs 46.2 | 2.05 (0.87 to 4.85) | 0.10 |
| P21b (≤ 3.5 vs > 3.5) | 55.6 vs 44.4 | 1.03 (0.49 to 2.15) | 0.95 |
| P27b (≤ 7.0 vs > 7.0) | 59.4 vs 40.6 | 2.23 (1.04 to 4.78) | 0.04 |
| p-ERKb (> 2.5 vs ≤ 2.5) | 57.8 vs 42.2 | 0.92 (0.37 to 2.26) | 0.85 |
| Pontinb (> 3.5 vs ≤ 3.5) | 46.7 vs 53.3 | 1.70 (0.76 to 3.80) | 0.20 |
| Snailb (> 3.5 vs ≤ 3.5) | 39.7 vs 60.3 | 1.08 (0.51 to 2.29) | 0.83 |
| Stathminb (≤ 7.0 vs > 7.0) | 57.8 vs 42.2 | 2.22 (1.04 to 4.75) | 0.04 |
| Survivinb (> 2.5 vs ≤ 2.5) | 57.8 vs 42.2 | 1.09 (0.52 to 2.30) | 0.82 |
| TIMP-2b (> 7.0 vs ≤ 7.0) | 53.1 vs 46.9 | 0.92 (0.44 to 1.93) | 0.82 |
| Twistb (> 2.5 vs ≤ 2.5) | 46.0 vs 54.0 | 0.76 (0.36 to 1.59) | 0.46 |
| EA-IgAd (≤ 1:40 vs > 1:40) | 74.2 vs 25.8 | 1.11 (0.50 to 2.45) | 0.80 |
| VCA-IgAd (≤ 1:160 vs > 1:160) | 41.9 vs 58.1 | 1.12 (0.51 to 2.47) | 0.78 |
| AERd (≤ 64.5% vs > 64.5%) | 52.5 vs 47.5 | 1.21 (0.55 to 2.67) | 0.64 |
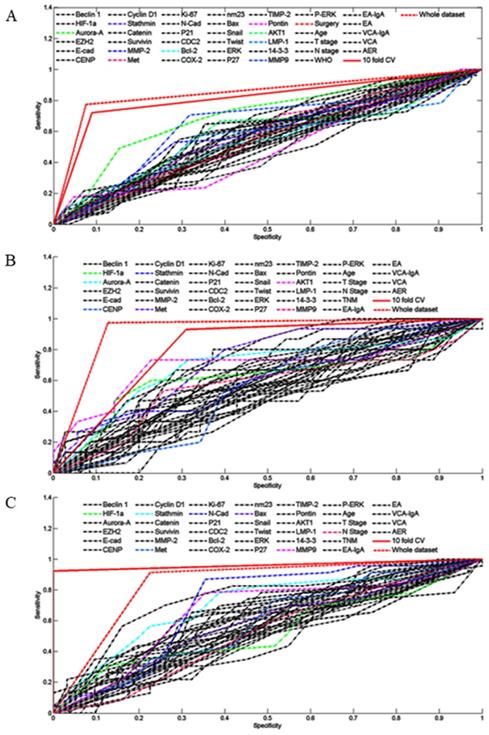
By using 10-fold cross-validation scheme, the sensitivity and specificity to predict 5-year recurrence pattern (with recurrence vs. without recurrence) on the training subsets were 91.0% and 72.0%, respectively. The overall performance on overall patient was satisfactory: PPV of 87.6%, NPV of 85.4%, sensitivity of 92.4%, specificity of 77.4%, and AUC of 92.2% (Fig. 1A, Table S2). In total, this 10-fold cross-validation scheme accurately identified the recurrence pattern for 118 patients, and the overall accuracy was 86.9%. Importantly, a significant RFS difference was detected between the subgroups that were identified as with recurrence and without recurrence. Specifically, the median RFS for high recurrence risk subgroup was 22.8 months compared with 61.7 months for the low recurrence risk subset (P < 0.001, Fig. 2A). As expected, this 10-fold cross-validation scheme was confirmed to be an independent prognostic factor to predict tumor recurrence (Table 4). Taken together, these results suggested that the 10-fold cross-validation decision tree model was indeed a powerful approach to predict the patient recurrence pattern individually.
Participant characteristics and association with recurrence free survival in IC/RT subgroup
| Characteristic | No. (%)a | Univariate, HR (95% CI) | P Value |
|---|---|---|---|
| Age, year (> 43 vs ≤ 43) | 43.5 vs 56.5 | 1.03 (0.44 to 2.46) | 0.94 |
| Gender (Male vs Female) | 78.3vs 21.7 | 0.82 (0.36 to 1.87) | 0.64 |
| T stage (T1-T2 vs T3-T4) | 10.1vs 89.9 | 2.17 (0.64 to 7.37) | 0.22 |
| N stage (N0-N1 vs N2-N3) | 43.5 vs 56.5 | 1.25 (0.53 to 2.96) | 0.62 |
| Overall stage (III vs IVa) | 46.4 vs 53.6 | 1.06 (0.44 to 2.56) | 0.90 |
| 14-3-3σb (> 7.0 vs ≤ 7.0) | 39.6 vs 60.4 | 0.70 (0.26 to 1.83) | 0.46 |
| AKT1b (> 5.0 vs ≤ 5.0) | 57.8 vs 42.2 | 0.45 (0.18 to 1.10) | 0.08 |
| Aurora-Ab (≤ 7.0 vs > 7.0) | 47.8 vs 52.2 | 2.13 (0.90 to 5.02) | 0.08 |
| Baxb (≤ 3.5 vs > 3.5) | 47.1 vs 52.9 | 0.97 (0.41 to 2.31) | 0.94 |
| Bcl-2b (> 3.5 vs ≤ 3.5) | 43.5 vs 56.5 | 1.06 (0.45 to 2.50) | 0.90 |
| Beclin 1b (≤ 8.0 vs > 8.0) | 55.9 vs 44.1 | 1.37 (0.55 to 3.40) | 0.50 |
| β-cateninb (> 5.0 vs ≤ 5.0) | 70.6 vs 29.4 | 1.53 (0.59 to 3.94) | 0.38 |
| CDC2b (≤ 5.0 vs > 5.0) | 44.9 vs 55.1 | 0.91 (0.39 to 2.15) | 0.83 |
| CD31c (≤ 379.1 vs > 379.1) | 55.6 vs 44.4 | 1.85 (0.62 to 5.52) | 0.27 |
| CD34c (≤ 380.5 vs > 380.5) | 60.9 vs 39.1 | 1.59 (0.53 to 4.77) | 0.40 |
| CENP-Hb (≤ 5.0 vs > 5.0) | 58.8 vs 41.2 | 1.41 (0.60 to 3.32) | 0.43 |
| C-Metb (≤ 5.0 vs > 5.0) | 54.4 vs 45.6 | 2.48 (1.00 to 6.14) | 0.05 |
| COX2b (≤ 5.0 vs > 5.0) | 46.4 vs 53.6 | 1.47 (0.61 to 3.55) | 0.39 |
| Cyclin D1b (≤ 3.5 vs > 3.5) | 49.3 vs 50.7 | 1.41 (0.60 to 3.33) | 0.43 |
| E-Cadherinb (> 3.5 vs ≤ 3.5) | 52.9 vs 47.1 | 0.62 (0.26 to 1.46) | 0.27 |
| ERKb (≤ 5.0 vs > 5.0) | 55.1 vs 44.9 | 1.74 (0.74 to 4.09) | 0.21 |
| EZH2b (≤ 8.5 vs > 8.5) | 50.7 vs 49.3 | 1.14 (0.48 to 2.71) | 0.76 |
| HIF-1αb (≤ 5.0 vs > 5.0) | 55.1 vs 44.9 | 2.49 (1.00 to 6.16) | 0.05 |
| Ki-67b (≤ 5.0 vs > 5.0) | 51.5 vs 48.5 | 1.46 (0.62 to 3.43) | 0.39 |
| LMP1b (> 5.0 vs ≤ 5.0) | 47.4 vs 52.6 | 1.16 (0.46 to 2.96) | 0.75 |
| MMP-2b (≤ 7.0 vs > 7.0) | 50.7 vs 49.3 | 1.63 (0.69 to 3.83) | 0.27 |
| MMP-9b (> 1.5 vs ≤ 1.5) | 53.8 vs 46.2 | 1.02 (0.41 to 2.57) | 0.96 |
| N-Cadherinb (≤ 5.0 vs > 5.0) | 55.2 vs 44.8 | 1.06 (0.45 to 2.49) | 0.90 |
| nm23-H1b (≤ 5.0 vs > 5.0) | 40.7 vs 59.3 | 1.23 (0.52 to 2.89) | 0.64 |
| P21b (≤ 3.5 vs > 3.5) | 44.9 vs 55.1 | 1.80 (0.76 to 4.28) | 0.18 |
| P27b (≤ 7.0 vs > 7.0) | 56.5 vs 43.5 | 1.82 (0.77 to 4.28) | 0.17 |
| p-ERKb (> 2.5 vs ≤ 2.5) | 47.2 vs 52.8 | 0.93 (0.36 to 2.46) | 0.89 |
| Pontinb (> 3.5 vs ≤ 3.5) | 52.3 vs 47.7 | 0.72 (0.31 to 1.69) | 0.45 |
| Snailb (> 3.5 vs ≤ 3.5) | 44.9 vs 55.1 | 0.68 (0.29 to 1.60) | 0.38 |
| Stathminb (≤ 7.0 vs > 7.0) | 53.6 vs 46.4 | 0.85 (0.35 to 2.04) | 0.71 |
| Survivinb (> 2.5 vs ≤ 2.5) | 43.5 vs 56.5 | 0.70 (0.28 to 1.73) | 0.44 |
| TIMP-2b (> 7.0 vs ≤ 7.0) | 46.4 vs 53.6 | 0.94 (0.40 to 2.22) | 0.89 |
| Twistb (> 2.5 vs ≤ 2.5) | 43.3 vs 56.7 | 1.54 (0.64 to 3.73) | 0.34 |
| EA-IgAd (≤ 1:40 vs > 1:40) | 61.2 vs 38.8 | 1.17 (0.45 to 3.05) | 0.75 |
| VCA-IgAd (≤ 1:160 vs > 1:160) | 37.3 vs 62.7 | 1.24 (0.50 to 3.03) | 0.64 |
| AERd (≤ 64.5% vs > 64.5%) | 50.0 vs 50.0 | 2.06 (0.85 to 4.99) | 0.11 |
Prediction of individual recurrence pattern on IC/CCRT subgroup by decision tree algorithm
Next, we asked whether this 10-fold cross-validation decision tree model would also be predictive in the IC/CCRT subgroup that only contained 64 locally advanced NPC patients (Table 2). After the FSS procedure, eight variables were shown to be highly prognostic, including HIF-1α, Aurora-A, Stathmin, c-Met, N-cadherin, Bax, MMP-9, and N stage. Similarly, this 10-fold cross validation-algorithm also had a powerful predictive efficacy to determine the individual patient recurrence pattern. The PPV, NPV, sensitivity, and specificity were 85.7%, 88.9%, 93.3% and 77.4%, respectively. Moreover, the AUC and overall accuracy to predict individual recurrence pattern reached to 91.3% and 86.8%, respectively (Fig. 1B, Table S2). Significantly, the survival analysis confirmed that the median RFS for the high recurrence risk subgroup was 24.5 months and 65.8 months for the low recurrence subset (P < 0.001, Fig. 2B). Multivariate analysis indicated that this 10-fold cross-validation decision tree algorithm was an independent prognostic factor to predict specific patient recurrence risk (Table 4).
ROC analysis plotted to individual recurrence pattern using tumor related molecular and clinicopathological variables, and three decision tree algorithms. The recurrence predictive efficacy of the tumor related molecular and clinicopathological variables, and the three decision tree algorithms in overall patients (A), IC/CCRT subgroup (B) and IC/RT subgroup (C).
Prediction of individual recurrence pattern on IC/RT subgroup by decision tree algorithm
We further applied the 10-fold cross-validation decision tree model to the IC/RT subset (Table 3). After the FSS selection, seven biomarkers, including of HIF-1α, Aurora-A, CENP-H, Stathmin, C-Met, AKT1and MMP-9, were recruited to construct the 10-fold cross-validation decision tree algorithm. Determined by this algorithm, the predictive efficacy was encouraging: the PPV, NPV, sensitivity, and specificity were 93.2%, 76.0%, 87.2% and 87.4%, respectively. The AUC and overall accuracy to predict individual recurrence pattern were 93.6 and 87.0%, respectively (Fig. 1C, Table S2). Moreover, Kaplan-Meier analysis showed that the high-risk recurrence subgroup identified by the decision tree had significantly shorter RFS than that of low risk subset (18.5 vs. 61.3 months, P < 0.001, Fig. 2C). Multivariate analysis demonstrated that this 10-fold cross validation decision tree algorithm was indeed an independent prognostic factor to predict patient recurrence individually (Table 4). Taken together, although validated for a small cohort (72 patients), these results confirmed that the 10-fold cross-validation decision tree algorithm still had a high power to predict the patient recurrence pattern individually.
Discussion
Molecular tumor biomarkers are useful prognostic factors to predict patient outcome that complement the Tumor-Node-Metastasis staging system. However, the use of a single tumor biomarker is limited due to its low predictive efficacy. Taken HER2 in breast cancer as an example, high HER2 expression correlates with a favourable outcome [28], and is targeted specifically by trastuzumab according to its expression level [29], but HER2 status has a limited efficacy to predict individual patient prognosis. By integrating the 21-gene signature, the patient recurrence pattern was determined individually for breast cancer [30]. Similarly, the multi-biomarkers and support vector machine (SVM) algorithm based data mining approach displayed a promising way to predict patient outcome individually for lung cancer and nasopharyngeal carcinoma [31,32]. After integrating multiple variables and training in one subgroup, the SVM algorithm accurately predicted the outcome for 83.5-91.8% patients, suggesting a potential method for predicting the tumor prognosis of individual patients [32].
Kaplan-Meier estimation of recurrence-free survival according to decision tree algorithm predicted recurrence pattern in overall patients (A), IC/CCRT subgroup (B), and IC/RT subgroup (C).
Multivariate cox proportional-hazards analysis in 3 decision tree algorithms
| Characteristic | Overall patients | IC/CCRT | IC/RT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | HR (95% CI) | P Value | ||||
| Aurora-A (≤ 7.0 vs > 7.0) | 0.29 (0.13 to 0.69) | 0.01 | 0.53 (0.11 to 2.61) | 0.43 | 0.31 (0.06 to 1.50) | 0.15 | |||
| AKT 1 (> 5.0 vs ≤ 5.0) | 2.74 (1.16 to 6.49) | 0.02 | 0.51 (0.15 to 1.72) | 0.28 | |||||
| Bcl-2 (> 3.5 vs ≤ 3.5) | 1.06 (0.46 to 2.43) | 0.90 | |||||||
| LMP 1 (> 5.0 vs ≤ 5.0) | 1.35 (0.63 to 2.87) | 0.44 | |||||||
| MMP-2 (≤ 7.0 vs > 7.0) | 0.76 (0.33 to 1.75) | 0.52 | |||||||
| MMP-9 (> 1.5 vs ≤ 1.5) | 2.23 (1.07 to 4.67) | 0.03 | 1.08 (0.34 to 3.44) | 0.89 | 2.86 (0.85 to 9.65) | 0.91 | |||
| Stathmin (≤ 7.0 vs > 7.0) | 0.86 (0.38 to 1.94) | 0.72 | 1.39 (0.33 to 5.91) | 0.65 | 0.71 (0.26 to 1.92) | 0.50 | |||
| Pontin (> 3.5 vs ≤ 3.5) | 0.87 (0.37 to 2.02) | 0.74 | |||||||
| C-Met (≤ 5.0 vs > 5.0) | 0.58 (0.27 to 1.24) | 0.16 | 0.27 (0.07 to 1.03) | 0.06 | 2.31 (0.76 to 7.04) | 0.14 | |||
| HIF-1α (≤ 5.0 vs > 5.0) | 0.36 (0.08 to 1.63) | 0.18 | 1.00 (0.34 to 2.94) | 0.99 | |||||
| Bax (≤ 3.5 vs > 3.5) | 2.37 (0.32 to 17.36) | 0.40 | |||||||
| N-Cadherin (≤ 5.0 vs > 5.0) | 0.64 (0.18 to 2.34) | 0.50 | |||||||
| N stage (N0-N1 vs N2-N3) | 0.48 (0.12 to 1.98) | 0.31 | |||||||
| CENP-H (≤ 5.0 vs > 5.0) | 2.17 (0.70 to 6.70) | 0.18 | |||||||
| Decision Tree 1 (1 vs 0) | 0.07 (0.03 to 0.16) | < 0.01 | |||||||
| Decision Tree 2 (1 vs 0) | 0.13 (0.04 to 0.44) | < 0.01 | |||||||
| Decision Tree 3 (1 vs 0) | 0.13 (0.03 to 0.68) | 0.02 | |||||||
Tumor recurrence and metastasis are the major forms of disease progression contributing to patient mortality. If recurrences were diagnosed at an earlier stage, the salvage treatment would achieve more favourable outcome. For recurrent T1-2 NPC, our and other studies have demonstrated that interstitial intensity-modulated brachytherapy-based re-radiation and nasopharyngectomy achieved more than 85.7% 3-year disease-free survival [33-36], which is essentially the same for those with primary T1-2 NPC [37]. Therefore, identifying the high-risk patients prior to their recurrence may provide the opportunity to earlier diagnosis and timely salvage treatment. Indeed, the breast cancer 21-gene signature recurrence prediction model initiated a novel approach to realize this goal by integrating multiple variables [38,39]. Importantly, the combined multiple-molecule algorithms are informative to predict patient recurrence pattern early and individually for other types of tumors, including NPC.
However, we have noticed that previously used SVM model was case-sensitive as its prognostic classification efficacy was prone to alteration by selecting different training cases, which was published at journal of PloS One [32]. Moreover, the SVM predictive performance was also easily distorted by using noisy variables. Here, we addressed these problems by employing a rigorous processing framework. This framework consist of two well-founded steps, a hybrid filter-wrapper FSS to select a concise yet informative biomarker panel and a prognostic classification through the AdaBoost algorithm [24]. In the FSS step, a recently reported model of LH-RELIEF was used to find the potential candidate variables [25]. The main advantage of LH-RELIEF is in its capability of featuring important estimation by utilizing the local approximation [27]. In other words, patients sharing similar pathological baseline and molecular phenotype were clustered to maximize a margin between their class assignments. This novel approach displayed an outstanding performance even when the feature was highly degraded by “noise”. The selected candidates were further scrutinized by wrapping them to the classification model to remove the molecules with negligible contributions to the classification performance. In this sense, the obtained biomarker panel was compact but possessed higher prognostic power. The well-selected variables were then fed into a classification model by decision tree rules and boosted with the AdaBoost technique to validate their performances. Such configuration was extensively used in machine learning community due to its powerful performance and independence on data distributions [24]. Indeed, our 10-fold cross-validation decision tree model proved that by selecting and integrating molecular biomarkers and clinicopathological variables, the individual recurrence pattern could be precisely predicted (overall accuracy 84.5-95.2%, Table S2).
This precision recurrence prediction algorithm also provided the rationale of determining a follow-up strategy individually for patients with locally advanced NPC. First, the parameters of molecule expression level and variable here integrated were all recruited at each patient's diagnosis. This means that the individual recurrence pattern may be determined at their diagnosis, and therefore would leave a nearly 2-year monitoring window for the algorithm identified recurrence high risk individuals to receive intensive follow-up (RFS for individuals with high-risk recurrence were 18.5-24.5 months, Fig. 2). Taken the high risk individual for example, the follow-up interval may be modified from three months to one month within the first 24.5 months after the completion of radiochemotherapy. More importantly, this intensive surveillance provides the opportunity to identify individual recurrence at an early stage, so that they may receive timely salvage treatment, such as interstitial intensity-modulated brachytherapy based re-radiation or nasopharyngectomy. Together, this decision tree algorithm provides a potential way to optimize posttreatment surveillance strategy and to earlier diagnose recurrence for salvage treatment to endemic NPC.
In recent years, intensity-modulated radiation therapy (IMRT) was employed to the locally advanced NPC and was a favorable therapeutic variate than 2D radiation. Moreover, IMRT was associated with a lower incidence of late toxicities comparing with 2D radiation. In our study, the prospective clinic trial was enrolled from August 2002 to April 2005, our center mainly carried out 2D treatment. In the 2010s, our center executed IMRT generally. In this study, we addressed the prognostic vale of molecular and clinicopathological variables. Significantly, all cases received the 2D radiation, indicating a uniform radiation effect between the two arms. Therefore, the radiotherapy approach had no difference to our results.
While our proposed algorithm has potential to precisely identify the individual patient recurrence pattern, the method may pose some limitations. For instance, patients in this study were originated from a one-center prospective randomized controlled phase III trial, and an outside testing subset should be used to further validate the predictive power of these decision tree algorithm classifiers. Technically, the IHC staining requires rigor of execution, and may present significant bias. A non-biased reverse phase protein array (RPPA) assay could be utilized to analyse tumor biomarker expression level in our ongoing and future validation studies.
Our molecular and clinicopathological variables combined with a 10-fold cross-validation decision tree algorithm demonstrate a high capacity to identify the patient recurrence pattern individually. This method provides the potential to identify the patient recurrence at an early stage and to improve the outcome by timely salvage treatment for recurrent NPC.
Supplementary Material
Supplementary methods and tables.
Acknowledgements
This work was supported by the Natural Science Foundation of China (No. 61771007 to HMC, No. 81572371 to XJF, No. 81872188 to XBW), International Centre for Genetic Engineering and Biotechnology Research Grant, China (No. CRP/CHIN16-04_EC to XBW), Guangdong Natural Science Foundation for Distinguished Young Scholar, China (No. 2014A030306016 to XBW), Guangdong Science and Technology Project, China (No. 2017B090901065 to XBW), the Special Support Planning Grant of Guangdong Province, China (No. 2015TQ01R562 to XBW), Natural Science Foundation of Guangdong Province, China (No. 2015A030313166 to XJF, 2016A030310187 to JX), Foundation for Pearl River Science & Technology Young Scholars of Guangzhou, China (No. 201610010059 to XJF).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wei WI, Sham JS. Nasopharyngeal Carcinoma. Lancet. 2005;365:2041-2054
2. Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1765-1777
3. Chua MLK, Wee JTS, Hui EP. et al. Nasopharyngeal carcinoma. Lancet. 2016;387:1012-1024
4. Chua DT, Ma J, Sham JS. et al. Long-term survival after cisplatin-based induction chemotherapy and radiotherapy for nasopharyngeal carcinoma: a pooled data analysis of two phase III trials. J Clin Oncol. 2005;23:1118-1124
5. Karam I, Huang SH, McNiven A. et al. Outcomes after reirradiation for recurrent nasopharyngeal carcinoma: North American experience. Head Neck. 2016;38(Suppl 1):E1102-1109
6. Chen KC, Yen TT, Hsieh YL. et al. Postirradiated carotid blowout syndrome in patients with nasopharyngeal carcinoma: a case-control study. Head Neck. 2015;37:794-799
7. Qiu S, Lin S, Tham IW. et al. Intensity-modulated radiation therapy in the salvage of locally recurrent nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2012;83:676-683
8. Wan XB, Jiang R, Xie FY. et al. Endoscope-guided interstitial intensity-modulated brachytherapy and intracavitary brachytherapy as boost radiation for primary early T stage nasopharyngeal carcinoma. PloS one. 2014;9:e90048
9. Tian YM, Guan Y, Xiao WW. et al. Long-term survival and late complications in intensity-modulated radiotherapy of locally recurrent T1 to T2 nasopharyngeal carcinoma. Head Neck. 2016;38:225-231
10. Farrell PJ. Can plasma Epstein-Barr virus DNA levels be used to monitor nasopharyngeal carcinoma progression? Nat Clin Prac Oncol. 2005;2:14-15
11. Rettig EM, Wentz A, Posner MR. et al. Prognostic implication of persistent human papillomavirus type 16 DNA detection in oral rinses for human papillomavirus-related oropharyngeal carcinoma. JAMA oncol. 2015;1:907-915
12. Tang LQ, Li CF, Li J. et al. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J Natl Cancer Inst. 2015:108
13. Albain KS, Barlow WE, Shak S. et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55-65
14. Paik S, Shak S, Tang G. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817-2826
15. Zhang JX, Song W, Chen ZH. et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295-1306
16. Scott JG, Berglund A, Schell MJ. et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. 2017;18:202-211
17. Huang PY, Zeng Q, Cao KJ. et al. Ten-year outcomes of a randomised trial for locoregionally advanced nasopharyngeal carcinoma: A single-institution experience from an endemic area. Eur J Cancer. 2015;51:1760-1770
18. Hong MH, Mai HQ, Min HQ. et al. A comparison of the Chinese 1992 and fifth-edition International Union Against Cancer staging systems for staging nasopharyngeal carcinoma. Cancer. 2000;89:242-247
19. Fan XJ, Wan XB, Huang Y. et al. Epithelial-mesenchymal transition biomarkers and support vector machine guided model in preoperatively predicting regional lymph node metastasis for rectal cancer. Br J Cancer. 2012;106:1735-1741
20. Xu J, Wan XB, Huang XF. et al. Serologic antienzyme rate of Epstein-Barr virus DNase-specific neutralizing antibody segregates TNM classification in nasopharyngeal carcinoma. J Clin Oncol. 2010;28:5202-5209
21. Wan XB, Fan XJ, Chen MY. et al. Elevated Beclin 1 expression is correlated with HIF-1alpha in predicting poor prognosis of nasopharyngeal carcinoma. Autophagy. 2010;6:395-404
22. Takemura A, Shimizu A, Hamamoto K. Discrimination of breast tumors in ultrasonic images using an ensemble classifier based on the AdaBoost algorithm with feature selection. IEEE Trans Med Imaging. 2010;29:598-609
23. Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507-2517
24. Srivastava A, Ghosh S, Anantharaman N. et al. Hybrid biogeography based simultaneous feature selection and MHC class I peptide binding prediction using support vector machines and random forests. J Immunol Methods. 2013;387:284-292
25. Cai H, Ruan P, Ng M. et al. Feature weight estimation for gene selection: a local hyperlinear learning approach. BMC Bioinformatics. 2014;15:70
26. Yetton BD, Niknazar M, Duggan KA. et al. Automatic detection of rapid eye movements (REMs): A machine learning approach. J Neurosci Methods. 2016;259:72-82
27. Freund Y, Schapire RE. A short introduction to boosting. J Jap Soc Artif Intell. 1999;14:771-780
28. Baselga J, Cortés J, Im SA. et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol. 2014;32:3753-3761
29. Tolaney SM, Barry WT, Dang CT. et al. Adjuvant paclitaxel and trastuzumab for node-negative, HER2-positive breast cancer. N Engl J Med. 2015;372:134-141
30. Sparano JA, Gray RJ, Makower DF. et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373:2005-2014
31. Zhu ZH, Sun BY, Ma Y. et al. Three immunomarker support vector machines-based prognostic classifiers for stage IB non-small-cell lung cancer. J Clin Oncol. 2009;27:1091-1099
32. Wan XB, Zhao Y, Fan XJ. et al. Molecular prognostic prediction for locally advanced nasopharyngeal carcinoma by support vector machine integrated approach. PloS One. 2012;7:e31989
33. Chen MY, Cao XP, Sun R. et al. Application of interstitial brachytherapy via parapharynx involvement transnasal approach to enhance dose in radiotherapy for nasopharyngeal carcinoma. Ai Zheng. 2007;26:513-518
34. You R, Zou X, Hua YJ. et al. Salvage endoscopic nasopharyngectomy is superior to intensity-modulated radiation therapy for local recurrence of selected T1-T3 nasopharyngeal carcinoma - A case-matched comparison. Radiother Oncol. 2015;115:399-406
35. Zou X, Han F, Ma WJ. et al. Salvage endoscopic nasopharyngectomy and intensity-modulated radiotherapy versus conventional radiotherapy in treating locally recurrent nasopharyngeal carcinoma. Head Neck. 2015;37:1108-1115
36. Chen MY, Wang SL, Zhu YL. et al. Use of a posterior pedicle nasal septum and floor mucoperiosteum flap to resurface the nasopharynx after endoscopic nasopharyngectomy for recurrent nasopharyngeal carcinoma. Head Neck. 2012;34:1383-1388
37. Su SF, Han F, Zhao C. et al. Long-term outcomes of early-stage nasopharyngeal carcinoma patients treated with intensity-modulated radiotherapy alone. Int J Radiat Oncol Biol Phys. 2012;82:327-333
38. Hornberger J, Alvarado MD, Rebecca C. et al. Clinical validity/utility, change in practice patterns, and economic implications of risk stratifiers to predict outcomes for early-stage breast cancer: a systematic review. J Natl Cancer Inst. 2012;104:1068-1079
39. Levine MN, Julian JA, Bedard PL. et al. Prospective evaluation of the 21-gene recurrence score assay for breast cancer decision-making in Ontario. J Clin Oncol. 2016;34:1065-1071
Author contact
![]() Corresponding authors: Jian Zheng, Department of Radiation Oncology, The Sixth Affiliated Hospital, Sun Yat-sen University, 26 Yuancun Erheng Road, Guangzhou 510065, China; E-mail: zhengj48sysu.edu.cn and Yanchun Lv, Department of Medical Radiology, Sun Yat-sen University Cancer Center; State Key Laboratory of Oncology in South China; Collaborative Innovation Center for Cancer Medicine, 651 Dongfeng Rd East, Guangzhou 510060, China; E-mail: lvychorg.cn
Corresponding authors: Jian Zheng, Department of Radiation Oncology, The Sixth Affiliated Hospital, Sun Yat-sen University, 26 Yuancun Erheng Road, Guangzhou 510065, China; E-mail: zhengj48sysu.edu.cn and Yanchun Lv, Department of Medical Radiology, Sun Yat-sen University Cancer Center; State Key Laboratory of Oncology in South China; Collaborative Innovation Center for Cancer Medicine, 651 Dongfeng Rd East, Guangzhou 510060, China; E-mail: lvychorg.cn

 Global reach, higher impact
Global reach, higher impact