Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(15):3560-3570. doi:10.7150/jca.28272 This issue Cite
Research Paper
MiR-506 Suppresses Colorectal Cancer Development by Inhibiting Orphan Nuclear Receptor NR4A1 Expression
1. Department of Cell Biology and Genetics and Key Laboratory of Molecular Biology in High Cancer Incidence Coastal Chaoshan Area of Guangdong Higher Education Institutes, Shantou University Medical College, Shantou 515041, China.
2. Department of Pathology and Central Laboratory, Shantou Central Hospital, Affiliated Shantou Hospital of Sun Yat-sen University, Shantou 515041, China.
3. Institute of Translational Medicine, Shenzhen Second People's Hospital, the First Affiliated H-ospital of Shenzhen University, Health Science Center, Shenzhen 518035, China.
4. Department of General Surgery, First Affiliated Hospital of Shantou University Medical College, Shantou 515041, China.
* Equal Contributors
Received 2018-7-2; Accepted 2019-5-1; Published 2019-6-9
Abstract
NR4A1 acts as an oncogene and plays an important role in colorectal cancer development and progression, but little is known about the regulatory mechanism of NR4A1 expression. MicroRNA (miRNA) is involved in the progression of various tumors, affecting proliferation, apoptosis or migration. We aimed to elucidate whether miRNA regulates NR4A1 expression and determine its underlying significance in colorectal cancer. By using the TargetScan database, we identified a miR-506 binding site in the NR4A1 3'-UTR. Examination of colorectal cancer tissues and cells revealed that NR4A1 mRNA and protein were up-regulated, while miR-506 expression was down-regulated. Spearman correlation analysis revealed that expression of NR4A1 mRNA was negatively correlated with miR-506 levels in colorectal cancer tissue. Further studies indicated that miR-506 decreased NR4A1 expression through directly targeting the NR4A1 mRNA 3'-UTR. Functional experiments showed that rescue of NR4A1 expression in cells reversed the inhibitory effects of miR-506 on proliferation, migration and invasion of colorectal cancer cells. In conclusion, miR-506 acts as a tumor suppressor and inhibits proliferation, migration and invasion in colorectal cancer cells partly through decreasing NR4A1 expression.
Keywords: Colorectal cancer, NR4A1, MiR-506, Proliferation, Migration, Invasion
Introduction
Colorectal cancer remains the third most common malignant tumor worldwide, and is a serious threat to human health. It is estimated that, there are 1,200,000 new cases diagnosed every year, and more than half will die from the cancer [1]. In addition, the five-year survival rate is merely 10%~15% for patients who suffer from metastatic colorectal cancer, but would be 40%~90% in the absence of metastasis [2]. Colorectal cancer is closely related with hereditary factors and lifestyle [3]. With the morbidity of colorectal cancer increasing yearly, it is extremely urgent to find safe and effective therapeutic targets.
MicroRNAs (miRNAs) are a family of endogenous non-coding RNAs, whose length ranges from about 18 to 25 nucleotides. At present, nearly 2,000 miRNAs have been discovered by researchers, with new species being constantly explored [4]. MiRNAs play an extensive role in regulating tumor development and progression, being involved in cell proliferation, differentiation and apoptosis. It is estimated that, more than 60% of genes in mammals are regulated by miRNAs [5].
The miR-506 gene is located on the X chromosome, and forms a gene cluster together with miR-507, miR-508, miR-509, miR-510, miR-513 and miR-514 [6]. The whole cluster is highly conserved among primates [7, 8]. Since the initial discovery of miR-506 by Bentwich et al. [6], researchers have found that the expression pattern of miR-506 is inconsistent in different tumor types. For example, miR-506 is down-regulated in ovarian cancer, hepatic carcinoma, glioblastoma, cervical cancer and colorectal cancer, and acts as a tumor suppressor [9-13], but Streicher et al. showed that the miR-506-514 cluster is up-regulated in melanoma and promotes tumor development as an oncogene [14]. These all reflect the complexity of miR-506 function.
Nuclear receptor subfamily 4 group A member 1 (NR4A1) is an immediate-early response gene, and can be quickly induced by many factors, such as hormones, growth factors and inflammatory factors. Large numbers of studies indicate that NR4A1 could act either as an oncogene or as a tumor suppressor. For example, it serves as an oncogene in lung cancer, breast cancer and colorectal cancer [15-17], but displays a tumor suppressor role in acute myeloid leukemia or metastatic ovarian cancer [18, 19]. Since NR4A1 exhibits a pro-oncogenic role in most cancers, it is a potential therapeutic target for clinical application, and studies have found that diindolylmethane analogs (C-DIMs) act strictly through nuclear NR4A1 to induce apoptosis in cancer cells and tumors [20, 21].
Our study examines the regulation of NR4A1 expression by miR-506 and its significance in colorectal cancer. We found miR-506 decreases NR4A1 expression, through directly targeting the NR4A1 mRNA 3'-UTR, to inhibit proliferation, migration and invasion of colorectal cancer cells, thus providing new evidence for the functional mechanism of miR-506 in colorectal cancer.
Materials and methods
Cell culture and clinical samples
Cells used in this study included colorectal cancer cell lines HCT116, SW480, and LoVo, and the normal colorectal epithelial cell line denoted FHC. HCT116 cells were cultured in McCoy's 5A medium (Gibco, USA), SW480 cells were cultured in RPMI-1640 medium (Hyclone, USA), LoVo cells were cultured in Ham's F-12K medium (Gibco, USA), and FHC cells were maintained in DMEM/F12 medium (Hyclone, USA). All media were supplemented with 10% fetal bovine serum (Hyclone, USA), and cells were cultured in a 37℃, as well as 5% CO2 humidified incubator.
Colorectal cancer tissues and matched adjacent normal tissues were collected from the First Affiliated Hospital of Shantou University Medical College, and the clinical characteristics of these CRC patients are shown in the Supplemental Table. All tissue samples used in our research were put into liquid nitrogen and snap-frozen, then stored at -80℃. All patients provided informed written consent in this study, and the study protocol was approved by the local ethics committees (Shantou University Medical College and the First Affiliated Hospital of Shantou University Medical College, China). All procedures were performed according to the Declaration of Helsinki and Good Clinical Practice guidelines.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from colorectal tissues and cells using RNAiso plus (Takara, Dalian, China). The mRNA was reverse transcribed with a PrimeScript RT reagent Kit with gDNA Eraser (Takara), and NR4A1 cDNA was isolated by PCR using a QuantiNovaTM SYBR Green PCR Kit (QIAGEN, Germany), while for miR-506, reverse transcription was performed using a MiRcute miRNA cDNA first strand synthesis kit (TIANGEN, Beijing, China) and compatible qPCR kit (TIANGEN). Both qPCRs were carried out on an Applied Biosystems 7500 real-time PCR system. For mRNA and miRNA detection, β-Actin and U6 snRNA were respectively taken as the endogenous control. The primers sequences were as follows (5'-3'):
NR4A1: ACGGCTACACAGGAGAGTTTGAGGTGGCTGAGGACGAGGAT.
β-Actin: CTGGAACGGTGAAGGTGACAAAGGGACTTCCTGTAACAATGCA.
miR-506: CGTGTAAGGCACCCTTCTGAGTAGA.
U6: CTCGCTTCGGCAGCACA.
Western blot
Colorectal tissues and cells were lysed with RIPA buffer containing 0.5% proteinase inhibitor cocktail (Millipore, USA), followed by centrifuging at 12,000 rpm for 10 min. The supernatant protein concentration was measured with a BCA protein assay kit (Beyotime, Shanghai, China). Then, 50 μg denatured protein samples were separated by 8% SDS polyacrylamide gel electrophoresis, and transferred to a nitrocellulose membrane. Next, the membrane was blocked using 5% non-fat powered milk for 1 h at room temperature. When blocking was complete, the membrane was washed twice with TBST (1‰ Tween), and incubated with primary antibody (β-Actin, Sigma; NR4A1, Santa Cruz) in a shaker at 4℃ overnight. Subsequently, the membrane was rinsed in TBST thrice, each time for 10 min, then probed with HRP-conjugated secondary antibody (goat anti-mouse IgG, Santa Cruz; goat anti-rabbit IgG, Santa Cruz) for 2 h at room temperature. Finally, blots were detected using chemiluminescent solution and exposed to film.
Cell transfection
MiR-506 mimics, miR-506 inhibitor and siRNA targeting NR4A1 (si-NR4A1), including their negative control were all ordered from GenePharma (Shanghai, China). When transfected, 75 nM mimics, 100 nM inhibitor and 40 nM si-NR4A1 were used. They were transiently transfected with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). For co-transfection, 75 nM miR-506 mimics and 3 μg pcDNA3.1-NR4A1 plasmid (Transheep, Shanghai, China) were added to cells. Moreover, FuGENE® HD Reagent (Promega, Madison, WI, USA) was used for plasmid transfection. The relevant sequences were as follows (5'-3'):
miR-506 mimics: UAAGGCACCCUUCUGAGUAGAUACUCAGAAGGGUGCCUUAUU.
miR-506 inhibitor: UCUACUCAGAAGGGUGCCUUA.
NR4A1 siRNA: CAGUGGCUCUGACUACUAUTTAUAGUAGUCAGAGCCACUGTT.
Luciferase reporter assay
For luciferase reporter assays, the full length of NR4A1 3'-UTR or its mutant form was cloned into the psiCHECK-2 vector (Transheep, Shanghai, China). In brief, HCT116 cells were inoculated in a 48-well plate, then co-transfected with miR-NC (or miR-506 mimics) and NR4A1 3'-UTR wild-type (or mutant) plasmid the next day. At 48 h after transfection, cells were rinsed in 1×PBS and lysed with 1× passive lysis buffer (PLB), then luciferase activity was detected using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's protocol.
Cell counting kit-8 (CCK-8) assay
The CCK-8 assay was conducted to determine cell viability. HCT116 cells were inoculated into 96-well plates at a density of 5000 cells per well, then were transfected the next day. At 1-3 days after transfection, the cell viability was measured daily using a CCK-8 assay kit (Dojindo Laboratories, Japan) following the manufacturer's instructions. OD values at 450 nm were measured by a microplate reader.
Cell cycle assay
Transfected HCT116 cells were harvested at 48 h, and pipetted gently into a single cell suspension. After washing with PBS, centrifugation and discarding of the supernatant, cells were fixed in 70% ethanol overnight at 4℃. After fixation, cells were washed twice in PBS, filtered through a 300-mesh nylon membrane, and then incubated for 30 min in a 37℃ waterbath following addition of 100 μl RNase A (Kaiji, Nanjing, China). Finally, cells were stained with propidium iodide (Kaiji, Nanjing, China) for 30 min at 4℃ in the dark. Cell cycle distribution was performed using a BD Accuri™ C6 Flow Cytometer (Becton-Dickinson, USA).
Colony formation assay
For the colony formation assay, 2000 transfected HCT116 cells were inoculated in 3 ml growth medium in each well of a 6-well plate, and incubated at 37°C for 2 weeks. Then cells were fixed with 4% paraformaldehyde, air dried and stained with crystal violet (Beyotime, Shanghai, China). Colonies with >50 cells were counted under a microscope.
Migration and invasion assays
Cell migration and invasion assays were both conducted using a transwell chamber (8-μm pore size, BD Biosciences, USA). In the migration assay, 1.0×105 transfected HCT116 cells were plated onto the top chamber, with the lower chamber containing 600 μl media supplemented with 20% FBS. After incubation for 48 h, cells were fixed with 4% paraformaldehyde, air dried and stained with crystal violet (Beyotime, Shanghai, China), then cells remaining on the top surface were gently removed with a cotton swab. Cells that had passed through the pore and adhered to the lower membrane surface were counted, from five randomly chosen fields, under an inverted microscope. For the invasion assay, the membrane was pre-coated with Matrigel and 2.5 ×105 transfected cells were added into the top chamber.
Statistical analysis
All data were obtained from at least three independent experiments and shown as means ± SEM. Graphing and statistical analyses were processed by GraphPad Prism 5.0 and SPSS version 19.0 software (SPSS Inc., Chicago, IL, USA). For cell experiments, the independent-sample t-test was used to compare differences between groups, while the paired-sample t-test was used to analyze the expression level of groups in tissue. Correlation between NR4A1 mRNA and miR-506 expression was determined using Spearman correlation analysis. Differences were considered to be statistically significant when the p-value was less than 0.05.
Results
NR4A1 is up-regulated in colorectal cancer tissues and cells
We initially detected NR4A1 mRNA and protein expression levels by qRT-PCR and western blot in colorectal cancer tissues and cells, and our results were in line with prior reports [23, 24]. Compared with normal FHC cells, expression of NR4A1 mRNA and protein levels were both up-regulated in the SW480, HCT116 and LoVo colorectal cancer cell lines (Figure 1A and 1B). Moreover, NR4A1 expression levels were higher in colorectal cancer tissues than in adjacent normal tissues (Figure 1C and 1D). Thus, NR4A1 expression is elevated in colorectal cancer tissues and cancer cell lines.
MiR-506 is down-regulated in colorectal cancer tissues and cells
We searched the online database TargetScan and found that NR4A1 had miR-506 binding sites in the 52nd to 58th base-pair of NR4A1 3'-UTR (Figure 2A). Based on its high conservation, and we further pursued a potential role for miR-506 in the regulation of NR4A1. Through qRT-PCR, we determined the miR-506 level in colorectal cancer tissues and cells. Expression of miR-506 was significantly down-regulated in both colorectal cancer cells (Figure 2B) and cancer tissues (Figure 2C).
NR4A1 expression negatively correlates with miR-506 level in colorectal cancer tissues
The above results suggested that expression of miR-506 is inversely correlated with NR4A1. To better confirm the relationship between their expression, we continued to analyze every colorectal cancer tissue, and showed that in 36 paired colorectal cancer tissues, for 28 cases both miR-506 and NR4A1 mRNA were inversely expressed (Figure 3A). Furthermore, Spearman correlation analysis revealed that NR4A1 expression was negatively correlated with miR-506 levels in colorectal cancer tissues (r= - 0.349, p= 0.037, n= 36; Figure 3B).
MiR-506 reduces NR4A1 expression through directly targeting the NR4A1 3'-UTR
In view of the negative correlation between expression of miR-506 and NR4A1 mRNA, we speculated that miR-506 might regulate NR4A1 expression. To verify our prediction, we separately transfected HCT116 cells with miRNA mimics and FHC cells with inhibitor, so as to overexpress or knockdown miR-506 level in these two cells. Based on qRT-PCR, miR-506 mimics increased expression of miR-506 significantly (Figure 4A), and inhibitor led to slight decreases in miR-506 levels (Figure 4D). Then we examined NR4A1 expression, and found that miR-506 mimics decreased both NR4A1 mRNA (Figure 4B) and protein expression (Figure 4C). In addition, miR-506 inhibitor obviously increased NR4A1 protein expression (Figure 4F), but had no influence on NR4A1 mRNA levels (Figure 4E). These results illustrate that miR-506 can reduce NR4A1 expression.
NR4A1 is up-regulated in colorectal cancer tissues and cells. A. Relative NR4A1 mRNA expression in FHC, SW480, HCT116 and LoVo cells as determined by qRT-PCR. β-Actin was used as an internal control. Error bars represent the SEM from 3 independent experiments. *p<0.05, **p<0.01 vs. FHC. B. NR4A1 protein levels in FHC, SW480, HCT116 and LoVo cells were determined by western blot. β-Actin was used as the internal control. C. Relative NR4A1 mRNA level in paired colorectal cancer tissues and adjacent normal tissues was detected by qRT-PCR. β-Actin used as internal control. n=50. D. Western blot analysis of protein expression with β-Actin as the internal control. Quantitation of protein levels was by densitometry. N=corresponding tumor-adjacent normal tissue, T=colorectal cancer tissue (n=42).
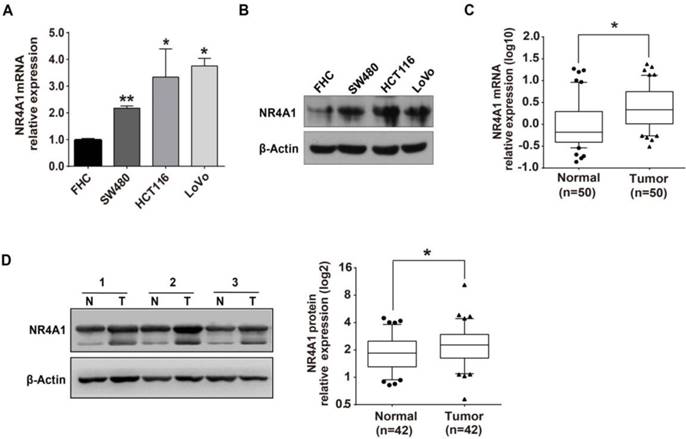
MiR-506 is down-regulated in colorectal cancer tissues and cells. A. Graph shows the binding site of miR-506 in the NR4A1 3'-UTR, as predicted by the TargetScan database. B. Relative miR-506 expression in FHC, SW480, HCT116 and LoVo cells as determined by qRT-PCR. U6 served as an internal control. Error bars represent the SEM from 3 independent experiments. **p<0.01 vs. FHC. C. Relative miR-506 expression in paired colorectal cancer tissues (T) and adjacent normal tissues (N) as determined by qRT-PCR. U6 served as an internal control (n=36).
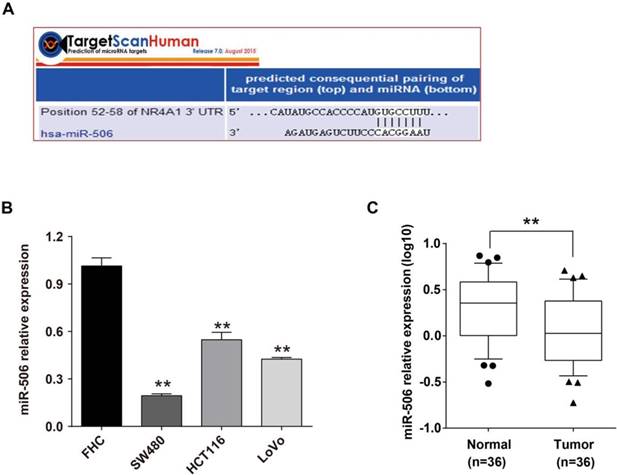
NR4A1 expression correlates with miR-506 level in colorectal cancer tissues. A. The graph revealed a one-to-one relationship between fold change of NR4A1 mRNA and miR-506 level. The ratio of NR4A1/miR-506 expression level in colorectal cancer tissue (T) to corresponding tumor-adjacent normal tissue (N) was measured to analyze the fold change for each patient. Green: NR4A1, red: miR-506 (n=36). B. Spearman correlation analysis demonstrated a negative relationship between NR4A1 expression and miR-506 level (n=36).
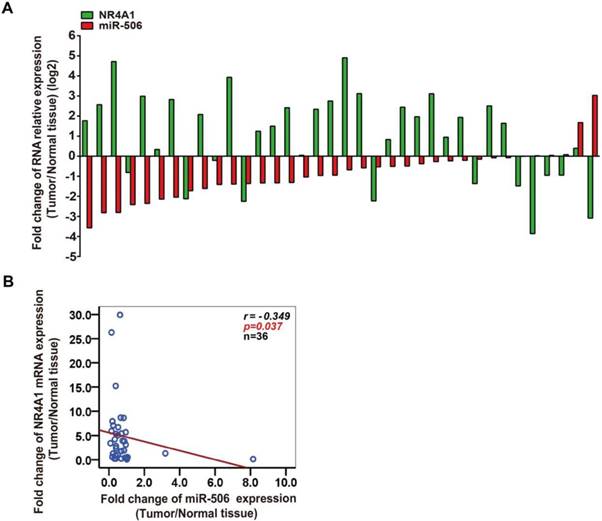
MiR-506 reduces NR4A1 expression through directly targeting the NR4A1 3'-UTR. A-C. HCT116 cells were treated with 75 nM miR-506 mimics for 48 h. QRT-PCR analysis of relative expression for miR-506 (A) and NR4A1 mRNA (B), U6 and β-Actin were the internal controls, respectively. Western blot analysis of NR4A1 protein expression with β-Actin as the internal control (C). Quantitation of protein levels was by densitometry. **p<0.01 vs. miR-NC. D-F. FHC cells were treated with 100 nM miR-506 inhibitor for 48 h. QRT-PCR analysis of relative expression for miR-506 (D) and NR4A1 mRNA (E), U6 and β-Actin were the internal controls, respectively. Western blot analysis of NR4A1 protein expression with β-Actin as the internal control. (F) Quantitation of protein levels was by densitometry. *p<0.05, **p<0.01 vs. NC. G. Putative binding site of miR-506 and the NR4A1 3'-UTR was predicted using the TargetScan database, and the mutant site is shown with blue characters. H. HCT116 cells were co-transfected with miR-NC (or miR-506 mimics) and the psiCHECK2 3'-UTR wild-type (or mutant) plasmid, and relative luciferase activity was examined 48 h later. **p<0.01 vs. miR-NC. All error bars represent the SEM from at least 3 independent experiments.
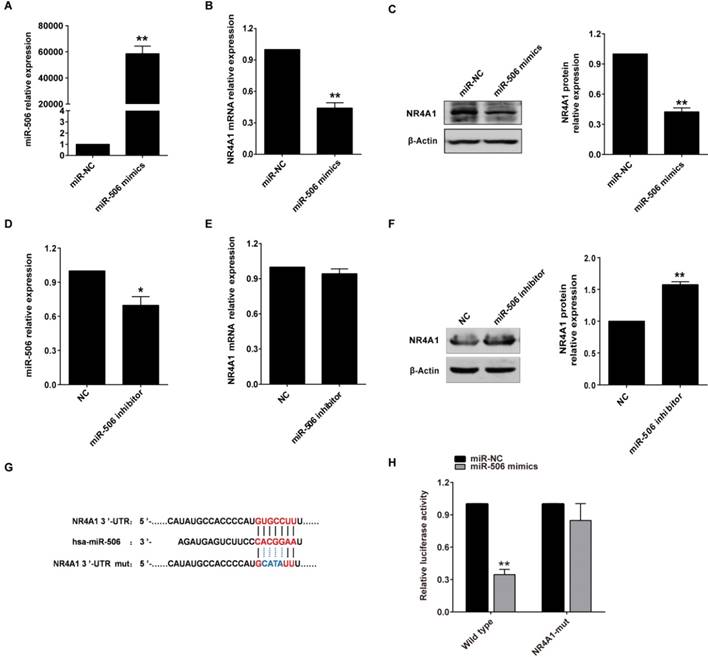
It is known that miRNAs play their role mainly via directly targeting 3'-UTR of the target genes, so we used a dual-luciferase reporter assay to validate whether this is also true for miR-506. Luciferase reporter genes, containing an NR4A1 3'-UTR with either a wild-type or mutated miR-506 binding site, were created (Figure 4G). Luciferase reporter assay showed that miR-506 could significantly decrease relative luciferase activity of luciferase mRNA containing the wild-type NR4A1 3'-UTR, but not the mutated 3'-UTR (Figure 4H). Therefore, miR-506 reduced NR4A1 expression through directly targeting the NR4A1 3'-UTR.
Reduced expression of NR4A1 inhibits proliferation, migration and invasion of colorectal cancer cells
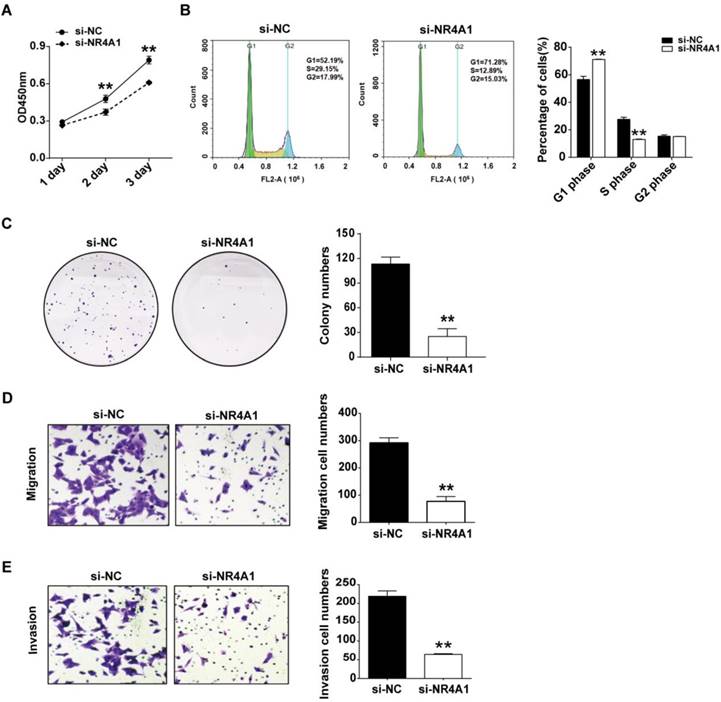
Several studies have reported that decreased NR4A1 expression inhibits migration and invasion of colorectal cancer cells. We transfected HCT116 cells using siRNA to knock down NR4A1 levels, and further studied its effect on cell function. CCK-8 assay showed that reduced NR4A1 expression resulted in a relative decrease of cell viability (Figure 5A). Cell cycle distribution was detected by flow cytometry, which showed that reduced NR4A1 expression could increase cell number in G1 phase and lower cell percentages in S phase, but had no effect on G2 phase, so reduced NR4A1 expression led to G1 arrest and inhibited cell proliferation (Figure 5B). Colony formation assay indicated that NR4A1 knockdown decreased the colony formation rate compared to the si-NC group (Figure 5C). In addition, transwell migration and invasion assays suggested that cell migration and invasion were both suppressed when NR4A1 expression was reduced (Figure 5D and 5E).
Overexpression of miR-506 inhibits proliferation, migration and invasion of colorectal cancer cells
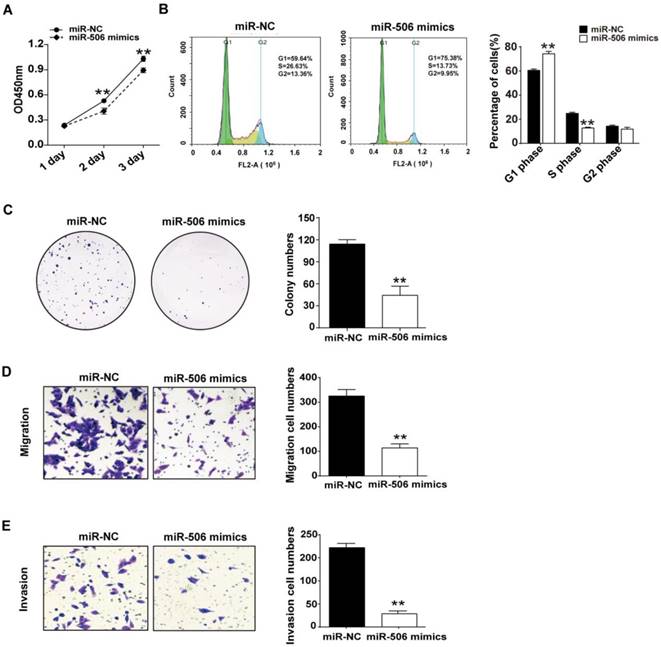
Next, we investigated the influence of overexpressed miR-506 on proliferation and migration of colorectal cancer cells. Ectopic expression was achieved by transfecting HCT116 cells with miR-506 mimics. CCK-8 assay indicated that overexpression of miR-506 gave rise to a significant decrease in cell viability (Figure 6A). Furthermore, cell cycle distribution analysis showed that overexpression of miR-506 could increase cell proportions in G1 phase, with a corresponding reduction in S phase, and no change in G2 phase, which also indicated that overexpressed miR-506 led to G1 arrest and inhibited cell proliferation (Figure 6B). Colony formation assay showed that miR-506 overexpression significantly reduced number of cell colonies (Figure 6C), and transwell assays revealed that ectopic expression of miR-506 led to inhibition of cell migration and invasion (Figure 6D and 6E).
Reduced expression of NR4A1 inhibits proliferation, migration and invasion of colorectal cancer cells. HCT116 cells were transfected with 40 nM si-NR4A1 (or si-NC). A. CCK-8 assay was used to assess cell viability for different days (1-3 day). B. At 48 h after transfection, cells were harvested and fixed in cold 70% ethanol at 4℃ overnight, then cell cycle distribution was determined via flow cytometry. C. 2000 transfected cells seeded in 6-well plates were cultured for 2 weeks, then colonies with >50 cells were counted under a microscope. D and E. Transwell migration (D) and invasion (E) assays were used to elucidate the migration and invasion of cells, respectively. ImageJ software was used for cell counting. All error bars represent the SEM from at least 3 independent experiments. **p<0.01 vs. si-NC.
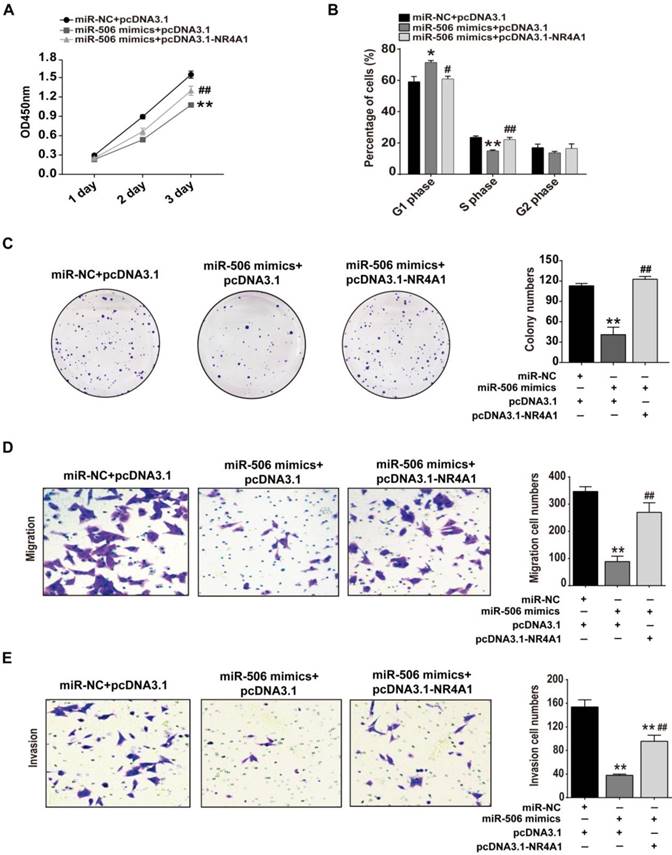
Restoration of NR4A1 expression alleviates miR-506-mediated inhibition of proliferation, migration and invasion of colorectal cancer cells
As mentioned above, proliferation, migration and invasion of colorectal cancer cells all were inhibited when NR4A1 was knocked down or miR-506 was overexpressed. To ensure that this effect was due to loss of NR4A1, we examined whether restoration of NR4A1 expression would reverse the inhibitory effects of miR-506 on cancer cells. Hence, we co-transfected NR4A1 expression plasmids and miR-506 mimics into HCT116 cells, and examined changes related to cell proliferation, migration and invasion. CCK-8, cell cycle detection, colony formation assay and transwell assays demonstrated that, compared with cells transfected with miR-506 mimics only, restoration of NR4A1 expression increased relative cell viability (Figure 7A), reduced miR-506-induced G1 arrest (Figure 7B), increased colony numbers (Figure 7C), and promoted the recovery of cell migration and invasion (Figure 7D and 7E), indicating the inhibitory effects of miR-506 were due to decreased NR4A1 expression.
Discussion
Radical surgical excision of early-detected colorectal cancer results in a high five-year survival rate, which can be up to 90%~95%. However, for most patients, metastasis has already occurred at the time of diagnosis, resulting in high mortality. So early diagnosis and treatment are of great importance. Expression of miRNA is abnormal in many cancers, and is closely related with tumor development and progression, including colorectal cancer. For instance, Yi et al. found that the miR-21 and miR-29 families are related to early diagnosis and prognosis of colorectal cancer [22].
Overexpression of miR-506 inhibits proliferation, migration and invasion of colorectal cancer cells. HCT116 cells were transfected with 75 nM miR-NC (or miR-506 mimics). A. CCK-8 assay was used to assess cell viability daily, on Days 1-3 after transfection. B. At 48 h after transfection, cells were harvested and fixed in cold 70% ethanol at 4℃ overnight, then cell cycle distribution was determined via flow cytometry. C. 2000 transfected cells were inoculated in 6-well plates and cultured for 2 weeks, then colonies with >50 cells were counted under a microscope. D and E. Transwell migration (D) and invasion (E) assays were used to elucidate the migration and invasion of cells, respectively. ImageJ software was used for cell counting. All error bars represent the SEM from at least 3 independent experiments. **p<0.01 vs. miR-NC.
Restored NR4A1 expression alleviates miR-506-mediated inhibition of proliferation, migration and invasion of colorectal cancer cells. HCT116 cells were co-transfected with miR-NC (or miR-506 mimics) and pcDNA 3.1-NR4A1 (or pcDNA 3.1). A. CCK-8 assay was used to assess cell viability daily, on Days 1-3 after transfection. B. At 48 h after transfection, Cells were harvested and fixed in cold 70% ethanol at 4℃ overnight, then cell cycle distribution was determined via flow cytometry. C. 2000 transfected cells were inoculated in 6-well plates and cultured for 2 weeks, then colonies with >50 cells were counted under a microscope. D and E. Transwell migration (D) and invasion (E) assays were used to elucidate the migration and invasion of cells, respectively. ImageJ software was used for cell counting. All error bars represent the SEM from 3 independent experiments. *p<0.05, **p<0.01 vs. miR-NC+pcDNA3.1; # p<0.05, ##p<0.01 vs. miR-506 mimics + pcDNA3.1.
Orphan nuclear receptor NR4A1 is up-regulated in colorectal cancer tissues and cells [23, 24], and can promote metastasis and invasion through regulating the MMP9/E-cadherin axis [17], β1-integrin [25] or forming a feed-forward loop with β-catenin under hypoxic conditions [26]. Lee and coworkers confirmed that NR4A1 knockdown could inhibit mTOR signaling or expression of survivin and led to inhibited colon cancer cell growth [21]. In our study, we also found that NR4A1 expression is increased in colorectal cancer tissues and cells, and proliferation, migration or invasion of colorectal cancer cells is inhibited upon NR4A1 knockdown.
Previously, few articles have involved miRNA relating to NR4A1. Results of our bioinformatic analysis show miR-506 binding sites exist in the NR4A1 3'-UTR. Furthermore, miR-506 is highly conserved. Zhang et al. demonstrated that miR-506 is down-regulated in colorectal cancer tissues [2]. Our qRT-PCR results also reveal that miR-506 expression is decreased in colorectal cancer tissues and cells. Spearman correlation analysis also shows a negative relationship between NR4A1 expression and miR-506 level. Thus, we hypothesized that miR-506 might regulate NR4A1. Further studies indicated that ectopic expression of miR-506 or reduced miR-506 level leads to corresponding decreases or increases in NR4A1 expression. One miRNA could interact with many target genes, which is true for miR-506. Previous studies show target genes of miR-506 are varied, such as YAP [10], Gli3 [12], COTL1 [27], SPON1 [28] and TCF3 [29]. Without exception, miR-506 regulates their expression through targeting the corresponding 3'-UTRs. We also demonstrated, by dual-luciferase reporter assay, that miR-506 reduces NR4A1 levels through directly targeting the NR4A1 3'-UTR, and the binding sites are specific.
MiR-506 participates in important cancer-related processes, such as cell growth, apoptosis, migration and invasion [30], and plays roles via interplaying with its target genes. Wu et al. reported that miR-506 inhibits proliferation and invasion of colorectal cells by regulating the TET family [13]. Chen et al. demonstrated that overexpression of miR-506 reduces global DNA methylation by targeting DNMT3B and DNMT1, thus restoring the expression of E-cadherin, MGMT and P16 and inhibiting colorectal cancer progression [31]. Paweł et al. revealed that elevated expression of miR-506 in peripheral blood is a potential molecular marker for early colorectal cancer [32].Through CCK-8, cell cycle, colony formation, and transwell migration and invasion assays, we confirmed that overexpression of miR-506 leads to inhibition of proliferation, migration and invasion of colorectal cancer cells. Our co-transfection results show that restoration of NR4A1 levels could alleviate the inhibitory effect caused by miR-506, indicating that miR-506 exerts its action on colorectal cancer cells at least partly by regulating NR4A1. Taken together, our study demonstrates that miR-506 decreases NR4A1 expression by directly targeting the NR4A1 3'-UTR, and inhibits proliferation, migration and invasion of colorectal cells as a tumor suppressor.
Abbreviations
NR4A1: nuclear receptor subfamily 4 group A member 1; miRNA: microRNA; 3'-UTR: 3'-untranslated region; PCR: polymerase chain reaction; qRT-PCR: quantitative real-time PCR; gDNA: genome DNA; qPCR: quantitative PCR; cDNA: complementary DNA; CCK-8: cell counting kit-8; DMSO: dimethyl sulfoxide; FBS: fetal bovine serum; OD: optical density; SEM: standard error of the mean; MMP9: matrix metalloproteinases-9; YAP: yes-associated protein; COTL1: coactosin-like protein 1; SPON1: spondin 1; TCF3: transcription factor 3; TET: ten-eleven translocation; MGMT: O-6-methylguanine-DNA methyltransferase; DNMT3B: DNA methyltransferase3B; DNMT1: DNA methyltransferase1; mTOR: mammalian target of rapamycin.
Supplementary Material
Supplementary table.
Acknowledgements
We thank Stanley Lin for critical reading of the manuscript. This work was supported by National Natural Science Foundation of China (NSFC, 81400616, 31770876, 81470441).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
2. Zhang Y, Lin C, Liao G, Liu S, Ding J, Tang F, Wang Z, Liang X, Li B. et al. MicroRNA-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2. Oncotarget. 2015;6:32586
3. Choi Y, Sateia HF, Peairs KS, Stewart RW. Screening for colorectal cancer. Semin Oncol. 2017;44:34-44
4. Friedlander MR, Lizano E, Houben AJ, Bezdan D, Banez-Coronel M, Kudla G, Mateu-Huertas E. et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014;15:R57
5. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105
6. Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genetics. 2005;37:766-770
7. Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612-617
8. Li J, Liu Y, Dong D, Zhang Z. Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol. 2010;27:671-683
9. Lei R, Xue M, Zhang L, Lin Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. OncoTargets and Therapy. 2017;10:35-46
10. Wang Y, Cui M, Sun BD, Liu FB, Zhang XD, Ye LH. MiR-506 suppresses proliferation of hepatoma cells through targeting YAP mRNA 3'UTR. Acta Pharmacol Sin. 2014;35:1207-1214
11. Luo Y, Sun R, Zhang J, Sun T, Liu X, Yang B. MiR-506 inhibits the proliferation and invasion by targeting IGF2BP1 in glioblastoma. Am J Transl Res. 2015;7:2007-2014
12. Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, Li J, Zhang YL. et al. MiR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717-725
13. Wu M, Zhang Y, Tang A, Tian L. MiR-506 inhibits cell proliferation and invasion by targeting TET family in colorectal cancer. Iran J Basic Med Sci. 2016;19:316-322
14. Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z. et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012;31:1558-1570
15. Lee SO, Andey T, Jin UH, Kim K, Singh M, Safe S. The nuclear receptor TR3 regulates mTORC1 signaling in lung cancer cells expressing wild-type p53. Oncogene. 2012;31:3265-3276
16. Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, Sheppard KA, Goumans MJ, Luwor RB. et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-beta signalling. Nat Commun. 2014;5:3388
17. Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu XX, Li JM, Wu H. Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis. 2014;35:2474-2484
18. Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730-735
19. Wilson AJ, Liu AY, Roland J, Adebayo OB, Fletcher SA, Slaughter JC, Saskowski J, Crispens MA. et al. TR3 modulates platinum resistance in ovarian cancer. Cancer Res. 2013;73:4758-4769
20. Lee SO, Li X, Khan S, Safe S. Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin. Ther. Targets. 2011;15:195-206
21. Lee SO, Li X, Hedrick E, Jin UH, Tjalkens RB, Backos DS, Li L, Zhang Y, Wu Q, Safe S. Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Molecular Endocrinology. 2014;28:1729-1739
22. Yi R, Li Y, Wang FL, Miao G, Qi RM, Zhao YY. MicroRNAs as diagnostic and prognostic biomarkers in colorectal cancer. World J Gastrointest Oncol. 2016;8:330-340
23. Cho SD, Yoon K, Chintharlapalli S, Abdelrahim M, Lei P, Hamilton S, Khan S, Ramaiah SK, Safe S. Nur77 Agonists Induce Proapoptotic Genes and Responses in Colon Cancer Cells through Nuclear Receptor-Dependent and Nuclear Receptor-Independent Pathways. Cancer Research. 2007;67:674-683
24. Wu H, Lin Y, Li W, Sun Z, Gao W, Zhang H, Xie L, Jiang F. et al. Regulation of Nur77 expression by β-catenin and its mitogenic effect in colon cancer cells. The FASEB Journal. 2011;25:192-205
25. Hedrick E, Lee SO, Safe S. The nuclear orphan receptor NR4A1 regulates β1-integrin expression in pancreatic and colon cancer cells and can be targeted by NR4A1 antagonists. Mol Carcinog. 2017;56:2066-2075
26. To SKY, Zeng W, Zeng J, Wong AST. Hypoxia triggers a Nur77-β-catenin feed-forward loop to promote the invasive growth of colon cancer cells. British Journal of Cancer. 2014;110:935-945
27. Guo S, Yang P, Jiang X, Li X, Wang Y, Zhang X, Sun B, Zhang Y, Jia Y. Genetic and epigenetic silencing of mircoRNA-506-3p enhances COTL1 oncogene expression to foster non-small lung cancer progression. Oncotarget. 2017;8:644-657
28. Dai W, Huang H, Hu M, Wang S, He H, Chen N, Li M. MicroRNA-506 regulates proliferation, migration and invasion in hepatocellular carcinoma by targeting F-spondin 1 (SPON1). American journal of cancer research. 2015;5:2697
29. Wang Y, Jiaqi C, Zhaoying C, Huimin C. MicroRNA-506-3p regulates neural stem cell proliferation and differentiation through targeting TCF3. Gene. 2016;593:193-200
30. Li J, Ju JF, Ni B, Wang HZ. The emerging role of miR-506 in cancer. Oncotarget. 2016;7:62778-62788
31. Chen ZH, Liu SJ, Tian L, Wu MH, Ai FY, Tang WL. et al. MiR-124 and miR-506 inhibit colorectal cancer progression by targeting DNMT3B and DNMT1. Oncotarget. 2015;6:38139-38150
32. Paweł K, Tomasz P, Tomasz O, Adam D, Joanna D, Dariusz K, Janusz M. Evaluation of miR-506 and miR-4316 expression in early and non-invasive diagnosis of colorectal cancer. Int J Colorectal Dis. 2017;32:1057-1060
Author contact
![]() Corresponding authors: Dongyang Huang, Bin Liang. E-mail: huangdyedu.cn and bliangedu.cn.
Corresponding authors: Dongyang Huang, Bin Liang. E-mail: huangdyedu.cn and bliangedu.cn.

 Global reach, higher impact
Global reach, higher impact