Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3593-3607. doi:10.7150/jca.30975 This issue Cite
Review
Associations of mRNA expression of DNA repair genes and genetic polymorphisms with cancer risk: a bioinformatics analysis and meta-analysis
1. Department of Pharmacology, School of Pharmacy, Liaoning Key Laboratory of Molecular Targeted Anti-Tumor Drug Development and Evaluation, China Medical University, Shenyang, 110122, P. R. China
2. Department of Anorectal Surgery, First Hospital of China Medical University, Shenyang, 110001, P. R. China
3. Department of Breast Surgery, First Hospital of China Medical University, Shenyang, 110001, P. R. China
#Contribute equally to this work.
Received 2018-10-26; Accepted 2019-5-16; Published 2019-6-9
Abstract
A systematical bioinformatics and meta-analysis were carried out to establish our understanding of possible relationships between DNA repair genes and the development of cancer. The bioinformatics analysis confirmed that lower XPA and XPC levels and higher XPD, XPF, and WRN levels were observed in 19 types of cancer, and subsequently results indicated that elevated XPA and XPC had a better impact on overall survival, however, higher XPD, XPF, and WRN showed worse influence on cancer prognosis. The meta-analysis included 58 eligible studies demonstrated that harboring XPA rs10817938, XPD rs238406 increased overall cancer risk, however, XPA rs2808668 SNP in overall cancer analysis and XPF rs3136038 in the digestive system remarkably reduced the cancer risk. Moreover, no correlation was investigated for XPC rs1870134, WRN rs1346044 and rs1801195. These suggest that the DNA repair gene was associated with carcinogenesis, and contribute to the prognosis, and the critical SNPs further involved in affecting cancer risk.
Keywords: DNA repair genes, bioinformatics, cancer, prognosis, meta-analysis
Introduction
Cancer continues being a growing public health problem across the world [1]. According to the Cancer Statistics 2018, approximately 1,735,350 new cancer cases, 609,640 death cases were reported worldwide [2]. Cancer exists as a complex disease in humans, which is due to the multifaceted interactions among multiple inherited genetic mutations as well as environmental exposures and lifestyle factors [3-5]. Accumulating evidence showed that common mutations or polymorphisms in DNA repair genes involved in altering protein function and capacity to repair damaged DNA, thus deficits in repair capacity which lead to genetic instability, pathogenesis and carcinogenesis, however, the results were inconsistent in distinct cancers, and need further to be elucidated and pay more attention [6].
There are less three major DNA repair pathways in humans including nucleotide excision repair (NER), base excision repair (BER), non-homologous end joining (NHEJ) etc, consisting of more than 150 genes [7]. The NER is among the classical general damage repair pathways in cells to remove a wide range of bulky DNA lesions induced by environmental mutagens, radiation, and certain chemicals [8, 9]. The molecular basis of XP has been attributed to mutations in seven XP proteins (A-through-G: XPA, XPB, XPC, XPD, XPE, XPF, and XPG) that are required for NER-mediated removal of DNA damage and XP-variant (XPV)[10]. The core XP proteins are involved in different steps of the DNA repair process. XPA protein is the sole recognition factor required for NER activities and is required for sensing DNA damage and initiating the repair process [11, 12]. The XPC binds to RAD23B to form the XPC-RAD23B complex, which is associated with DNA repair initiation in the NER pathway [13]. XPD acts as a 5'→3' ATP-dependent dominant that creates a DNA strand opening surrounding the adducted bases in the NER pathway [14]. XPF is the endonucleases that perform the dual incisions to release the damaged strand and allow resynthesize using the non-damaged strand as a template [15]. Furthermore, WRN protein is one of the members of the RecQ helicase family, which also plays a crucial role in DNA replication and the maintenance of genome stability in non-homologous end joining (NHEJ) DNA repair, homologous recombination repair, and non-homologous end joining (NHEJ) pathway [16]. Inactivating mutations or genetic variations in DNA repair genes can modify an individual's capacity to repair damaged DNA, and thus lead to the clinical disorder xeroderma pigmentosum (XP) and a predisposition to the development of cancers [17, 18].
To date, among the core DNA repair genes, there are more than 1000 coding-region SNPs (cSNPs) (http://www.ncbi.nlm.nih.gov/projects/SNP) found. Among those cSNPs, a few potential functional SNPs such as located in the 5'-untranslated regions (5'UTR) or Exon sites of the candidate genes could change the DNA repair capacity by regulating the transcriptional activity or protein expression, thereby playing critical roles in altering individual's susceptibility to cancer. Previous accumulating studies provided evidence about the association of SNPs in DNA repair gene with cancer risk, and most widely studied polymorphism including XPA rs2808668 [19-26], rs10817938 [21, 25, 26], XPC rs1870134 [21, 22, 26-31], XPD rs238406 [32-54], XPF rs3136038 [39, 55-63], WRN rs1801195 [64-67], rs1346044 [64, 65, 68-75]. The SNPs XPA rs10817938, XPA rs2808668 and XPF rs3136038 are all the polymorphisms in the site of the gene promoter. The SNP rs10817938 is located in the 5'-untranslated region (UTR), which is a T to C substitution distant from the transcriptional start site (TSS) about -2718bp, meanwhile, rs2808668 is a T to C substitution located -514bp from the TSS within XPA gene, which have been reported novel promoter SNPs in the XPA loci associated with cancer risk and development including hepatic cancer (HCC) [26], breast cancer [19], gastric cancer [24], oral squamous cell carcinoma (OSCC) [25]. Furthermore the SNP rs3136038 is near the 5' end of XPF gene (also known as ERCC4, excision repair cross-complementation group 4 gene) exhibits remarkably associated with the susceptibility of variety of cancer such as breast cancer [55, 63], lung cancer [56], ESCC [61], gastric cancer [39], etc. The other selected candidate SNPs XPC rs1870134 (Leu16Met), XPD rs238406 (Arg156Arg), WRN rs1346044 (Cys1367Arg) and WRN rs1801195 (Leu1074Phe) are all located in the Exon and Intron sites of the corresponding genes. Recent studies have shown that polymorphisms at Exon 1 (C>G, rs1870134) of XPC gene, Exon 6 (A>C, rs238406) of XPD gene, Exon 34 (C>T, rs1346044) and Exon 26 (T>G, rs1801195) of WRN gene have been correlated with several types of cancer including breast and prostate cancers [29, 41, 45, 46, 48, 50, 64, 66, 69, 70, 75], however, these results were inconclusive. Thus, considering the critical role of these genetic variations in the DNA repair genes, and understanding the association between these SNPs and cancer susceptibility is urgently required.
In this study, a comprehensive bioinformatics analysis based on Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) data and meta-analysis was expanded to 7 SNPs of DNA repair genes included 58 eligible case-control studies for evaluating the cancer risk and providing more precise estimation of these associations. We are convinced this review will have important implications in elucidating critical interconnections between DNA repair genes expression, SNPs and cancer risk, and improving the basic understanding of the composition, regulation and function of the DNA repair pathway, eventually paving the way for the development of novel cancer risk evaluation biomarker.
Materials and Methods
Online public dataset analyses
In the present meta-analysis, online tools including the Oncomines (https://www.oncomine. org/resource/login.html) and the Gene Expression across Normal and Tumors Tissue (GENT, http://medical-genome.kribb.re.kr/GENT/) were used to compare the expression of the DNA repair genes between a variety of cancers and their normal controls tissues based on Array Express (http://www.ebi.ac.uk/microarrayas/ae/) and GEO datasets. Furthermore, the association of the candidate genes expression with prognosis was assessed by dataset available in Prediction of Clinical Outcomes from Genomic Profiles (PRECOG, http://precog.stanford.edu). The survival Z-scores for each gene and cancer type were provided by the PRECOG website, and the Log-rank test and HR ratio and 95%CI was established by GEO and Array Express datasets. In addition, genes alterations of these DNA repair genes were represented including mutation, fusion, amplification, deep deletion by using cBioPortal tool based on TCGA datasets.
Literature research
The present meta-analysis was conducted based on the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analysis statement. A comprehensive literature search was assessed independently by three authors (S.Q.L, X.Y.H., and W.Y.Q.) in seven electronic databases: PMC database, PubMed database, the China National Knowledge Infrastructure (CNKI), the Web of Science, Embase, WanFang Date, BIOSIS Preview, and ClinicalKey. All the searched eligible original studies and review articles were reviewed carefully to identify the relevant articles by using the following search terms “rs1346044, Cys1367Arg” or “rs1801195, Leu1074Phe” or “rs2808668” or “rs10817938” or “rs1870134, Leu16Met” or “rs3136038” or “rs238406, Arg156Arg” and “polymorphism or SNP or single nucleotide polymorphism or variation or mutation” and “cancer or carcinoma or tumor or neoplasm”, (the search was updated on May 13, 2018). This search was limited to these articles with English or Chinese language, and the results were examined and compared by a third reviewer (Y. L.W.).
Data extraction
In this meta-analysis, included publications were eligible if they fulfilled the following criteria: 1) the design of case-control study or cohort study; 2) investigated the association of XPA, XPC, XPD, XPF, and WRN genetic polymorphisms with the cancer risk; 3) provided sufficient genotypic and/or allelic information for estimating the odds ratio (OR) with 95% confidence intervals (CIs); 4) the samples size of cases or controls were ≥20. Excluding the following criteria: (1) animal studies, case reports, reviews, and unpublished results; (2) no case-control study; (3) other mutations and other diseases; (4) lack of adequate information. The data was extracted from each publication in the collection criterion by T.S., Z. K.W, and X.F.W. independently as follows: first author, publication year, ethnicity, country, cancer type, control source (population-based controls, or hospital-based controls based on randomized and prospective), genotyping method, the total number of genotyped cases or controls, and the number of each genotype for cases and controls with each SNP for cancer risk assessment.
Statistical analysis
Stata 12.0 software (StataCorp LP, College Station, USA) and Open Meta-Analyst (http://www.cebm. brown. edu/openmeta/) were used to perform all of the statistical analyses in this meta-analysis. The Hardy-Weinberg equilibrium (HWE) was estimated by using the Chi-square test or Fisher's exact test (P<0.05 represented significant deviation) for each control-study. The pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were estimated to assess the strength of association between these 7 SNPs and cancer risk. The pooled estimated ORs and CIs were determined by Z-test based on homozygote model, additive model, dominant model, and recessive model (P<0.05 represented statistically significant). The heterogeneity between-study was assessed across all eligible comparisons by using χ2-based Cochran's Q-test (significant for P<0.10).The random-effects model (DerSimonian-Laird method) was chosen if there is statistical heterogeneity. Otherwise the fixed-effects model (Mantel-Haenszel method) was used if the studies were homogeneous. I2 statistics were established from 0% to 100%, which the heterogeneity irrespective of the number of studies (I2of 25%-55%, 55%-75%, or >75% represented low, moderate, high heterogeneity). The sensitivity meta-analysis was implemented by omitting each data to reflect the influence of individual datasets to heterogeneity. Publication bias was evaluated by the Egger's and Begg-Matzumdar linear regression tests using asymmetry of the funnel plot (significant for P<0.10).
Results
The mRNA expression of the DNA repair pathway genes in cancers
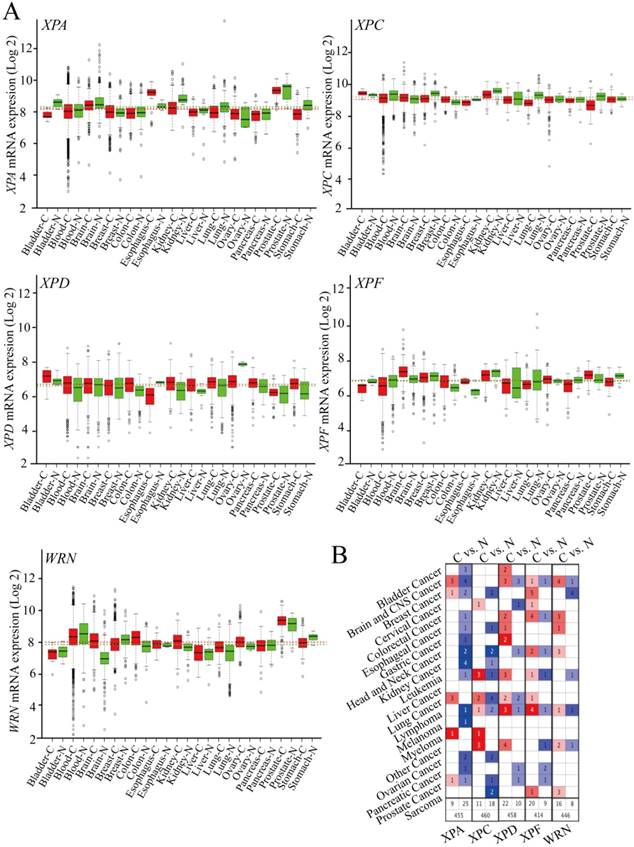
To identify the potential role of DNA repair pathway genes mRNA expression in different cancers, we used two techniques to estimate associations between these included genes (XPA, XPC, XPD, XPF, WRN) and mRNA expression. (i) mRNA expression in 13 types of cancers from RNA sequencing dataset platform (GENT, http://medical-genome.kribb.re.kr/GENT/) of tumor samples based on GEO and ArrayExpress datasets was compared to adjacent normal tumor tissues. We found decreased expression of XPA in bladder cancer, kidney cancer, liver cancer, lung cancer, prostate cancer, and stomach cancer; XPC in blood system cancer, breast cancer, esophageal cancer, kidney cancer, lung cancer and prostate cancer. Elevated expression of XPD in bladder cancer, blood system cancer, breast cancer, colorectal cancer, kidney cancer and liver cancer, lung cancer, pancreas cancer, and gastric cancer; XPF in brain cancer, colorectal cancer, esophageal cancer, liver cancer, prostate cancer and WRN in brain cancer, colorectal cancer, esophageal cancer, kidney cancer, lung cancer, ovarian cancer, and prostate cancer was observed in comparison to normal controls (Figure 1 A). (ii) The information of the DNA repair pathway genes expression was freely available in Oncomines online platform (https://www.oncomine.org/) based on TCGA datasets. Then, those genes were evaluated in selected 19 types of cancers datasets and the normal controls, and also found decreased XPA and XPC expression occurred in majority of tumors, however, XPD, XPF, and WRN genes exhibit increased expression in most of the tumor datasets (Figure 1 B). Such imbalance expression suggests that those DNA repair genes act as oncogene or tumor suppressor gene in tumorigenesis and progression and maybe exhibit a distinct prognostic impact on survival.
The relative mRNA expressions of the DNA repair genes in cancer tissues and normal samples (A).The association of these genes expression with cancer, the redder of the square, and the more related with cancer (B).
The associations of the DNA repair pathway genes expression with cancer prognosis
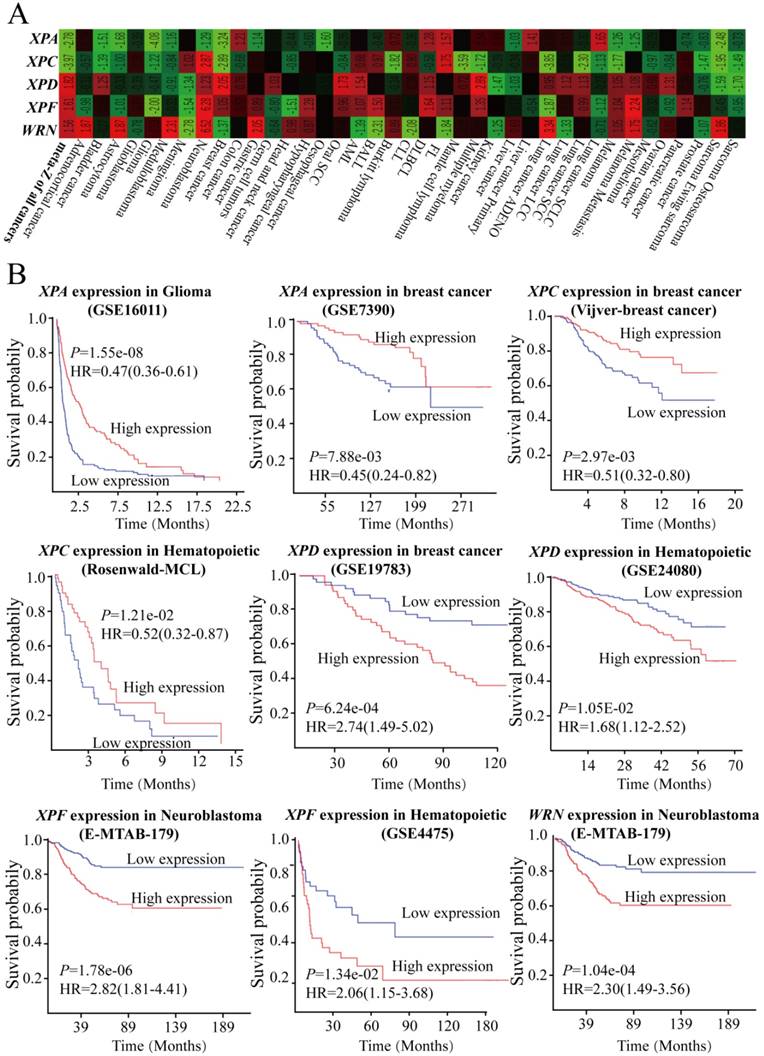
To further explore the impact of mRNA expression of DNA repair genes among those included cancers on clinical outcomes, we used dataset available in Prediction of Clinical Outcomes from Genomic Profiles (PRECOG, http://precog.stanford.edu) based on GEO and Array Express datasets. It was noted that higher expression of XPA and XPC gene was associated with better overall survival (OS), and the meta survival Z-scores for all cancer were -2.78 and -3.97, respectively (Figure 2A). The log-rank test and multivariate Cox proportional hazards regression model analysis further proved that higher expression of XPA gene had a better prognosis compared with lower group in glioma cancer (Log-rank test: P<0.0001, HR=0.47, 95%CI=0.36-0.61) and breast cancer (Log-rank test: P<0.0001, HR=0.45, 95%CI=0.24-0.82) (Figure 2B). Furthermore, increased XPC expression showed significantly protective influence on overall survival including in breast cancer (Log-rank test: P<0.0001, HR=0.51, 95%CI=0.32-0.80) and hematopoietic cancer (Log-rank test: P<0.0001, HR=0.52, 95%CI=0.32-0.87) (Figure 2B). However, inversely results were determined that higher expression of XPD, XPF, and WRN gene had a worse impact on OS for all cancers, and the meta Z-scores for all cancer were 1.82, 1.61, and 1.56, respectively (Figure 2 A). When compared with higher expression group, lower expression of XPD, XPF, and WRN gene showed longer overall survival time for patients with breast cancer, hematopoietic cancer, and neuroblastoma (Log-rank test: P<0.0001) (Figure 2B). These data states that the altered mRNA expression levels of these DNA repair genes might have a distinct correlation with the clinical outcomes.
The characters of the genes dataset altered in cancer samples
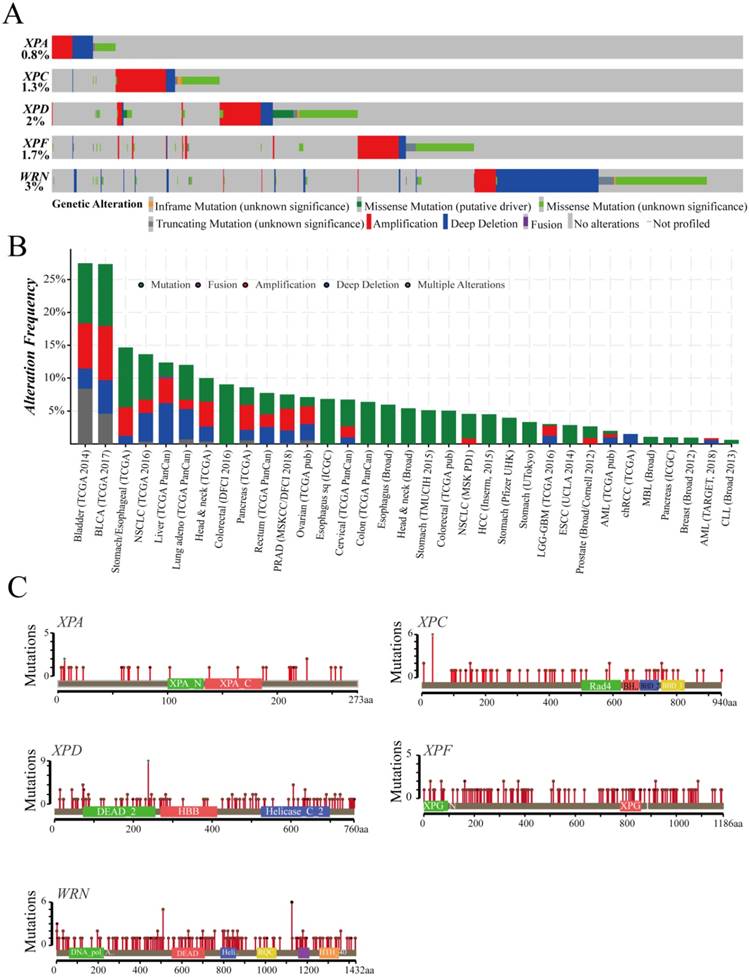
These gene alterations of XPA, XPC, XPC, XPF, WRN were displayed including mutation, fusion, amplification, deep deletion by using cBioPortal online tool based on TCGA database(Case Set: 63658 patients/65690 samples). The detail cancer type, mutation fusion amphomdel and alteration frequency of those genes are shown in Figure 3. Although the total genetic alteration levels of the candidate genes were not very high only from 0.8%-3% (Figure 3A), the alteration frequency showed an higher levels in 33 types of cancers, especially in bladder cancer, BCLA, which were more than 25% (Figure 3B). Furthermore, a higher level of duplicate polymorphisms or somatic mutations such as missense, truncating, inframe occurred, which includes 46 SNPs or mutations in XPA gene, 91 mutations in XPC gene, 227 mutations in XPD gene, 210 mutations in XPF gene, and 267 mutations in WRN gene (Figure 3C).These datas suggested that those DNA repair genes alterations probably closely related to the mRNA and protein expression, subsequently involved in affecting the prognosis of cancer patients.
Meta-analysis of the DNA repair genes with cancer risk
Figure S1 summarizes a flowchart presenting the literature review of study identification, inclusion, exclusion. With the search strategy, a total of 380 published articles was extracted and determined for cancer risk assessment from PMC database, PubMed, CNKI, CbmWeb, WanFang Date, BIOSIS Preview. After manually screening abstracts and texts of the included studies, 324 were exempted for 24 lack of adequate information, 54 with other mutations or diseases, 13 abstracts, meta or review, and 233 duplicated publication or overlapping with other publications for further evaluation. Then update 2 studies in Feb 1, 2019. Finally, 58 studies were met the inclusion criteria, 4 studies evaluated the association of XPA rs10817938 with cancer risk, 8 reports determined the SNP rs2808668 of XPA gene and cancer susceptibility, 8 publications were XPC rs1870134 polymorphism, 10 reports studied XPF rs3136038 polymorphism, 24 reports determined XPD rs238406 SNP, 4 reports and 11 reports analyzed WRN rs1801195 and rs1346044, respectively. The distribution of genotypes in the controls for the SNPs of DNA repair genes were in the HWE, except for these 6 publications of Gao, C et al. [25] for XPA rs2808668, Sun, K for et al. [73] for WRN rs1346044, Liu, Y for et al. [61] for XPF rs3136038, Chang, J.S. et al. [42]; Szczur, K.P. et al. [49]; Miercla, A[50] for XPD rs238406 (Table S1). In this final meta-analysis, XPA rs10817938 included 1775 cases and 2156 controls, as well as XPA rs2808668 included 2616 cases and 3099 controls. Meanwhile, XPC rs1870134, XPD rs238406, and XPF rs3136038 contained 4987 cases and 6193 controls; 6999 cases and 8652 controls; 5247 cases and 5607 controls, respectively. Moreover, cases and controls for WRN rs1801195 and rs1346044 were 3161 and 3142, 6538 and 7657, respectively. The types of cancers chiefly include breast cancer, lung cancer, gastric cancer, hepatocellular cancer, laryngeal cancer, colorectal cancer and esophageal squamous cell cancer (ESCC), prostate cancer, bladder cancer, glioma, basal cell cancer. The ethnicity of the included studies is Asian, Caucasian, Mix and genotyping method includes TaqMan assay, sequencing and PCR-RFLP method. The essential characteristics for all studies were presented in Table S1.
GEO and Array Express datasets shows the DNA repair genes had an impact on clinical outcomes by using of meta survival Z-scores (A) and log-rank test and multivariate Cox proportional hazards regression model analysis (B).
cBioPortal software exhibits the characteristic of the DNA repair genes in the total genetic alteration (A), the alteration frequency (B), and duplicate polymorphisms or somatic mutations (C) based on TCGA database (Case Set: 63658 patients/65690 samples).
XPA rs10817938, rs2808668 and cancer risk analysis
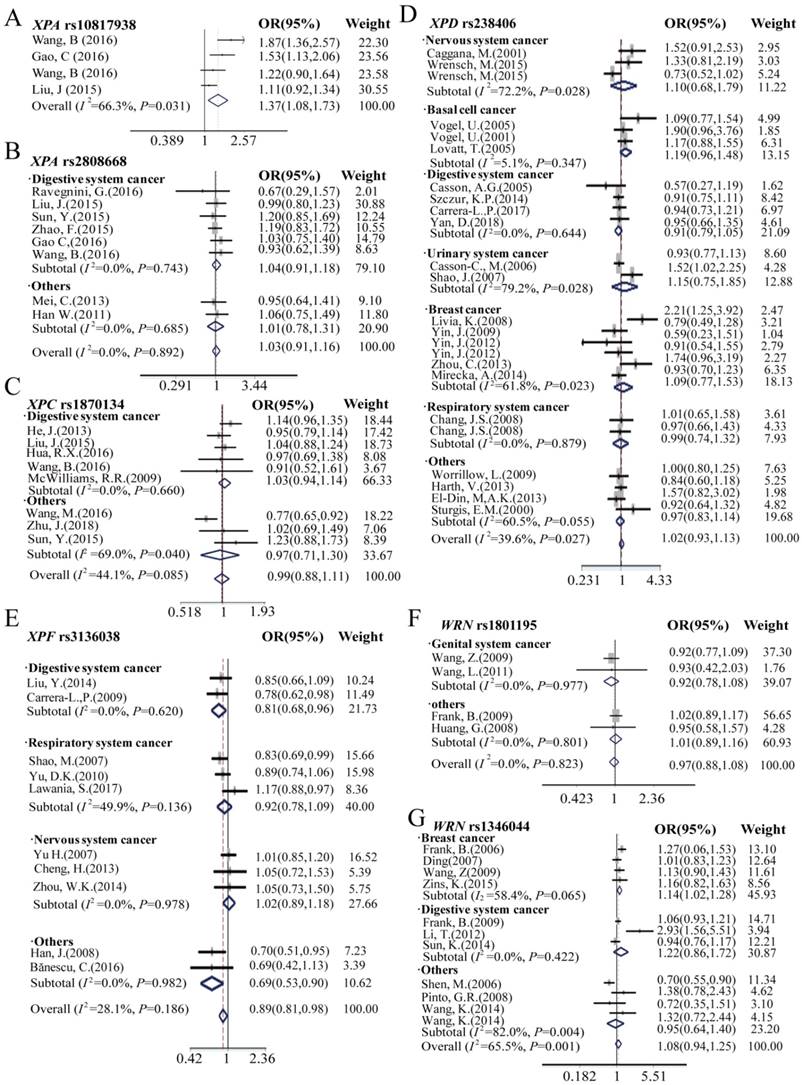
The meta-analysis results of the XPA rs10817938, rs2808668 polymorphism and cancer risk are shown in Table 1, Table S2, and Figure 4, 5, S3, S4. Overall, harboring XPA rs10817938 homozygous CC genotype, C allele, and CC/CT genotype in dominant model showed significant association with increased cancer risk [CC vs.TT in homozygous model: OR (95%CI) = 1.68 (1.02- 2.76), P = 0.04; C vs. T allele in additive model: OR (95%CI) = 1.20 (1.04-1.38), P = 0.01; and CC/CT vs. TT in dominant model: 1.37 (1.09-1.74), P = 0.008] (Figure 4, 5,S3, S4A). Although no association with cancer risk observed in XPA rs2808668 SNP with overall analysis, subgroup analysis showed significant decreased cancer risk except for digestive system cancer [CC vs.TT in homozygous model: OR (95%CI) = 0.71 (0.52-0.98), P = 0.04; C vs. T allele in additive model: OR (95%CI) = 0.84 (0.72-0.99), P = 0.03; CC vs.TT/TC in recessive model: 0.63 (0.49-0.82), P<0.0001] (Figure 4, 5, S3, S4B).
XPC, XPD, and XPF polymorphism and cancer risk analysis
The effect of XPC, XPD, and XPF polymorphism on cancer risk in overall and subgroup analysis was presented in Table 1, Table S2 and Figure 4, 5, S3, S4: C, D, and E. In the overall analysis, the SNP rs238406 of XPD gene was associated with an increased overall cancer risk, however, XPF rs3136038 showed significantly decreased cancer risk, and no significant association was determined in XPC rs1870134 polymorphism. SNP rs238406 AA genotype or A allele carriers showed significantly increased overall cancer risk [AA vs.CC in homozygote model: OR (95%CI) = 1.20 (1.02-1.41), P = 0.03; A vs. C allele in additive model: OR (95%CI) = 1.08 (1.00-1.18), P=0.04; and AA vs. CC/CA in recessive model: OR (95%CI) = 1.21 (1.05- 1.40), P=0.007].
Meta-analysis of the association between genetic polymorphisms of DNA repair pathway and cancer risk by dominant and recessive models
| Variables | Pz* | Dominant OR(95%CI) | Phet# | I2#(%) | Pz* | Recessive OR(95%CI) | Phet# | I2#(%) |
|---|---|---|---|---|---|---|---|---|
| XPA rs10817938 | CC/CT VS. TT | CC VS. CT/TT | ||||||
| Digestive system† | 0.008 | 1.37 (1.08,1.73) | 0.03 | 66.3 | 0.06 | 1.60 (0.98,2.62) | 0.211 | 33.5 |
| Overall | 0.008 | 1.37 (1.08,1.73) | 0.03 | 66.3 | 0.06 | 1.60 (0.98,2.62) | 0.211 | 33.5 |
| XPA rs2808668 | CC/CT VS. TT | CC VS. TT/TC | ||||||
| Digestive system† | 0.61 | 1.04 (0.91,1.18) | 0.74 | 0.0 | 0.03 | 1.18 (1.02,1.37) | 0.91 | 0.0 |
| Others | 0.96 | 1.01 (0.78,1.31) | 0.69 | 0.0 | 0.000 | 0.63 (0.49,0.82) | 0.33 | 0.0 |
| Overall | 0.64 | 1.029 (0.91,1.16) | 0.89 | 0.0 | 0.94 | 1.01 (0.80,1.37) | 0.006 | 64.3 |
| XPC rs1870134 | CC/CG VS. GG | CC VS. GG/GC | ||||||
| Digestive system† | 0.45 | 1.03 (0.94,1.14) | 0.66 | 0.0 | 0.56 | 0.93 (0.71,1.20) | 0.21 | 34.2 |
| Others | 0.79 | 0.96 (0.71,1.30) | 0.04 | 69.0 | 0.66 | 0.88 (0.51,1.53) | 0.09 | 58.8 |
| Overall | 0.84 | 0.99 (0.88,1.11) | 0.09 | 44.1 | 0.46 | 0.92 (0.74,1.15) | 0.15 | 36.4 |
| XPD rs238406 | AA/AC VS.CC | AA VS. CC/CA | ||||||
| Nervous system§ | 0.70 | 1.10 (0.68,1.79) | 0.03 | 72.2 | 0.33 | 1.36 (0.72,2.57) | 0.01 | 77.5 |
| Basal cell cancer | 0.11 | 1.19 (0.96,1.48) | 0.35 | 5.1 | 0.03 | 1.30 (1.03,1.65) | 0.68 | 0.0 |
| Digestive system† | 0.20 | 0.91 (0.79,1.05) | 0.64 | 0.0 | 0.08 | 1.15 (0.98,1.35) | 0.42 | 0.0 |
| Urinary system£ | 0.56 | 1.15 (0.75,1.85) | 0.03 | 79.2 | 0.68 | 1.24 (0.45,3.45) | 0.000 | 93.4 |
| Genital system¥ | 0.62 | 1.09 (0.77,1.53) | 0.02 | 61.8 | 0.06 | 1.36 (0.98,1.87) | 0.08 | 49.3 |
| Respiratory system¶ | 0.94 | 0.99 (0.74,1.32) | 0.88 | 0.0 | 0.03 | 1.65 (1.04,2.60) | 0.99 | 0.0 |
| Others | 0.72 | 0.97 (0.83,1.14) | 0.06 | 60.5 | 0.64 | 1.08 (0.78,1.51) | 0.06 | 60.5 |
| Overall | 0.65 | 1.03 (0.93,1.13) | 0.04 | 37.0 | 0.007 | 1.21 (1.05,1.40) | 0.000 | 61.0 |
| XPF rs3136038 | TT/TC VS. CC | TT VS. CC/CT | ||||||
| Respiratory system¶ | 0.34 | 0.92 (0.78,1.09) | 0.17 | 49.9 | 0.96 | 0.94 (0.59,2.08) | 0.01 | 77.1 |
| Nervous system§ | 0.75 | 1.02 (0.89,1.18) | 0.98 | 0.0 | 0.19 | 0.83 (0.61,1.11) | 0.34 | 7.4 |
| Digestive system† | 0.02 | 0.81 (0.68,0.96) | 0.62 | 0.0 | 0.03 | 0.70 (0.52,0.96) | 0.28 | 12.8 |
| others | 0.01 | 0.69 (0.53,0.90) | 0.98 | 0.0 | 0.74 | 1.06 (0.76,1.48) | 0.18 | 44.7 |
| Overall | 0.02 | 0.89 (0.81,0.98) | 0.19 | 28.1 | 0.32 | 0.88 (0.73,1.07) | 0.05 | 47.6 |
| WRN rs1801195 | GT/TT VS. GG | TT VS. GG/GT | ||||||
| Genital system¥ | 0.30 | 0.92 (0.78,1.08) | 0.98 | 0.0 | 0.001 | 1.44 (1.17,1.78) | 0.49 | 0.0 |
| Others | 0.85 | 1.01 (0.89,1.16) | 0.80 | 0.0 | 0.81 | 0.98 (0.84,1.15) | 0.69 | 0.0 |
| Overall | 0.63 | 0.97(0.88,1.08) | 0.82 | 0.0 | 0.21 | 1.17 (0.91,1.51) | 0.03 | 66.9 |
| WRN rs1346044 | CC/CT VS.TT | CC VS. TT/TC | ||||||
| Breast system | 0.02 | 1.14 (1.02,1.28) | 0.07 | 58.4 | 0.03 | 1.37 (1.03,1.83) | 0.40 | 0.0 |
| Digestive system† | 0.26 | 1.22 (0.86,1.72) | 0.42 | 0.0 | 0.35 | 0.62 (0.23,1.68) | 0.004 | 82.0 |
| Others | 0.78 | 0.95 (0.64,1.40) | 0.004 | 82.0 | 0.57 | 0.87 (0.55,1.39) | 0.79 | 0.0 |
| Overall | 0.27 | 1.08 (0.94,1.25) | 0.001 | 65.5 | 0.97 | 1.01 (0.72,1.41) | 0.01 | 58.1 |
*Pz: the significance of the pooled OR was determined by Z-test, and P <0.05 was considered as statistically significant. # Phet and I2 were calculated by Chi square-based Q-test.
¶Respiratory system cancer: lung cancer; £Urinary system cancer: bladder cancer; †Digestive system cancer: gastric cancer, ESCC, hepatocellular cancer, colorectal cancer, laryngeal cancer, OSCC, pancreastic cancer; §Nervous system cancer: glioma; ¥Genital system cancer: breast cancer, prostate cancer.
Forest plots manifests the relationship of cancer risk with polymorphism of XPA rs10817938 (A), XPA rs2808668 (B), XPC rs1870134 (C), XPD rs238406 (D), XPF rs3136038 (E), WRN rs1801195(F) and rs1346044(G) in overall analysis and stratification analysis under dominant model. Estimates of OR (95%CIs) are plotted with a box and a horizontal line for each study. ◇pooled ORs (95% CIs).
Forest plots displays the association of cancer risk with polymorphism of XPA rs10817938 (A), XPA rs2808668 (B), XPC rs1870134 (C), XPD rs238406 (D), XPF rs3136038 (E), WRN rs1801195(F) and rs1346044(G) in overall analysis and stratification analysis under recessive model. Estimates of OR (95%CIs) are plotted with a box and a horizontal line for each study. ◇, pooled ORs (95% CIs).
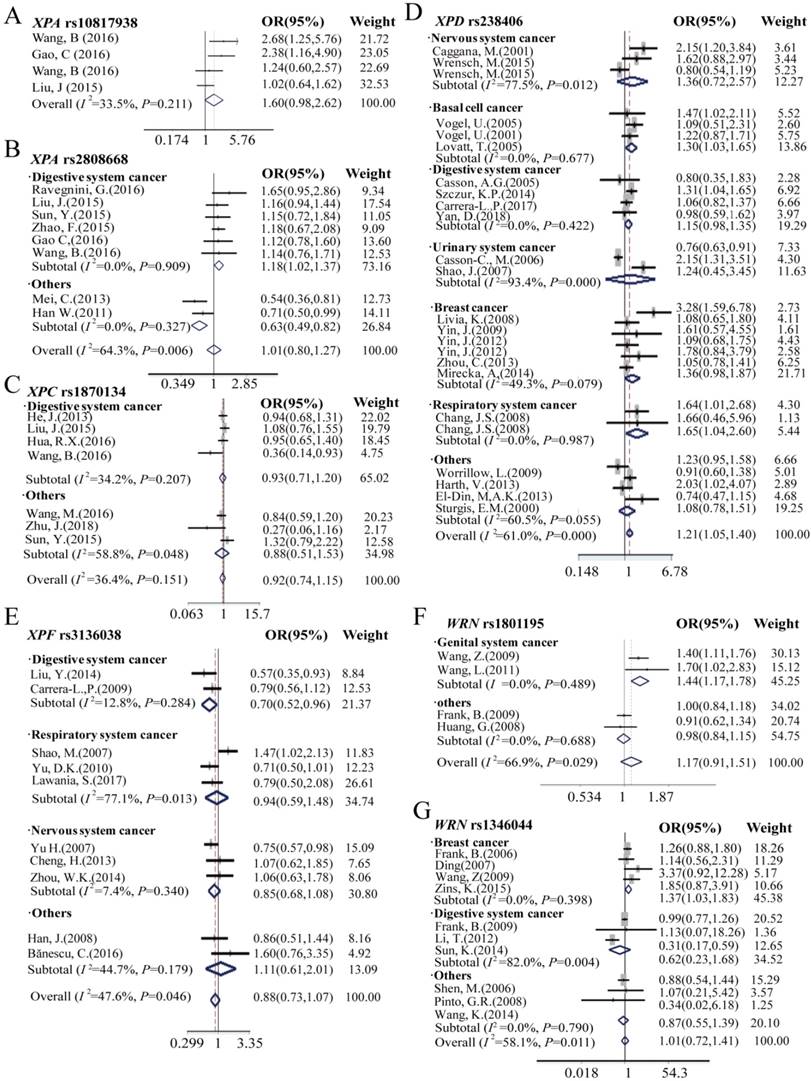
Because of the relatively higher heterogeneities (I2 = 61.1) of rs238406, the further stratification analysis was assessed by using nervous system, basal cell cancer, digestive system, urinary system, genital system and respiratory system cancer, and subsequently, we found significant impact on cancer risk in the subgroup of basal cell cancer [AA vs.CC in homozygote model: OR (95%CI) = 1.38 (1.05-1.81), P = 0.02; A vs. C allele in additive model: OR (95%CI) = 1.18 (1.03- 1.35), P = 0.02; and AA vs. CC/ CA in recessive model: OR (95%CI) = 1.30 (1.03-1.65), P = 0.03], however no association observed with cancer risk in the other groups. Furthermore, only patients with prostate cancer carrying XPC rs1870134 C allele or CC/CG genotype in the dominant model are associated with decreased cancer risk [C vs. G allele in additive model: OR (95%CI) = 0.82 (0.72- 0.95), P = 0.007; CC/CG vs.GG in dominant model: 0.77 (0.65-0.92), P = 0.004]. Moreover, XPF rs3136038 polymorphism was associated with decreased overall cancer risk in the patients with additive model [OR (95%CI) = 0.91 (0.86-0.96), P = 0.001, Table 1] and dominant model [OR (95%CI) = 0.89 (0.81-0.98), P = 0.02, Table S2]. More importantly, similar results were found in the stratification group of digestive system cancer patients who harboring TT genotype [OR (95%CI) = 0.64 (0.48-0.87), P = 0.004], T allele in additive model [OR (95%CI) = 0.82 (0.72- 0.94), P = 0.003], dominant model [OR (95%CI) = 0.81 (0.68-0.96), P=0.02] and recessive model [OR (95%CI) = 0.70 (0.52-0.96), P = 0.025] significantly decreased cancer risk, however, there was no association observed in other subgroups.
WRN rs1801195, rs1346044 and cancer risk analysis
Although no significant association between polymorphisms of WRN rs1801195, rs1346044 and cancer risk in overall meta-analysis results was found, the further stratification analysis showed significantly increased cancer risk in the genital system subgroup patients with rs1801195 genotype and breast cancer with rs1346044 genotype, as shown in Table 1, Table S2 and Figure 4, 5, S3, S4: F and G. For SNP rs1801195, genital system cancer patients correlated with increased cancer risk when carrying TT genotype [OR (95%CI) = 1.37 (1.08-1.73), P=0.009], T allele [OR (95%CI) = 1.15 (1.02-1.29), P = 0.02], and recessive model [OR (95%CI) = 1.44 (1.17-1.78), P = 0.001]. Moreover, breast cancer patients with rs1346044 CC genotype [OR (95%CI) = 1.47 (1.10-1.97), P = 0.010], C allele [OR (95%CI) = 1.15 (1.04-1.26), P = 0.005], dominant model[OR (95%CI) = 1.14 (1.02-1.28), P = 0.018] and recessive model [OR (95%CI) = 1.37 (1.03-1.83), P = 0.03] significantly correlated with elevated cancer susceptibility.
Heterogeneity and sensitivity analysis
In this meta-analysis, no significant heterogeneities were found in the overall analysis of XPA rs10817938, rs2808668, XPC rs1870134, XPF rs3136038, and WRN rs1801195. However, the moderate heterogeneity was observed when all the studies were analyzed for all the cases of XPD rs238406 under the homozygous model (I2 = 61.1%) and WRN rs1346044 under the homozygous model (I2 = 61.7%) (Table 1, Table S2). After stratification analysis, the heterogeneity of SNP rs238406 was found accused of the nervous system cancer and urinary system cancer, which is available in the higher I2 value, 82.9% and 93.2%, respectively. For another SNP rs1346044, the heterogeneity exists the subgroup of digestive system cancer (I2 = 82.3%), and other subgroups showed no heterogeneity (Figure 4, 5, S3, S4 G).
Publication bias analysis
In this included meta-analysis, the Begg's funnel plot and Egger's test were determined to evaluate the publication bias of the studies. We found no significantly publication bias was observed for all the dominant models of the seven SNPs (XPA rs10817938: P=0.289; XPA rs2808668: P=0.673; XPC rs1870134: P= 0.763; XPD rs238406: P=0.09, XPF rs3136038: P=0.245; WRN rs1801195: P=0.722; and WRN rs1346044: P=0.966) by the Egger's test. The funnel plots shapes showed obvious symmetry, which were obtained for the association of the seven SNPs (XPA rs10817938: P=0.734; XPA rs2808668: P=0.902; XPC rs1870134: P= 0.711; XPD rs238406: P=0.107, XPF rs3136038: P=0.152; WRN rs1801195: P=0.497; and WRN rs1346044: P=0.929) (Figure S2). The data indicated that no publication bias might be under a significant influence on the observed effect of SNPs located at DNA repair genes on susceptibility of cancer as assessed.
Discussion
In humans, DNA repair systems act fundamental function in maintenance of cellular genomic integrity in facing the environmental insults, DNA replication errors, and their interactions, thereby the dysregulation lead to a significantly alteration in individual susceptibility to cancer [7]. Previous accumulating studies established that aberrant expression of the core DNA repair genes such as XPs and WRN can modulate the capability of DNA damage and/or repair and subsequently contribute to carcinogenesis [76-79]. In our bioinformatics analysis, we confirmed that the critical proteins of DNA damage pathway was significantly associated with carcinogenesis, and lower XPA and XPC mRNA levels, higher XPD, XPF, and WRN expression levels were observed in majority of included 19 types of cancers compared with the normal control tissues. These data suggested that downregulated oncogenes of XPA, XPC or upregulated tumor suppressor genes such as XPD, XPF, and WRN may be involved in promoting carcinogenesis via impairment of DNA repair. Since distinct expression levels of candidate genes are the essence of affecting cancer prognosis, we further found that elevated expression of XPA and XPC had a better impact on cancer survival time, in contrast, higher expression of XPD, XPF, and WRN showed significantly worse influence on overall survival in overall meta Z-scores analysis. More importantly, these genes alteration frequency displayed higher levels in 33 types of cancers. So we hypothesized that the genetic variations of DNA repair core genes, especially in the 5'UTR (XPA rs10817938 and rs2808668, XPF rs3136038) or Exon sites (XPC rs1870134, XPD rs238406, WRN rs1346044 and rs1801195) may play a critical role in altering protein function and capability to repair damaged DNA, thus affecting the cancer risk.
NER is a multiple protein mediated sequential DNA damage repair process, and XPA, a 32kDa zinc-finger DNA-binding protein, is the sole recognition factor required its damage verification and initiation in NER activities [13]. Recent studies reported that the potentially functional SNPs in the XPA gene, especially in the transcription factors binding sites (TFBS) such as novel SNPs rs10817938 and rs2808668, which located at the XPA prompter region and may be involved in the cancer risk[19-26, 29]. For instance, Gao et al. [25] reported that the risk effect of rs10817938 T to C substitution not only had a strong impact on transcription activity, XPA mRNA and protein expression, but also obviously showed a significant gene-environment interaction with the smoking status of the patients. In the present meta-analysis, 1775 cases and 2156 controls included for SNP rs10817938, a significantly increased cancer risk was observed harboring CC, CC/CT genotype, or C allele on the overall analysis as well as the stratification analysis in the digestive system cancers. However, no significant association was observed between rs2808668 and cancer susceptibility in the overall analysis, and 8 studies were included with 2616 cases and 3099 controls. Thereafter stratified analyses showed that carrying XPA rs2808668 homozygous CC genotype or C allele was associated with significantly decreased cancer risk in the subgroup of including lung cancer and breast cancer except for digestive system cancer. Recent studies reported that XPA rs2808668 can interact with environmental factor such as drinking in gastric cancer, and clinicopathological parameters such as tumor size, metastatic status at onset and mitotic index in gastrointestinal stromal cancer [21, 24]. These findings highlight an obvious divergence between the SNP rs2808668 or rs10817938and cancer risk, although these SNPs were all in the XPA promoter region, which might be partially explained by environmental factors or clinicopathological parameters interacted with genetic variants synergistically contributing to the carcinogenesis.
XPC, a 106 kDa protein, is another primary damage recognition factor required for the global genome NER repair pathway, which can form a tight heterodimeric complex with HR23B, and to be indispensable for the stability of each protein as well as damage recognition function of XPC [13]. Following the initial damage recognition step, XPD, an 87 kDa protein, is an ATP-dependent helicase and serves as the dominant helicase in NER repair pathway [14]. Next, endonuclease is required for NER steps, XPF, a 103 kDa protein, is structure-specific nuclease that recognizes the DNA junction 5' to the adducted base(s) [15]. Thus, these XPs proteins act their function in the different steps of NER process including damage recognition, opening of the DNA surrounding the damaged base, and removal of the damaged oligonucleotides. In this pooled meta-analysis, 8 studied in 4987 cases and 6193 controls for XPC rs1870134, 24 studies with 6999 cases and 8652 controls for XPD rs238406, and 10 studies of 5247 cases and 5607 controls for XPF rs3136038 polymorphisms were recruited. A significantly increased overall cancer risk was observed in the patients with XPD rs238406 AA homozygous or A allele, but occurring the moderate heterogeneities, stratification analysis was subsequently performed and found a significant impact on susceptibility only in basal cell cancer, and no association was observed in the other subgroups, this indicated that the heterogeneities may be results from the basal cell cancer. For XPF rs3136038 polymorphism, a reduced overall cancer risk was noted in the additive model. In the subgroup analysis, harboring rs3136038 TT genotype, dominant model or recessive model significantly reduced cancer risk in the digestive system cancer, however, no association was observed in the other subgroup. These results hint that genetic variants involved in the carcinogenesis dependent on the cancer type-specificity such as cancer subgroup based on organ systems. In addition, no significant association was determined in XPC rs1870134 polymorphism and cancer susceptibility. Only one report demonstrated that XPC rs1870134 polymorphism is a cancer risk biomarker for prostate cancer [29], which suggested more and further research about this association is necessary.
Werner syndrome protein (WRN) is an important RecQ helicase that recognizes specific DNA structures and participates in DNA repair, replication, and genome stability through different pathways [16]. Overexpression of WRN is found in several cancer cell lines, and depletion of WRN protein induces cell death in these cells [77]. It is assumed that WRN genetic variants such as rs1346044 and rs1801195 may correlate with the susceptibility to cancers [64-75]. In our pooled meta-analysis, 5 studied of 3162 cases and 3142 controls for SNP rs1801195, 11 studies with 6538 cases and 7657 controls for SNP rs1346044 were included. Despite the fact that no association observed between overall cancer risk and these 2 SNPs, further stratification analysis found that increased cancer risk in the genital system patients with rs1801195 TT genotype and breast cancer with rs1346044 CC genotype. Both of the rs1346044 and rs1801195are located at Exon region of WRN gene, which contains a T to C substitution for rs1346044 (amino acids change: Cys1367Arg), and a T to G substitution for rs1801195 (amino acids change: Leu1074Phe). It is plausible that the amino acids substitution can change the tertiary structure, which results in the direct binding of BRCA1 and then alter the function of the WRN protein in facilitating the carcinogenesis [80, 81], which only partially explains the cancer risk associated with these SNPs because of not all types of cancer susceptibility observed, and further studies are needed.
Regardless of the fact that no previous bioinformatics analysis and meta-analysis to date that systematically investigated the correlation between the seven SNPs of the DNA damage response genes and risk of cancer. Some limitations are taken into account. First, although overall recruited eligible studies sample is relatively large, only 4 studies were included for the SNPs XPA rs10817938 and WRN rs1801195, thus the results cannot be rule out the possibility of fortuitous considering the limited small size for these two SNPs. Second, in the present review, all the 58 studies included were searched available in the languages of Chinese or English, which may lead to the main source of publication bias because of publications of other languages including the relevant SNPs studies. Third, seven SNPs located in the 5'UTR or Exon of the DNA repair genes were selected, however, the SNPs of 3'UTR or Intron region was not included. Therefore, limited SNPs were not sufficient to elucidate most genetic information of DNA repair process, more SNPs should be included, and interaction with other factors such as environmental factor should be investigated in the future analysis.
In conclusion, this included 33 types of cancers bioinformatics analysis, XPA and XPC mRNA expression reduced, XPD, XPF, and WRN expression increased based on the GEO dataset. Further meta Z-scores prognosis analysis displayed that overexpression of XPA or XPC had a better impact on overall survival time, nevertheless, elevated XPD, XPF, and WRN expression showed a worse influence on prognosis. The meta-analysis included the latest publications exhibit that harboring SNPs XPA rs10817938 and XPD rs238406 were significantly associated with increased overall cancer risk, however, polymorphisms of XPA rs2808668 in overall cancer analysis and XPF rs3136038 in the digestive system remarkably reduced the susceptibility of cancer. In addition, no relationship to the overall cancer risk was observed in SNPs of XPC rs1870134, WRN rs1346044 and rs1801195 to data included literature. Therefore, these findings provide the most valid cancer prevalence estimate to date, which is a necessary foundations piece for further research in this field.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 31828005, 81872905, 81603149,81601370), National Natural Science Foundation of China and Liaoning joint fund key program (No.U1608281), Liaoning Revitalization Talents Program (XLYC1807155), Liaoning Province Scientific Research Foundation (2014226033), Shenyang S&T Projects (17-123-9-00, Z18-4-020), the Key Laboratory Foundation from Shenyang S&T Projects (F16-094-1-00), Key Laboratory Foundation from Liaoning Province (No.LS201617).
Author contributions
H.W., S.L., and M. W. conceived the study and edited the paper. S.L., H.W., X.H., W. Q., Y. W., T. S., and Z. W. searched and collected the data. X. W., S. L., D. X., Y. L., and S. G. performed the table and statistical analysis. H.W., S.L., and M. W. interpreted data and wrote the manuscript. H.Z., W. Y., and M. L. critically revised the manuscript. All authors approved the final version of the manuscript.
Ethical Approval and Informed Consent
All analyses were based on previous published studies, thus no ethical approval and patient consent are required.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends-An Update. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2016;25:16-27
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7-30
3. Costello LC, Franklin RB. The genetic/metabolic transformation concept of carcinogenesis. Cancer metastasis reviews. 2012;31:123-30
4. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
5. Vineis P, Wild C. Global cancer patterns: causes and prevention. Lancet. 2014;383:549-57
6. Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nature reviews Cancer. 2016;16:35-42
7. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD. et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell reports. 2018;23:239-54.e6
8. Hoeijmakers JH. DNA damage, aging, and cancer. The New England journal of medicine. 2009;361:1475-85
9. Terabayashi T, Hanada K. Genome instability syndromes caused by impaired DNA repair and aberrant DNA damage responses. Cell Biology and Toxicology. 2018;34:337-50
10. Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environmental and molecular mutagenesis. 2010;51:520-6
11. Sugitani N, Sivley RM, Perry KE, Capra JA, Chazin WJ. XPA: A key scaffold for human nucleotide excision repair. DNA repair. 2016;44:123-35
12. Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949-56
13. Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harbor perspectives in biology. 2013;5:a012609
14. Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L. et al. Structure of the DNA repair helicase XPD. Cell. 2008;133:801-12
15. Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA repair. 2011;10:781-91
16. Monnat RJ Jr. Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Seminars in cancer biology. 2010;20:329-39
17. Williams K, Sobol RW. Mutation research/fundamental and molecular mechanisms of mutagenesis: special issue: DNA repair and genetic instability. Mutation research. 2013;743-744:1-3
18. Koberle B, Koch B, Fischer BM, Hartwig A. Single nucleotide polymorphisms in DNA repair genes and putative cancer risk. Archives of toxicology. 2016;90:2369-88
19. Han W, Kim KY, Yang SJ, Noh DY, Kang D, Kwack K. SNP-SNP interactions between DNA repair genes were associated with breast cancer risk in a Korean population. Cancer. 2012;118:594-602
20. Mei C, Hou M, Guo S, Hua F, Zheng D, Xu F. et al. Polymorphisms in DNA repair genes of XRCC1, XPA, XPC, XPD and associations with lung cancer risk in Chinese people. Thoracic cancer. 2014;5:232-42
21. Liu J, Sun L, Xu Q, Tu H, He C, Xing C. et al. Association of nucleotide excision repair pathway gene polymorphisms with gastric cancer and atrophic gastritis risks. Oncotarget. 2016;7:6972-83
22. Sun Y, Tan L, Li H, Qin X, Liu J. Association of NER pathway gene polymorphisms with susceptibility to laryngeal cancer in a Chinese population. International journal of clinical and experimental pathology. 2015;8:11615-21
23. Zhao F, Shang Y, Zeng C, Gao D, Li K. Association of single nucleotide polymorphisms of DNA repair genes in NER pathway and susceptibility to pancreatic cancer. International journal of clinical and experimental pathology. 2015;8:11579-86
24. Ravegnini G, Nannini M, Simeon V, Musti M, Sammarini G, Saponara M. et al. Polymorphisms in DNA repair genes in gastrointestinal stromal tumours: susceptibility and correlation with tumour characteristics and clinical outcome. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:13413-23
25. Gao C, Wang J, Li C, Zhang W, Liu G. A Functional Polymorphism (rs10817938) in the XPA Promoter Region Is Associated with Poor Prognosis of Oral Squamous Cell Carcinoma in a Chinese Han Population. PloS one. 2016;11:e0160801
26. Wang B, Xu Q, Yang HW, Sun LP, Yuan Y. The association of six polymorphisms of five genes involved in three steps of nucleotide excision repair pathways with hepatocellular cancer risk. Oncotarget. 2016;7:20357-67
27. He J. Polymorphismis in nucleotide excision repair genes and gastric cancer risk [Doctor Thesis]. Fudan University. 2013
28. Hua RX, Zhu ZJ, Shen GP, Zhu J, Zhang SD, Xue WQ. et al. Polymorphisms in the XPC gene and gastric cancer susceptibility in a Southern Chinese population. OncoTargets and therapy. 2016;9:5513-9
29. Wang M, Li Q, Gu C, Zhu Y, Yang Y, Wang J. et al. Polymorphisms in nucleotide excision repair genes and risk of primary prostate cancer in Chinese Han populations. Oncotarget. 2017;8:24362-71
30. McWilliams RR, Bamlet WR, Cunningham JM, Goode EL, de Andrade M, Boardman LA. et al. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res. 2008;68:4928-35
31. Zhu J, Fu W, Jia W, Xia H, Liu GC, He J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Molecular therapy Nucleic acids. 2018;12:854-60
32. Sturgis EM, Zheng R, Li L, Castillo EJ, Eicher SA, Chen M. et al. XPD/ERCC2 polymorphisms and risk of head and neck cancer: a case-control analysis. Carcinogenesis. 2000;21:2219-23
33. Caggana M, Kilgallen J, Conroy JM, Wiencke JK, Kelsey KT, Miike R. et al. Associations between ERCC2 polymorphisms and gliomas. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2001;10:355-60
34. Vogel U, Hedayati M, Dybdahl M, Grossman L, Nexo BA. Polymorphisms of the DNA repair gene XPD: correlations with risk of basal cell carcinoma revisited. Carcinogenesis. 2001;22:899-904
35. Lovatt T, Alldersea J, Lear JT, Hoban PR, Ramachandran S, Fryer AA. et al. Polymorphism in the nuclear excision repair gene ERCC2/XPD: association between an exon 6-exon 10 haplotype and susceptibility to cutaneous basal cell carcinoma. Human mutation. 2005;25:353-9
36. Vogel U, Olsen A, Wallin H, Overvad K, Tjonneland A, Nexo BA. Effect of polymorphisms in XPD, RAI, ASE-1 and ERCC1 on the risk of basal cell carcinoma among Caucasians after age 50. Cancer detection and prevention. 2005;29:209-14
37. Wrensch M, Kelsey KT, Liu M, Miike R, Moghadassi M, Sison JD. et al. ERCC1 and ERCC2 polyrnorphisms and adult glioma. Neuro-oncology. 2005;7:495-507
38. Casson AG, Zheng Z, Evans SC, Veugelers PJ, Porter GA, Guernsey DL. Polymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinoma. Carcinogenesis. 2005;26:1536-41
39. Carrera-Lasfuentes P, Lanas A, Bujanda L, Strunk M, Quintero E, Santolaria S. et al. Relevance of DNA repair gene polymorphisms to gastric cancer risk and phenotype. Oncotarget. 2017;8:35848-62
40. Shao J, Gu M, Xu Z, Hu Q, Qian L. Polymorphisms of the DNA gene XPD and risk of bladder cancer in a Southeastern Chinese population. Cancer genetics and cytogenetics. 2007;177:30-6
41. Kipikasova L, Wolaschka T, Bohus P, Baumohlova H, Bober J, Blazejova J. et al. Polymorphisms of the XRCC1 and XPD genes and breast cancer risk: a case-control study. Pathology oncology research: POR. 2008;14:131-5
42. Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP Jr. et al. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. International journal of cancer. 2008;123:2095-104
43. Harth V, Schafer M, Abel J, Maintz L, Neuhaus T, Besuden M. et al. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. Journal of toxicology and environmental health Part A. 2008;71:887-97
44. Worrillow L, Roman E, Adamson PJ, Kane E, Allan JM, Lightfoot TJ. Polymorphisms in the nucleotide excision repair gene ERCC2/XPD and risk of non-Hodgkin lymphoma. Cancer epidemiology. 2009;33:257-60
45. Yin J, Liang D, Vogel U, Chang Y, Liu Z, Yue L. et al. The Polymorphism of DNA Repair Gene ERCC2/XPD Arg156Arg and Susceptibility to Breast Cancer in a Chinese Population. Biochemical genetics. 2009;47:582-90
46. Yin J, Wang C, Liang D, Vogel U, Yue L, Liu J. et al. No evidence of association between the synonymous polymorphisms in XRCC1 and ERCC2 and breast cancer susceptibility among nonsmoking Chinese. Gene. 2012;503:118-22
47. El-Din MA, Khorshied MM, El-Saadany ZA, El-Banna MA, Reda Khorshid OM. Excision repair cross-complementing group 2/Xeroderma pigmentousm complementation group D (ERCC2/XPD) genetic variations and susceptibility to diffuse large B cell lymphoma in Egypt. International journal of hematology. 2013;98:681-6
48. Zhou C, Xie LP, Lin YW, Yang K, Mao QQ, Cheng Y. Susceptibility of XPD and hOGG1 genetic variants to prostate cancer. Biomedical reports. 2013;1:679-83
49. Paszkowska-Szczur K, Scott RJ, Gorski B, Cybulski C, Kurzawski G, Dymerska D. et al. Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the Polish population. Molecular biology reports. 2015;42:755-64
50. Mirecka A, Paszkowska-Szczur K, Scott RJ, Gorski B, van de Wetering T, Wokolorczyk D. et al. Common variants of xeroderma pigmentosum genes and prostate cancer risk. Gene. 2014;546:156-61
51. Michalska MM, Samulak D, Jablonski F, Romanowicz H, Smolarz B. The R156R ERCC2 polymorphism as a risk factor of endometrial cancer. Tumor Biology. 2016;37:2171-6
52. Romanowicz H, Michalska MM, Samulak D, Malinowski J, Szaflik T, Bienkiewicz J. et al. Association of R156R single nucleotide polymorphism of the ERCC2 gene with the susceptibility to ovarian cancer. European journal of obstetrics, gynecology, and reproductive biology. 2017;208:36-40
53. García-Closas M, Malats N, Real FX, Welch R, Kogevinas M, Chatterjee N. et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiology and Prevention Biomarkers. 2006;15:536-42
54. Yan D, Liang XH, Ding W, Xu XJ, Wang XY. Contribution of DNA repair xeroderma pigmentosum group D genotypes to pancreatic cancer risk in the Chinese Han population. Genetics and molecular biology. 2018;41:18-26
55. Shao M, Ma H, Wang Y, Xu L, Yuan J, Wang Y. et al. Polymorphisms in excision repair cross-complementing group 4 (ERCC4) and susceptibility to primary lung cancer in a Chinese Han population. Lung cancer (Amsterdam, Netherlands). 2008;60:332-9
56. Han J, Haiman C, Niu T, Guo Q, Cox DG, Willett WC. et al. Genetic variation in DNA repair pathway genes and premenopausal breast cancer risk. Breast cancer research and treatment. 2009;115:613-22
57. Yu D-K, Wu C, Tan W, Lin D-X. Functional XPF polymorphisms associated with lung cancer susceptibility in a Chinese population. Frontiers of Medicine in China. 2010;4:82-9
58. Yu H, Liu Z, Huang YJ, Yin M, Wang LE, Wei Q. Association between single nucleotide polymorphisms in ERCC4 and risk of squamous cell carcinoma of the head and neck. PloS one. 2012;7:e41853
59. Cheng HB, Xie C, Zhang RY, Hu SS, Wang Z, Yue W. Xeroderma pigmentosum complementation group f polymorphisms influence risk of glioma. Asian Pacific journal of cancer prevention: APJCP. 2013;14:4083-7
60. Zhou WK, Huang LY, Hui L, Wang ZW, Jin BZ, Zhao XL. et al. Association of polymorphisms of the xeroderma pigmentosum complementation group F gene with increased glioma risk. Genetics and Molecular Research. 2014;13:3826-31
61. Liu Y, Cao L, Chang J, Lin J, He B, Rao J. et al. XPF-673C>T polymorphism effect on the susceptibility to esophageal cancer in Chinese population. PloS one. 2014;9:e94136
62. Banescu C, Iancu M, Trifa AP, Dobreanu M, Moldovan VG, Duicu C. et al. Influence of XPC, XPD, XPF, and XPG gene polymorphisms on the risk and the outcome of acute myeloid leukemia in a Romanian population. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:9357-66
63. Lawania S, Sharma S, Singh N, Behera D. XPF polymorphism toward lung cancer susceptibility and survival in patients treated with platinum-based chemotherapy. Future oncology (London, England). 2018;14:1071-89
64. Wang Z, Xu Y, Tang J, Ma H, Qin J, Lu C. et al. A polymorphism in Werner syndrome gene is associated with breast cancer susceptibility in Chinese women. Breast cancer research and treatment. 2009;118:169-75
65. Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Colorectal cancer and polymorphisms in DNA repair genes WRN, RMI1 and BLM. Carcinogenesis. 2010;31:442-5
66. Wang L, Kaku H, Huang P, Xu K, Yang K, Zhang J. et al. Single nucleotide polymorphism WRN Leu1074Phe is associated with prostate cancer susceptibility in Chinese subjects. Acta medica Okayama. 2011;65:315-23
67. Huang G, Feng J, Hao S, Li D, Wang K, Wang L. et al. CASP8, XRCC1, WRN, NF2, and BRIP1 Polymorphisms Analysis Shows Their Genetic Susceptibility for Meningioma Risk and the Association with Tumor-Related Phenotype in a Chinese Population. World neurosurgery. 2018;114:e883-e91
68. Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS. et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Human genetics. 2006;119:659-68
69. Wirtenberger M, Frank B, Hemminki K, Klaes R, Schmutzler R, Wappenschmidt B. et al. Interaction of Werner and Bloom syndrome genes with p53 in familial breast cancer. Carcinogenesis. 2006 p. 1655-60
70. Ding SL, Yu JC, Chen ST, Hsu GC, Shen CY. Genetic variation in the premature aging gene WRN: a case-control study on breast cancer susceptibility. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:263-9
71. Pinto GR, Yoshioka FK, Clara CA, Santos MJ, Almeida JR, Burbano RR. et al. WRN Cys1367Arg SNP is not associated with risk and prognosis of gliomas in Southeast Brazil. Journal of neuro-oncology. 2008;90:253-8
72. Li T, Suo Q, He D, Du W, Yang M, Fan X. et al. Esophageal Cancer Risk is Associated with Polymorphisms of DNA Repair Genes MSH2 and WRN in Chinese Population. Journal Of Thoracic Oncology. 2012;7:448-52
73. Sun K, Gong A, Liang P. Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:1549-59
74. Wang K, Wang L, Feng J, Hao S, Tian K, Wu Z. et al. WRN Cys1367Arg polymorphism is not associated with skull base chordoma. Biomedical reports. 2014;2:521-4
75. Zins K, Frech B, Taubenschuss E, Schneeberger C, Abraham D, Schreiber M. Association of the rs1346044 Polymorphism of the Werner Syndrome Gene RECQL2 with Increased Risk and Premature Onset of Breast Cancer. International journal of molecular sciences. 2015;16:29643-53
76. Welsh C, Day R, McGurk C, Masters JR, Wood RD, Köberle B. Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. International journal of cancer. 2004;110:352-61
77. Opresko PL, Cheng W-H, von Kobbe C, Harrigan JA, Bohr VA. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791-802
78. Wu Y, Chang JT, Cheng Y, Wu T, Chen C, Lee H. Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene. 2007;26:4761
79. Ding H, Xu J-j, Huang Y, Du F-t, Zhang J-x. XPD could suppress growth of HepG2. 2.15 and down-regulate the expression of hepatitis B virus x protein through P53 pathway. Biochemical and biophysical research communications. 2012;419:761-7
80. Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML. et al. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic acids research. 2006;34:2751-60
81. von Kobbe C, Bohr VA. A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949-1092. Journal of cell science. 2002;115:3901-7
Author contact
![]() Corresponding author: Minjie Wei, MD, PhD. Department of Pharmacology, School of Pharmacy, Liaoning Key Laboratory of Molecular Targeted Anti-Tumor Drug Development and Evaluation, China Medical University, No.77 Puhe Road, Shenyang North New Area, Shenyang, Liaoning Province, 110122, P.R. China. Tel: +86-13358866401; E-mail: mjweiedu.cn
Corresponding author: Minjie Wei, MD, PhD. Department of Pharmacology, School of Pharmacy, Liaoning Key Laboratory of Molecular Targeted Anti-Tumor Drug Development and Evaluation, China Medical University, No.77 Puhe Road, Shenyang North New Area, Shenyang, Liaoning Province, 110122, P.R. China. Tel: +86-13358866401; E-mail: mjweiedu.cn

 Global reach, higher impact
Global reach, higher impact