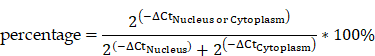Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3608-3617. doi:10.7150/jca.31832 This issue Cite
Research Paper
AC138128.1 an Intronic lncRNA originating from ERCC1 Implies a Potential Application in Lung Cancer Treatment
1. Dept. of Toxicology, School of Public Health, China Medical University, Shenyang, P.R. China
2. Dept. of Thoracic Surgery Ward 2, The first Hospital of China Medical University, Shenyang, P.R. China
3. Dept. of Thoracic Surgery, Liaoning Cancer Hospital & Institute, Shenyang, P.R. China
Received 2018-11-27; Accepted 2019-5-8; Published 2019-6-9
Abstract
Lung cancer is one of the most devastating tumors with a high incidence and mortality worldwide. Polymorphisms and expression of ERCC1 commonly predicted the occurrence and prognosis of lung cancer. However, few studies have focused on long non-coding RNAs related to ERCC1 though some studies reminded the importance of its post-transcriptional regulation. In the present study, an intronic lncRNA AC138128.1 originated from ERCC1 was firstly identified in microarray chip and database, and its possibility as a novel biomarker to predict lung cancer treatment was further discussed. Firstly, the qRT-PCR data showed that AC138128.1 expression was much lower in lung cancer comparing with its para-cancer tissues, which further analyzed by ROC curve. Similarly, the difference was also verified in 16HBE, A549 and LK2 cells. Then AC138128.1 expression was found to have an increasing trend in a dose or time-dependent manner after cisplatin treatment. Finally, the subcellular distribution of AC138128.1 reminded that AC138128.1 was mainly expressed in the nucleus. Interestingly a positive relationship between AC138128.1 and ERCC1 expression was only found in cancer tissues, which reminded AC138128.1 may be involved in the regulation of ERCC1. Therefore, as a preliminary exploration of the lncRNA originated from ERCC1, the present study suggested AC138128.1 is of potential value in predicting platinum analogue benefit in lung cancer.
Keywords: AC138128.1, intronic lncRNA, ERCC1, lung cancer, platinum analogue
Introduction
Lung cancer is one of the most devastating tumors with a high incidence and mortality worldwide. [1]. It is now widely accepted that cancer, particularly lung cancer, results from the interaction of environmental and genetic factors. A large amount of epidemiological evidence indicates that the occurrence of lung cancer is affected by environmental factors such as exposure to polycyclic aromatic hydrocarbons due to smoking and air pollution [2, 3]. Previous studies on biomarker associated with the susceptibility of lung cancer also confirmed the significance of genetic factors [4]. Thus, it is necessary to identify valid biomarkers for predicting the risk and prognosis of lung cancer.
DNA repair genes have long been considered as critical environmental response genes. Excision repair cross-complementing rodent repair deficiency, complementation group 1 (ERCC1), an endonuclease as a key member of the nucleotide excision repair (NER) pathway, is vital to the repair of DNA damage, particularly DNA adducts or double-stranded breaks [5, 6]. For many years, the polymorphisms and expression of ERCC1 have been widely examined in the oncogenesis of lung cancer [7, 8] and also become a common biomarker to predict platinum analogue benefit [9]. Our previous study showed that a polymorphism site in the 3′ untranslated region (UTR) of ERCC1 was associated with BPDE-DNA (7,8-dihydroxybenzo(a)pyrene-9,10-oxide) adduct levels, an indicator closely related to tumorigenesis [10]. Recent studies revealed that the inconsistency of ERCC1 mRNA and its protein level could be affected by post-transcriptional regulation [11, 12]. Therefore, epigenetic such as multiple microRNAs regulation is involved in numerous key mechanisms in the process of tumorigenesis and development [13, 14]. Besides microRNAs, long non-coding RNAs (lncRNAs) have received much more attention recently. LncRNAs are longer than 200 nucleotides (nt), abundant in the human genome, and tissue- and cell-specific [15]. Recent transcriptome studies of human cancers suggest that lncRNA expression is the most common transcriptional alteration [16]. LncRNAs can be classified as intergeneric, intronic, sense, antisense, or bidirectional according to their position in the genome [17]. The distribution of lncRNAs in the genome or subcellular location can determine their function [18, 19].
Till now no studies have examined ERCC1- related lncRNAs. In the present study, we screened the non-coding RNAs in the upstream and downstream 100k nts region of ERCC1 gene through microarray chips. Combined with a non-coding RNA database, a lncRNA AC138128.1 originated from ERCC1 was confirmed. Moreover, it has been reported that AC138128.1 is down-regulated in gastric cancer and liver cancer [20, 21]. However, whether AC138128.1 is associated with the risk or prognosis of lung cancer is unknown. In order to clarify the implication of lncRNA AC138128.1as a possible biomarker in lung cancer, we first detected the expression of lncRNA AC138128.1 in the cancer and its para-cancer tissues. Next, a tumor chemo-sensitivity assay was performed to measure the sensitivity to cisplatin in vitro. 16HBE, A549, and LK2 cells were used to further confirmed the previous results. Meanwhile the subcellular distribution of AC138128.1 was detected in 16HBE and A549 cells in order to explore its function. In addition, the correlation between AC138128.1 and ERCC1gene was further analyzed based on the mRNA levels. As a preliminary and unique exploration of the lncRNA originated from ERCC1, the present study will contribute to find a novel biomarker to predict the platinum analogue benefit in lung cancer.
Materials and methods
Patient information and sample collection
This study was conducted according to the Declaration of Helsinki. The Institutional Review Board of China Medical University approved the study and informed consent was obtained from all participants prior to the study. A total of 84 lung cancer patients from The First Hospital of China Medical University (Shenyang, China) and Liaoning Cancer Hospital and Institute (Shenyang, China) were screened. Pathologic and histological grade were assessed according to criteria set by World Health Organization. The basic information including age, sex, smoking status, and drinking status was collected. Additionally, tumor size, lymphatic metastasis, distant metastasis, chemotherapy or radiotherapy history, and TNM staging (the newest 7th edition of the TNM staging system) were also investigated. Fresh cancer tissue and its matched adjacent tissue samples from the 64 patients fully informed were collected during surgery and then transferred into liquid nitrogen for RNA extraction. It is worth noting that cancer tissues and its para-cancer tissue from 20 patients were also used to perform the tumor chemo-sensitivity assay by cisplatin treatment.
Tumor chemo-sensitivity assay
The fresh cancer tissues used for the chemo-sensitivity assay were immediately divided into two parts. One part was used for pathological examination and the other part was used to isolate single cells to test cisplatin susceptibility. The protocol for the tumor chemo-sensitivity assay was as follows. Fresh tissues were washed with normal saline containing 1% Penicillin-Streptomycin Solution, and then necrotic tissue was removed. Next, cut into mud in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 (Gibco, Grand Island, NY, USA) and the mixture passed through a 125-μm nylon membrane. The flow-through was centrifuged for 3 min at 1000 g. Cells were cultured in a 96-well plate containing medium with 10% fetal bovine serum. After 24h, the cells were exposed to different concentrations of cisplatin (Sigma, St. Louis, MO, USA) and the IC50 was tested by the MTT assay at 490 nm. Finally, the sensitivity of different tumor tissues to cisplatin was calculated.
RNA extraction, reverse transcription, and quantitative PCR
Total RNA was extracted from tissue samples using the GeneJET RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol. Briefly, 30 mg of tissue was placed in liquid nitrogen and ground thoroughly with a mortar and pestle. The tissue powder was immediately transferred into a microcentrifuge tube containing Lysis Buffer and β-mercaptoethanol, followed by vertexing to mix thoroughly. Protein K was added and mixed by vertexing, and the sample was incubated at 15-25°C for 10 min. After adding ethanol, the lysate was transferred to a purification column inserted into a collection tube. Wash buffer was used to wash the purification column twice. Finally, RNA was eluted by adding nuclease-free water. RNA purity was evaluated by UV spectroscopy (A260/A280 > 1.9 and A260/A230 > 2.0) using a Nanodrop One (Thermo Fisher Scientific). The RNA was immediately used for reverse transcription (RT-PCR). The RT-PCR reaction was conducted using the PrimeScriptTM RT reagent Kit with DNA Eraser (Takara, Shiga, Japan). The 20-μL RT-PCR reaction was incubated at 42℃ for 2 min to remove DNA, and then incubated for 30 min at 37℃ and 5 s at 85℃, after which the temperature was maintained at 4℃. For quantitative PCR, AceQ qPCR SYBR Green Master Mix (Vazyme Biotech, Nanjing, China) was used. The 20-μL reaction mix consisted of 10 μL 2 × AceQ qPCR SYBR Green Master Mix, 0.4 μL forward and reverse primers, 7.8 μL nuclease-free water, and 2 μL RT-PCR reaction products. The primers used in this study are shown in Table 1. The qRT-PCR cycling conditions were as follows: denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Relative gene expression was calculated by using the 2-ΔCt method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the endogenous control for expression normalization.
The sequence of primers
| Gene | Sequence (3'-5') | |
|---|---|---|
| GAPDH | Forward | TGTTGCCATCAATGACCCCTT |
| Reverse | CTCCACGACGTACTCAGCG | |
| AC138128.1 | Forward | GGCAACAGTGCAGAACCAAG |
| Reverse | CCGTTTTCACTTGTCTCCTTTCT | |
| ERCC1 | Forward | AAGATGTGTATCCTGGCCGACTG |
| Reverse | TCTGCTCATAGGCCTTGTAGGTCTC | |
| U6 | Forward | TGCTCGCTTCGGCAGCACAT |
| Reverse | CTTGCGCAGGGGCCATGCTA | |
| MT-RNR1 | Forward | CCTCCCCAATAAAGCTAAAA |
| Reverse | GCTATTGTGTGTTCAGATAT |
Cell cultures and treatment
The human normal bronchial epithelial cell line 16HBE cell, kindly provided by Professor Wen Chen (Sun Yat-Sen University, Guangzhou, China), was cultured in minimum essential medium (HyClone, Logan, UT, USA). A549 and LK2 cells were purchased from the Cell Bank of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China), and were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 and Dulbecco's Modified Eagle Medium respectively. All cells were cultured with 10% fetal bovine serum (Biological Industries, Kibbutz BeitHaemek, Israel) in flasks at 37℃ in a humidified atmosphere with 5% CO2.
Each cell line was seeded at 3 × 105 cells/well in a 6-well plate. After 24 h, the cells were treated with different concentrations of cisplatin for 12, 24, and 48 h. Following incubation, RNA was extracted from the cells as described above.
Affymetrix microarray chips
Total RNA from A549 and 16HBE cells treated by 0, 2 and 4 mg/L cisplatin for 24h were hybridized with Affymetrix Human Transcriptome Assay 2.0 (Affymetrix, Santa Clara, CA, USA). And each chip was repeated once. All detection service was performed by OE Biotech's (Shanghai, China). The method of data standardization has been described previously [22]. The upstream and downstream 100k nts of ERCC1 gene is from 45876341 to 46078414 in chromosome 19 (GRCh37). In this region, there are 9 lncRNAs, the cluster heatmap was generated by MeV (Multiple Experiment Viewer) software v4.0.
Nuclear and cytoplasmic fractions
Nuclear and cytoplasmic fractions of RNA were prepared using the PARIS™ Kit (Ambion® Thermo Fisher Scientific) according to the manufacturer's protocol. Briefly, 16HBE cells in good condition were dispersed into single-cell suspensions using 0.25% trypsin with EDTA. Next, 106 cells were collected and washed in cold PBS by centrifugation at 2000 rpm for 5 min. The samples were disrupted in 400 μL ice-cold Cell Disruption Buffer and gently resuspended by pipetting, followed by incubation for 3 min on ice. The samples were immediately centrifuged for 3 min at 4°C at 500×g, and the supernatant containing the cytoplasmic fraction was transferred into a new microcentrifuge tube; the precipitate contained the nuclear fraction. Finally, RNA was extracted using the GeneJET RNA Purification Kit following the manufacturer's protocol. The RT-PCR reaction and qRT-PCR were conducted as described in part 3. U6, an snRNA expressed in the nucleus, and MT-RNR1, a mitochondrial genome-encoded RNA expressed in the cytoplasm, were used to evaluate the nuclear and cytoplasmic fractions.
Statistical analysis
All statistical analysis was performed using SPSS software version 19.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism software version 6.0 (GraphPad Software, Inc., San Diego, CA, USA). Gene expression levels in cancer and para-cancer tissues were calculated by logarithmic transformation of 2-ΔCt (ln(2-ΔCt)) to ensure that the expression data showed a normal distribution and improve the power of the test. Paired t-test was used to analyze gene expression levels in cancer and para-cancer tissue. Pearson's correlation was used to analyze the correlation between AC138128.1 and ERCC1 expression. The subcellular distribution of RNA in the nuclear and cytoplasmic fractions was analyzed as described previously [23] using the following formula:
Significance values were set at P-value of < 0.05.
Results
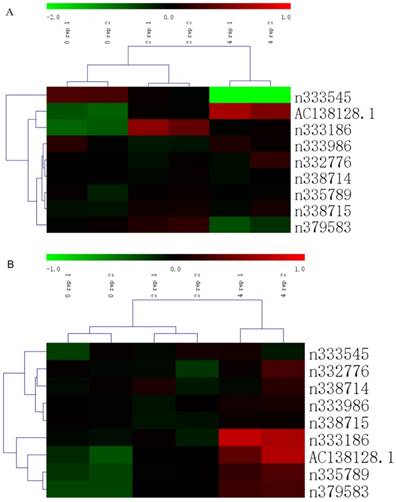
Heatmap of lncRNA in upstream and downstream 100k nts of ERCC1
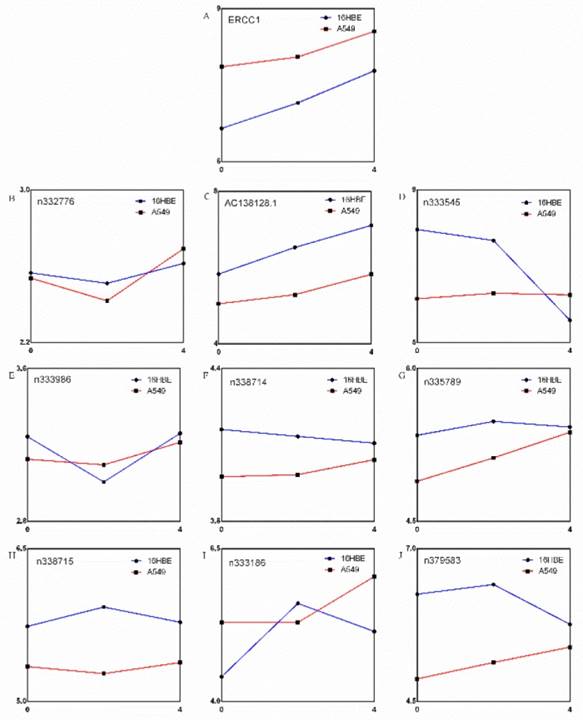
There are 9 lncRNAs in upstream and downstream 100,000 nucleotides (nts) of ERCC1 according to our gene chip results (Figure 1A and B). The expression of ERCC1 gene showed an increasing trend following the dose of cisplatin in both A549 and 16HBE cell (Figure 2A). Bioinformatics perspective believes that the expression levels of the two genes are correlated, suggesting that the function of two genes may be correlated each other. The expression of these 9 lncRNAs are changed differently, but only one lncRNA's (AC138128.1) expression has a similar trend to ERCC1 gene and much higher in 16HBE cell (Figure 2 B-J).
The cluster heatmap of lncRNAs treated by different cisplatin concentration in upstream and downstream of ERCC1 gene. Red color indicates high expression level and green color indicates low expression level. A. The heatmap of 16HBE cell. B. The heatmap of A549 cell.
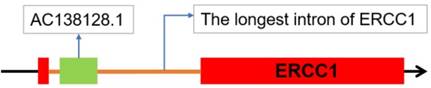
Position of A138128.1 in the chromosome
The lncRNA AC138128.1, with a length of 1981 nts, is in chromosome 19 45473082- 45475156. This lncRNA exists in the longest intron of ERCC1 (Figure 3).
Two noncoding RNA databases, LNCipedia and NONCODE, confirm the noncoding potential of AC138128.1 as an intronic lncRNA.
Relationship between AC138128.1 expression and patient information
The study population consisted of 64 lung cancer patients, including 42 males and 22 females; patient ages ranges from 42 to 76 (mean, 59.88; median, 62). 39 patients are previous smokers, while 25 patients are never-smokers or had quit smoking for more than 15 years. 51 patients consumed alcohol. 33 are diagnosed with adenocarcinoma, and 31 are diagnosed with squamous cell carcinoma. Tumor sizes are 1-9 cm (mean, 3.64 cm; median, 3.5 cm). 29 patients show lymphatic metastasis and 21 patients have distant metastasis. 48 patients are in stages Ⅰ or Ⅱ and 16 patients have received radiotherapy or chemotherapy. AC138128.1 expression in cancer ranges from 0.001 to 8.535 (mean, 0.570; median, 0.131). The patients are divided into low and high expression group by median. However, no significant differences are observed between expression of AC138128.1 and patient information (Table 2).
AC138128.1 expression in cancer and para-cancer tissues
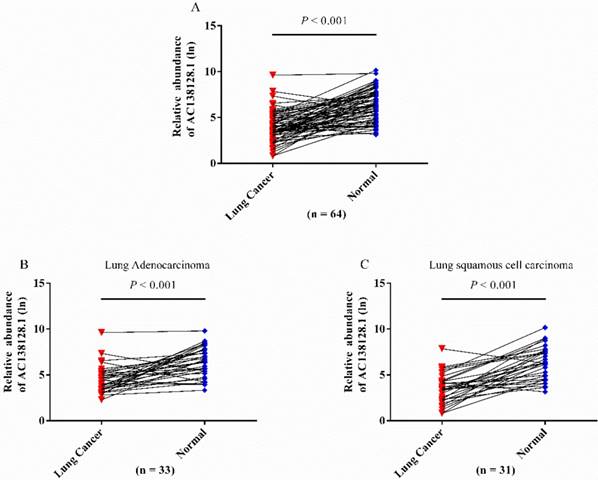
We detected the expression of lncRNA AC138128.1 in cancer and its matched para-cancer tissues by qRT-PCR. In 64 pairs of tissues, AC138128.1 is significantly downregulated in cancer tissues (Figure 4A). This downregulation is observed in both lung adenocarcinoma and lung squamous cell carcinoma (Figure 4 B and C).
ROC curve of AC138128.1 in lung cancer
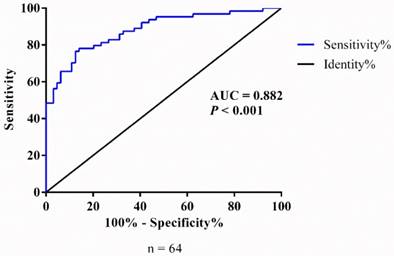
In order to further analyze the difference of AC138128.1 expression between cancer and its matched para-cancer tissues, a receiver operating characteristic (ROC) curve was performed by grouping the expression of AC138128.1. The area under the ROC curve is 0.852 (P < 0.001 95%CI: 0.787 - 0.918) (Figure 5), which reminds that AC138128.1 can be of value to distinguish the lung cancer and para-cancer tissue.
AC138128.1 expression in 16HBE, A549, and LK2 and cisplatin treatment
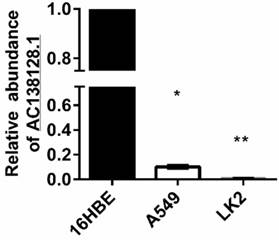
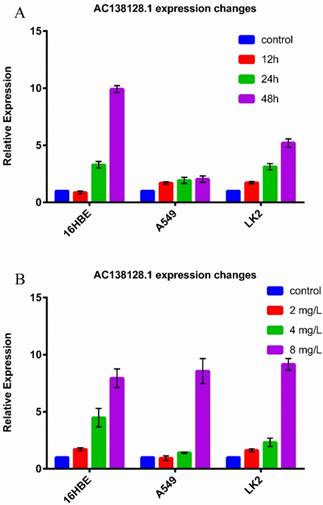
The expression of AC138128.1 in 16HBE, A549, and LK2 cell lines was detected. 16HBE is a normal bronchial epithelial cell line, A549 is a lung adenocarcinoma cell line, and LK2 represents lung squamous cell carcinoma. We found that the expression of AC138128.1 is much lower in the A549 and LK2 cell lines compared with 16HBE (both P < 0.05), as shown in Figure 6, which is consistent with the result of cancer tissue. We treated each cell line with 2, 4, and 8 mg/L cisplatin for 24 h or with 4 mg/L cisplatin for 12, 24, and 48 h, respectively. The results show that the expression of AC138128.1 is time-dependent and dose-dependent, as shown in Figure 7A and B. Similarly, the above results are also confirmed in our Affymetrix microarray chips.
Correlative analysis of AC138128.1 expression and cisplatin sensitivity
The characteristics of the lung cancer patients in the chemo-sensitivity assay are shown in Table 3.
MTT assay was used to evaluate the tumor chemo-sensitivity to cisplatin, and its correlation with ERCC1 level and AC138128.1 expression was also analyzed. The range of AC138128.1 expression (1×103) in cancer is 0.031-20.977 (median, 1.133) and the IC50 is 5.985-680.388 μg/mL (median, 42.14 μg/mL). In para-cancer tissues, AC138128.1 expression (1×103) ranges from 0.001 to 989.62 (median, 81.023) and the IC50 ranges from 13.002 to 6780.1 μg/mL (media, 254.98 μg/mL). The results indicate that AC138128.1 expression has a negative relationship with the sensitivity of cisplatin in cancer tissue (rs = 0.493, P = 0.027) while no similar significance is found in its para-cancer tissue (rs = -0.179, P = 0.450). Like ERCC1 expression different AC138128.1 level also reflects the distinction of sensitivity to cisplatin especially in cancer tissues (Table 4).
The expression of 9 noncoding RNAs and ERCC1, and their trend with increasing dose of cisplatin. A. The expression of ERCC1 gene. B~J. Expression of 9 noncoding RNAs.
AC138128.1's position in the chromosome. AC138128.1, located in 19q13, is in the longest intron of ERCC1 gene.
The relative abundance of AC138128.1 in cancer and para-cancer tissues. A, different expression in 64 pairs of lung cancer patients. AC138128.1 is down-regulated in cancer tissues compared with its adjacent para-cancer tissues. B & C, AC138128.1 is down-regulated in cancer tissues compared with its adjacent para-cancer tissues both in lung adenocarcinoma (n = 33) and lung squamous cell carcinoma (n = 31). Paired t test is used for statistical analysis and all the P value is less than 0.001.
Correlation between AC138128.1 expression and patients' data
| Variables | Total Number n = 64 (%) | AC138128.1 relative expression | χ2 | P value | |
|---|---|---|---|---|---|
| High expression n | Low expression n | ||||
| Sex | 3.206 | 0.064 | |||
| Man | 42 (65.6) | 15 | 27 | ||
| Woman | 22 (34.4) | 13 | 9 | ||
| Age Group | 3.004 | 0.069 | |||
| ≤ 60 | 33 (51.6) | 11 | 22 | ||
| > 60 | 31 (48.4) | 17 | 14 | ||
| Smoking | 1.135 | 0.210 | |||
| Yes | 39 (60.9) | 15 | 24 | ||
| No | 25 (39.1) | 13 | 12 | ||
| Drinking | 0.185 | 0.457 | |||
| Yes | 51 (79.7) | 23 | 28 | ||
| No | 13 (20.3) | 5 | 8 | ||
| Pathologic Type | 0.621 | 0.296 | |||
| Adenocarcinoma | 33 (51.6) | 16 | 17 | ||
| Squamous Cell Carcinoma | 31 (48.4) | 12 | 19 | ||
| Size of Tumor | 0.009 | 0.563 | |||
| > 3.6 | 27 (42.2) | 12 | 15 | ||
| ≤ 3.6 | 37 (57.8) | 16 | 21 | ||
| Lymphatic metastasis | 0.025 | 0.537 | |||
| Yes | 29 (45.3) | 13 | 16 | ||
| No | 35 (54.7) | 15 | 20 | ||
| Distant Metastasis | 0.190 | 0.432 | |||
| Yes | 21 (32.8) | 10 | 11 | ||
| No | 43 (67.2) | 18 | 25 | ||
| TNM Staging | 0.339 | 0.384 | |||
| Ⅰ & Ⅱ | 48 (75.0) | 20 | 28 | ||
| Ⅲ & Ⅳ | 16 (25.0) | 8 | 8 | ||
| Radiotherapy or chemotherapy | 0.339 | 0.384 | |||
| Yes | 16 (25.0) | 8 | 8 | ||
| No | 48 (75.0) | 20 | 28 | ||
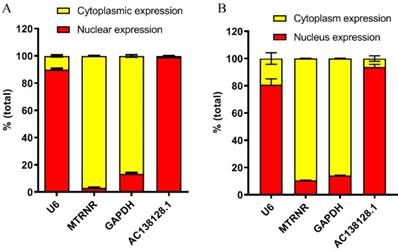
Subcellular distribution of AC138128.1
The subcellular distribution of lncRNA can strongly suggest its function [15, 16]. Therefore, the subcellular distribution of AC138128.1 was detected by RNA nucleus and cytoplasmic distribution kit in 16HBE cells and A549 cells. By analyzing the expression of the U6, MT-RNR1, and GAPDH genes, we found that the cytoplasm and nucleus distributions were clearly distinguished. In 16HBE cell, the results show that nearly 90% of U6 is distributed in the nucleus, while 95% of MT-RNR1 is distributed in the cytoplasm (Figure 8A). Interestingly, more than 95% of AC138128.1 is found in the nucleus (Figure 8A). For A549 cell, nearly 80% of U6 is distributed in the nucleus, while 90% of MT-RNR1 is distributed in the cytoplasm (Figure 8B). Interestingly, more than 90% of AC138128.1 is found in the nucleus (Figure 8B). Similarly, more than 90% of AC138128.1 is found in the nucleus (Figure 8B).
The characteristics of the lung cancer patients for chemo-sensitivity assay
| Variables | Number (n=19) | Percentage (%) |
|---|---|---|
| Sex | ||
| Man | 8 | 40.0 |
| Woman | 12 | 60.0 |
| Age Group | ||
| ≤ 60 | 8 | 40.0 |
| > 60 | 12 | 60.0 |
| Smoking | ||
| Yes | 12 | 60.0 |
| No | 8 | 40.0 |
| Drinking | ||
| Yes | 6 | 30.0 |
| No | 14 | 70.0 |
| Pathologic Type | ||
| Adenocarcinoma | 8 | 40.0 |
| Squamous Cell Carcinoma | 12 | 60.0 |
| Size of Tumor (cm) | ||
| > 5 | 16 | 80.0 |
| ≤ 5 | 4 | 20.0 |
| Lymphatic metastasis | ||
| Yes | 12 | 60.0 |
| No | 8 | 40.0 |
| TNM Staging | ||
| Ⅰ & Ⅱ | 11 | 55.0 |
| Ⅲ & Ⅳ | 9 | 45.0 |
Association between AC138128.1 expression and cisplatin sensitivity
| Position | AC138128.1(1×103) Median (Q1- Q3) | IC50 (μg/ml) Median (Q1- Q3) | rs valuea | P value |
|---|---|---|---|---|
| Cancer | 1.133 (0.253- 8.091) | 42.14 (16.40- 106.10) | 0.493 | 0.027 |
| Para-Cancer | 8.102 (1.661- 4.113) | 254.99 (37.99- 1351.60) | -0.179 | 0.450 |
a: Spearman's rank correlation
The ROC curve of AC138128.1. Data based on the expression of AC138128.1 in cancer and its adjacent para-cancer tissues. The area under the ROC curve (AUC) is 0.852 (P< 0.001).
Association of AC138128.1 with ERCC1 expression
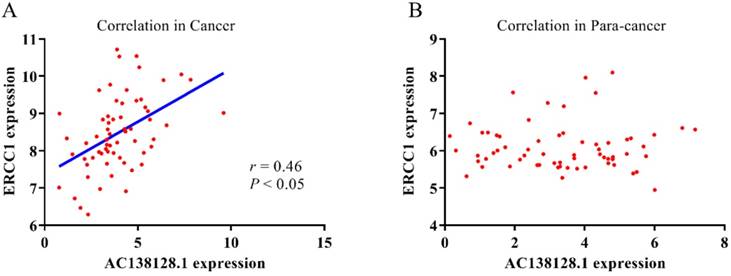
Because AC138128.1 is an intronic lncRNA mostly present in the nucleus, it may be involved in the transcriptional regulation of its neighboring genes. We analyze the relationship between the expression of AC138128.1 and ERCC1 gene (Figure 9 A & B). In cancer tissues, AC138128.1 and ERCC1 show a strong correlation (r = 0.46, P < 0.05). However, there is no significant relationship between these two genes in para-cancer tissues.
The relative expression of AC1381281 in 16HEB, A549 and LK2 cell. Using 16HBE as a reference, AC138128.1 expression in A549 and LK2 cells was significantly reduced. *: P< 0.05, **: P< 0.01.
A, 16HBE, A549 and LK2 cell treated with 4mg/L cisplatin for 12h, 24h and 48h respectively and then detected the expression of AC138128.1 by qRT-PCR. Control cells were also treated with cisplatin for 0h. B, 16HBE, A549 and LK2 cell treated with 2, 4 and 8mg/L cisplatin for 24h respectively and then detected the expression of AC138128.1 by qRT-PCR. Control cells were treated with phosphate buffer saline for 24h.
The subcellular distribution of AC138128.1. A. The subcellular distribution of AC138128.1 in 16HBE cell. B. The subcellular distribution of AC138128.1 in A549 cell. U6, a shRNA, exists in nucleus mainly. MT-RNR1, mitochondrial genome-encoded gene, locates in cytoplasm mainly. And the GAPDH exists in both nucleus and cytoplasm, but much more in cytoplasm. As for AC138128.1 mostly present in nucleus.
Discussion
ERCC1 is a key endonuclease in the nucleotide excision repair system for maintaining genome stability, which mediates DNA- and protein-protein interactions to play an important role in DNA damage induced by genotoxic agents. As we have known, ERCC1 has been extensively studied to evaluate the sensitivity of cells to platinum analogue. Over the past two decades, ERCC1 protein level measured by immunohistochemistry (IHC) appears to be a predictive marker of platinum-based chemotherapy. However, till now no clinical study can validate the interests of ERCC1 as a valid biomarker. Actually, ERCC1 has a variety of isoforms, and not all these isomers have DNA repair capabilities. Several studies have found the problems such as the reliability of ERCC1 antibodies and the presence of non-functional ERCC1 isoforms. As we have known the ERCC1-202 isoform was considered to be the only one to show the function of DNA repair [24, 25]. Therefore, IHC analysis with the use of currently available ERCC1 antibodies do not specifically detect the unique functional ERCC1 isoform [8]. Moreover, it is reported that both ERCC1 expression and polymorphisms are not enough to evaluate the sensitivity of cells to platinum [26]. Additionally, some researches on the 3' UTR function and post-transcriptional regulation of ERCC1 have also been concerned in recent years [14, 27]. The previous study also found that the efficiency of repairing DNA adduct was affected by a polymorphism located in ERCC1 3'UTR and ERCC1 was mainly regulated by the post-transcriptional regulation of miRNA [10, 28]. So it is necessary to explore some certain non-coding RNAs related to the post-transcriptional regulation of ERCC1 as a more sensitive or unique biomarker.
LncRNA AC138128.1, located in the longest intron of ERCC1, is found to have a lower expression in gastric cancer and hepatocellular carcinoma [20, 21]. However, whether AC138128.1 is associated with lung cancer is still uncertain; we attempted to design a study to clarify the linkage. Consistently with the previous report in gastric cancer and hepatocellular carcinoma, AC138128.1 level in lung cancer tissues was much lower than their para-cancer tissue. Interestingly, the ROC curve supports the value of the lncRNA AC138128.1 to distinguish AC138128.1 expression in cancer and para-cancer tissues (area under the curve > 0.8), which further confirmed its lower expression in lung cancer tissues. Additionally, AC138128.1 expression in 16HBE, A549, and LK2 cells was also detected. Similar to those in lung cancer tissue, AC138128.1 expression in 16HBE, a normal bronchial epithelial cell line was higher than that in the other lung cancer cell lines.
The relationship between AC138128.1 with ERCC1 expression. The correlation between AC138128.1 expression and ERCC1 expression in cancer tissues (A) (cancer, r = 0.46, P<0.05.) and in para-cancer tissues (B).
As known ERCC1 expression in tumor cells can determine the resistance to platinum drugs [29]. Due to AC138128.1 located in in the genome of ERCC1 genes, we wonder if AC138128.1 plays an important role in DNA damage caused by genotoxic substances such as cisplatin, like the ERCC1 gene. In the present study, the 3 cell lines including 16HBE, A549, and LK2 were treated by cisplatin. As we expected, the expression of AC138128.1 increased in both time-dependent and dose-dependent manners, which reminded AC138128.1 was sensitive to genotoxic agents as cisplatin. It suggested that AC138128.1 might response rapidly to the stress of DNA damage after cisplatin treatment due to its defective translation step. So, we speculated that comparing with ERCC1, AC138128.1 may become a more sensitive and unique biomarker to predict the benefit of platinum treatment.
The subcellular localization of a protein can usually remind its possible function, for example, nuclear factor-κB plays a role in transcriptional regulation in the nucleus. Similarly, the subcellular localization of a lncRNA can also suggest its function. ANRIL, an lncRNA in the nucleus, can interact with PRC2 to mediate trimethylation of lysine 27 on histone 3 and then inhibit transcription at the initial step [30]. Till now many lncRNAs were found to function in the nucleus as previously reported [31, 32]. We thereupon examined the expression of AC138128.1 in the nucleus and cytoplasm of 16HBE and A549 cells. Surprisingly, AC138128.1 expression was mainly present in the nucleus, which reminded it can play a role in transcriptional regulation. Additionally, AC138128.1 is an intronic lncRNA and may function in cis, meaning that it may regulate its adjacent genes. Therefore, the expression of AC138128.1 and ERCC1 were detected. And then their correlation at the mRNA level in cancer tissues but not in para-cancer tissues was found. Some reports and our previous study confirmed that numerous alternative splicing events occur in ERCC1, which means different spliceosomes of ERCC1 expressed differently in cancer and para-cancer tissues [22, 33, 34]. The difference of the association between AC138128.1 and ERCC1 expression in cancer and para-cancer tissues may be due to the changes in the alternative splicing isoform of ERCC1 expression in cancer and para-cancer. However, deep explorations about the effect of ERCC1 splicing isoform on AC138128.1 function need to be clarified in the future.
In conclusion, our present data showed that AC138128.1, an intronic lncRNA originated from ERCC1, may cooperate with ERCC1 and have a unique function, and become a potential biomarker to predict platinum treatment of lung cancer. Although as a preliminary work of a battery of profound studies a further functional research of AC138128.1 is necessary, our current study may contribute to understand the function of AC138128.1 and the regulation of ERCC1 gene in the occurrence and prognosis of lung cancer.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81773470) and National Natural Science Foundation of Liaoning Province (20170540991).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X. et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555-e67
2. Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435-51
3. Ceppi M, Munnia A, Cellai F, Bruzzone M, Peluso MEM. Linking the generation of DNA adducts to lung cancer. Toxicology. 2017;390:160-6
4. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-58
5. Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG. et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811-22
6. Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1-9
7. Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol. 2012;9:144-55
8. Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S. et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med. 2013;368:1101-10
9. Postel-Vinay S, Soria JC. ERCC1 as Predictor of Platinum Benefit in Non-Small-Cell Lung Cancer. J Clin Oncol. 2017;35:384-6
10. Yu T, Liu Y, Lu X, Xiao S, Cai Y, Jin C. et al. Excision repair of BPDE-adducts in human lymphocytes: diminished capacity associated with ERCC1 C8092A (rs3212986) polymorphism. Arch Toxicol. 2013;87:699-709
11. Friboulet L, Barrios-Gonzales D, Commo F, Olaussen KA, Vagner S, Adam J. et al. Molecular Characteristics of ERCC1-Negative versus ERCC1-Positive Tumors in Resected NSCLC. Clin Cancer Res. 2011;17:5562-72
12. Rosell R, Cuello M, Cecere F, Santarpia M, Reguart N, Felip E. et al. Treatment of non-small-cell lung cancer and pharmacogenomics: where we are and where we are going. Curr Opin Oncol. 2006;18:135-43
13. Smid D, Kulda V, Srbecka K, Kubackova D, Dolezal J, Daum O. et al. Tissue microRNAs as predictive markers for gastric cancer patients undergoing palliative chemotherapy. Int J Oncol. 2016;48:2693-703
14. Zhao L, Ren Y, Tang H, Wang W, He Q, Sun J. et al. Deregulation of the miR-222-ABCG2 regulatory module in tongue squamous cell carcinoma contributes to chemoresistance and enhanced migratory/invasive potential. Oncotarget. 2015;6:44538-50
15. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775-89
16. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y. et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018;33:706-20 e9
17. St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239-51
18. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
19. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62
20. Chen X, Sun J, Song Y, Gao P, Zhao J, Huang X. et al. The novel long noncoding RNA AC138128.1 may be a predictive biomarker in gastric cancer. Med Oncol. 2014;31:262
21. Liu WT, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL. et al. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014;20:6314-21
22. Zhang G, Xue P, Cui S, Yu T, Xiao M, Zhang Q. et al. Different splicing isoforms of ERCC1 affect the expression of its overlapping genes CD3EAP and PPP1R13L, and indicate a potential application in non-small cell lung cancer treatment. Int J Oncol. 2018;52:2155-65
23. Wei Y, Li L, Wang D, Zhang CY, Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem. 2014;289:10270-5
24. Wang X, Zhu X, Zhang H, Fan X, Xue X, Chen Y. et al. ERCC1_202 Is A Prognostic Biomarker in Advanced Stage Non-Small Cell Lung Cancer Patients Treated with Platinum-Based Chemotherapy. J Cancer. 2017;8:2846-53
25. Friboulet L, Postel-Vinay S, Sourisseau T, Adam J, Stoclin A, Ponsonnailles F. et al. ERCC1 function in nuclear excision and interstrand crosslink repair pathways is mediated exclusively by the ERCC1-202 isoform. Cell Cycle. 2013;12:3298-306
26. Jokic M, Vlasic I, Rinneburger M, Klumper N, Spiro J, Vogel W. et al. Ercc1 Deficiency Promotes Tumorigenesis and Increases Cisplatin Sensitivity in a Tp53 Context-Specific Manner. Mol Cancer Res. 2016;14:1110-23
27. Panou V, Vyberg M, Weinreich UM, Meristoudis C, Falkmer UG, Roe OD. The established and future biomarkers of malignant pleural mesothelioma. Cancer Treat Rev. 2015;41:486-95
28. Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG. et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207-20
29. Westerveld A, Hoeijmakers JH, van Duin M, de Wit J, Odijk H, Pastink A. et al. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425-9
30. Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662-74
31. Arnes L, Liu Z, Wang J, Carlo Maurer H, Sagalovskiy I, Sanchez-Martin M. et al. Comprehensive characterisation of compartment-specific long non-coding RNAs associated with pancreatic ductal adenocarcinoma. Gut. 2018
32. Miao Y, Ajami NE, Huang TS, Lin FM, Lou CH, Wang YT. et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun. 2018;9:292
33. Kuo MS, Adam J, Dorvault N, Robin A, Friboulet L, Soria JC. et al. A novel antibody-based approach to detect the functional ERCC1-202 isoform. DNA Repair (Amst). 2018;64:34-44
34. Sun Y, Li T, Ma K, Tian Z, Zhu Y, Chen F. et al. The impacts of ERCC1 gene exon VIII alternative splicing on cisplatin-resistance in ovarian cancer cells. Cancer Invest. 2009;27:891-7
Author contact
![]() Corresponding author: Dr. Xiaobo Lu, Dept. of Toxicology, School of Public Health, China Medical University, No.77 Puhe Road, Shenyang North New Area, 110122, Shenyang, Liaoning Province, P. R. China. Tel: +86 024 31939077; E-mail: xbluedu.cn
Corresponding author: Dr. Xiaobo Lu, Dept. of Toxicology, School of Public Health, China Medical University, No.77 Puhe Road, Shenyang North New Area, 110122, Shenyang, Liaoning Province, P. R. China. Tel: +86 024 31939077; E-mail: xbluedu.cn

 Global reach, higher impact
Global reach, higher impact