Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3698-3705. doi:10.7150/jca.32680 This issue Cite
Research Paper
RMP promotes the proliferation and radioresistance of esophageal carcinoma
1. Department of Cell Biology, School of Medicine, Soochow University, Suzhou, Jiangsu, China
2. Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
3. Department of Cardiothoracic surgery, Suzhou Science & Technology Town Hospital, Suzhou, Jiangsu, China
* These authors contributed equally to this work.
Received 2018-12-30; Accepted 2019-5-16; Published 2019-6-9
Abstract
RMP is a RNA polymerase II Subunit RPB-5 associated protein shown to act as an oncogene in several cancer. However, the mechanism of the involvement of RMP in esophageal cancer (EC) remains unclear. We analyzed RMP expression in EC cell lines and EC tissues. The connection between RMP and clinical pathological features of EC was also elucidated. To investigate the role of RMP in EC, We performed CCK-8 assay to evaluate cell proliferation, and Annexin V/PI double-staining to evaluate cell apoptosis. Effect of RMP on tumor progression in nude mouse models was assessed by measurement of volume and weight of tumors. Expression of RMP, CEA and CA199 in vivo were measured by Inmunohistochemical staining. First of all, our study showed that RMP was highly expressed in EC cell lines (compared with normal cells) and tumor tissues (compare with corresponding normal tissues). Then, we found that RMP was bound up with the status of nodal and T stage which indicating that RMP may be related to the growth and malignant degree of EC. Moreover upregulation of RMP could contribute to tumor growth in vitro and vivo. In addition, the results also showed that overexpression of RMP could significantly reduce the susceptibility to radiotherapy. Taken together, all these further suggested that RMP would play a chance-promoting in EC which may provide us a powerful goal for gene targeting treatment of esophageal cancer.
Keywords: RMP, EC, tumor growth, radiotherapy, gene targeting treatment
Introduction
Esophageal cancer is one of the most common malignancies worldwide (1). It kills about 386,000 people each year (2). There are two main subtypes of esophageal cancer, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), with ESCC being the most frequent type of esophageal malignancy (3). Despite the rapid advancement in diagnosis and therapy for EC, the average 5-year overall survival has remained at 10-20% because of the proliferation and invasion of cancer cells (4). Therefore, to improve the survival rate and the life quality of EC patients, it is urgent for us to have a good understanding of the molecular mechanisms underlying the development of EC.
RMP is a RPB5-associated protein. The RMP gene was first isolated and cloned from a human HepG2 cDNA library more than a decade ago (5). The unconventional prefoldin RPB5 interactor (URI), an alternative form of RMP, was shown to participate in a nutrient-related signaling pathway that is required for gene expression (6). More recently, URI has been shown to interact with the tumor suppressor protein parafibromin, a component of the PAF1 complex (7). URI-1 is the RMP homolog in Caenorhabditis elegans. It has been demonstrated that URI-1 is required to maintain genome stability by playing an important function in controlling cell cycle (8). Uri, the RMP homolog in Drosophila, has also been shown to be required for normal development by playing essential roles in transcriptional regulation and genome integrity maintenance (9). Dr Wei team has shown previously that RMP associates with RPB5, suppressing transcriptional activation via HBx. In addition, they have shown that the overexpression of HBx releases the inhibitory effect of RMP on transcriptional activation (10, 11). RMP/URI was amplified and overexpressed in tissues and cell lines of human ovarian carcinomas (12). Moreover, RMP has shown to be an oncogene in cervical cancer, endometrioid adenocarcinoma and multiple myeloma (13-15).
In our study, we have detected that RMP was highly expressed in EC tissues and cell lines. Furthermore, we found that RMP was related to the growth and malignant degree of esophageal cancer and overexpression of RMP could reduce the susceptibilities of EC cells to irradiation. In addition, we have shown that promote tumor growth in vitro and vivo. Therefore, our study indicates that targeting RMP could be a promising treatment for EC patients.
Materials and methods
Cell culture
Three EC cell lines (ECa-109, TE-1 and EC-9706), and a normal human esophageal epithelial cell line (HEEC) were maintained in DMEM containing 10% fetal bovine serum (Invitrogen, USA ) at 37°C with 95% air and 5% CO2.
Human tissue specimens
A total of 96 pairs of EC and adjacent non-tumor tissues were collected by the Department of Cardiothoracic Surgery, the First Affiliated Hospital of Soochow University between the year of 2011 and 2015. The tissues were immediately snap-frozen in liquid nitrogen. The patients had not been pretreated with chemotherapy or radiotherapy prior to surgery without other additional criteria. Clinicopathological data were obtained by reviewing their pathology records. Both tumor and corresponding normal tissues were histologically examined by pathologists. The distances between the corresponding adjacent normal tissues and EC lesions were at least 3-4 cm. The use of the tissues for all assays was obtained with informed consent and this project was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University.
RNA extraction and quantitative Real-time PCR
Total RNA was isolated from the cell lines and human tissues with the Trizol (Invitrogen, USA). Using a RMP specificity qRT-PCR detection kit (Stratagene, USA), qRT-PCR assays were performed on an ABI 7500 fast real-time PCR system (Applied, Biosystems) according to the manufacturer's instructions. The experiment was repeated three times. Primers for RMP were 5'-TCCGAATAAATACTGGAAAG-3' and 5'-AAGGCTCTGTAAATGTCTGC-3'. Primers for Bax were 5'-TTTTGCTTCAGGGTTTCATC-3' and 5'-GACACTCGCTCAGCTTCT TG -3'. Primers for Bcl-2 were 5'- GGT GGG AGGGAG GAAGAA-3' and 5'- CGC AGA GGCATCACATCG -3'. Primers for GAPDH were 5'-GAC CTG ACC TGC CGT CTA-3' and 5'-AGG AGTGGGTGTCGCTGT-3'.
For Real-Time PCR data analysis, cycle threshold (Ct) difference (ΔCt) was used for making comparisons between samples. The relative mRNA expression (FAM196B/ GAPDH) was determined by the 2 -ΔΔCt method.
Plasmid construction
All the plasmids were obtained from the laboratory of the Department of Cell Biology at Soochow University. The plasmids pGPU6/Neo-SCR, pGPU6/Neo-RMPi for RMP depletion and pFlagCMV4-RMP for RMP overexpression were constructed as described previously (16).
Cells transfection
Cells were divided into four groups according to the treatment and named as follows: Unt group (untreated), Vector group (transfected with pGPU6/Neo-SCR), RMPo group (transfected with pFlagCMV4-RMP) and RMPi group (transfected with pGPU6/Neo-RMPi). Plasmids (3-4 µg) were transfected into cells at 37˚C for 48h using Opti-MEM (Gibco, USA) and Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer's instructions. The cells (1x106 cells/ml) were fixed in 4% paraformaldehyde for 10 min at room temperature. The transfection efficiency was visualized by using an inverted fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany) at magnification, x200. The protein expression of ARTS was confirmed by western blotting.
Western blot
Total protein was extracted from cultured cells by using RIPA Lysis Buffer (Beyotime, Shanghai, China). The protein concentration was determined through the BCA Protein Assay Kit (Bio-Rad, USA). Equal doses protein from each sample was applied to SDS-PAGE gels and then transferred onto PVDF membranes. After blocking with 5% non-fat milk in TBST for 2h, the membranes were incubated overnight at 4°C with diluted primary antibody. Followed by washing with TBST solution three times, the membranes were incubated with the secondary antibody at room temperature (RT) while shaking on a rotary for 2 h. The signals were visualized using ECL reagents (Pierce). The primary antibodies included SNX1 (1:400, SantaCruz, USA) andβ-actin (1:1000, SantaCruz, USA). β-actin was used as an internal control.
Annexin V/PI double-staining assay
Apoptosis was analyzed by flow cytometry using Annexin V/PI double staining. Firstly ECa-109 or TE-1 cells were seeded in six-well plates. After transfected for 24h, cells of Unt group, Vector group, RMPo group and RMPi group were cultured in the presence of irradiation (6Gy) respectively for 48h. All the cells were collected and washed twice with cold PBS, and the cells were resuspended in 100ul of binding buffer and incubated with Annexin v/PI double staining solution (Sigma-Aldrich, USA) at 37°C in the dark for 15 min. The number of apoptotic and necrotic cells were calculated by flow cytometry (Becton-Dickinson Co, NJ, USA) and analyzed by Cell Quest Software.
Cell proliferation assay and tumor formation assay in nude mouse models
For analysis of cell proliferation, cells were seeded into 96-well plates at 2x103 cells per well. Cells were incubated with Cell Counting Kit-8 (CCK8, Dojindo, Japan) according to the manufacturer's protocol. The proliferation rate was measured at 0, 24, 48, and 72h after transfection. Absorbance was then determined at 450nm using a microplate reader (Bio-Rad, USA). Five-week BALB/c nude mice were purchased from Experimental Animal Center of Soochow University, and all animal protocols were approved by the Institutional Animal Care and Treatment Committee of Soochow University. Tumor cells in the logarithmic phase were collected, washed twice with PBS, and resuspended at a concentration of 2x107cells per milliliter of PBS. Then 0.1 ml suspending cells were subcutaneously injected into the armpit of each female nude mouse. The tumor volumes were examined every two days. After two weeks, animals were sacrificed and solid tumor tissues were removed and weighed.
Immunohistochemistry detection
Tissues from nude mice were subjected to formalin fixation, paraffin embedding and sectioning for immunohistochemistry assays as describe previously, The slides were blocked for 30 min in PBST containing 3% BSA (PBST-BSA) at 37°C and incubated overnight at 4°C with mouse monoclonal antibodies (1:500 in PBST-BSA). Slides were then washed 3 times in PBST (10 min at RT) and incubated for 2 h with the secondary antibody (anti-mouse HRP, 1:200) in PBST at RT. After incubation, slides were washed three times (10 min each) in PBST and mounted with 3 μl of Vectashield for further analysis. Sections were examined at high power (x 400) under a standard light microscope. Cell staining was regard as positive if nuclear was homogeneous stained or 10% or more of cytoplasm was heterogeneously stained.
Statistical analysis
All data are presented as the mean values±standard deviations (mean±SD). Statistical analysis was performed using the SPSS software(version 21.0). Differences were evaluated using Student's t test. A value of P<0.05 was considered statistically significant.
Results
RMP expression is elevated in cancer cells and EC tissues
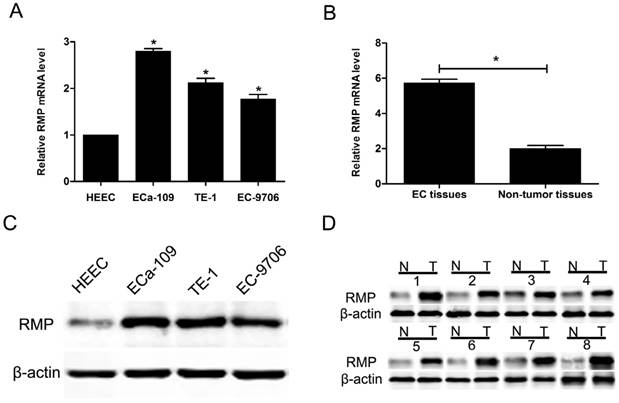
The expression of RMP in three EC cell lines (ECa-109, TE-1 and EC-9706) and a normal human esophageal epithelial cell line (HEEC) was analyzed by qRT-PCR. The results showed that RMP mRNA levels in three EC cell lines were higher than that in normal cells (Figure 1A). Then, we detected the expression of RMP mRNA in 96 EC tissues and adjacent non-tumor tissues. RMP mRNA was significantly higher expressed in EC tissues compared with corresponding normal tissues (Figure 1B). Then we performed western blot to analyze RMP protein levels in four different cell lines, EC tissues and corresponding non-tumor tissues. The results showed that RMP protein expression was significantly increased three EC cell lines compared with the normal cells (Figure 1C). Analogously, the expression of RMP protein in EC tissues from 96 patients was much higher than that in the matched non-tumor tissues (Figure 1D). Therefore, RMP expression is elevated in cancer cells and EC tissues.
The connection between RMP and clinical pathological features of EC
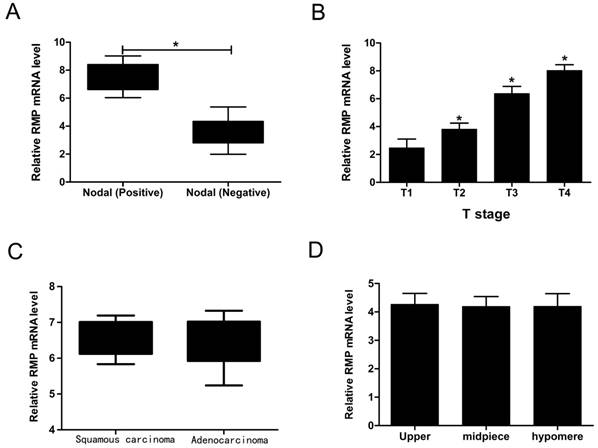
We have implied that RMP was increased both in mRNA level and protein level. In order to explore the connection between RMP and clinical pathological features of EC, we summarized the patients' detail information as shown in Table 1. Then we performed qRT-PCR to study whether the EC's four clinical characteristics containing nodal status, T stage, tumor types and tumor location were relevant to RMP. Interestingly, the RMP mRNA level was up-regulated in Nodal (Positive) compared with in Nodal (Negative) (Figure 2A). What's more, the RMP mRNA level was increased along with the T stage rose (Figure 2B). In contrast, RMP mRNA levels were no distinction in tumor types as well as in tumor locations (Figure 2C and D). So these data suggested that RMP was bound up with the status of nodal and T stage. In addition, it was also indicated that RMP may be related to the growth and malignant degree of EC.
RMP promotes EC cell growth in vitro
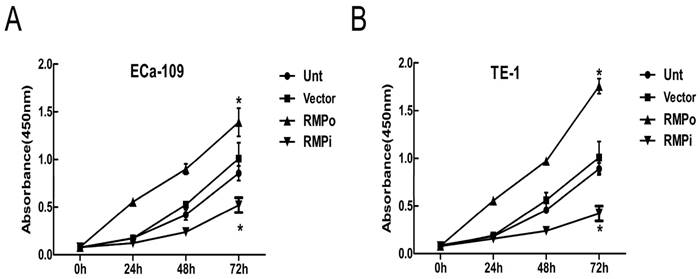
To explore the effects of RMP on EC cells, we performed CCK-8 assays to further analyze the effect of RMP on cell proliferation. The effect of RMP on the cell proliferation was examined in ECa-109 and TE-1. Obviously, the growth of cancer cells was promoted by the over-expression of RMP (RMPo), while depletion of RMP by RNA interference (RMPi) distinctly reduced the proliferation of cancer cells, compared with the cells untransfected (Unt) or transfected with vector alone (Vector) (Figure 3A and B). Therefore, the results suggested that RMP could promote EC cell growth in vitro.
RMP expression is elevated in cancer cells and EC tissues. (A) RMP mRNA levels in three EC cell lines (ECa-109, TE-1 and EC-9706) and normal cell line (HEEC); (B) RMP mRNA levels in EC tissues and the matched non-tumor tissues; (C) RMP protein levels in three EC cell lines (ECa-109, TE-1 and EC-9706) and normal cell line (HEEC). (D) RMP protein levels in EC tissues and the matched non-tumor tissues.
The connection between RMP and clinical pathological features of EC. (A) RMP mRNA levels in positive nodal and negative nodal. (B) RMP mRNA levels with the T stage rising. (C) RMP mRNA levels in different tumor types. (D) RMP mRNA levels in different tumor locations.
The clinical information of 96 EC patients.
| Clinical characteristics | EC(n=96) |
|---|---|
| Gender | |
| Male | 52 |
| Female | 44 |
| Age ate diagnosis | |
| Mean±SD | 61±14 |
| Median (range) | 57(38-84) |
| T-stage | |
| T1 | 19 |
| T2 | 39 |
| T3 | 21 |
| T4 | 17 |
| Nodal status | |
| positive | 50 |
| negative | 46 |
| Tumor types | |
| Squamous carcinoma | 62 |
| Adenocarcinoma | 34 |
| Tumor location | |
| Upper | 18 |
| Midpiece | 43 |
| Hypomere | 35 |
Upregulation of RMP enhances the radioresistance of EC cells
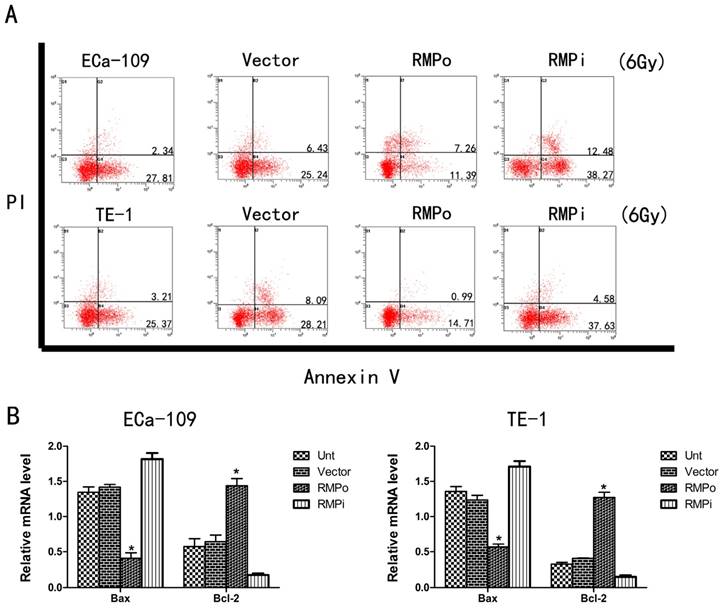
We further investigated the effect of RMP on the susceptibility of EC cells to radiotherapy. To study this phenomenon, we performed apoptosis assays to observe the apoptosis of EC cells that were treated with irradiation (6Gy). Compared with the Unt and Vector groups, RMPo group had a significantly decreased apoptosis rate whereas RMPi group increased the apoptosis rate dramatically (Figure 4A). Furthermore, we implemented qRT-PCR to explore the mRNA level of Bax (apoptosis related gene) and Bcl-2 (antiapoptosis related gene). As a result, overexpression of RMP could reduce the Bax mRNA level while increase the Bcl-2 mRNA level (Figure 4B). So these results indicated that RMP could promote radioresistance of EC cells through regulating the expression of apoptosis related genes and antiapoptosis related genes.
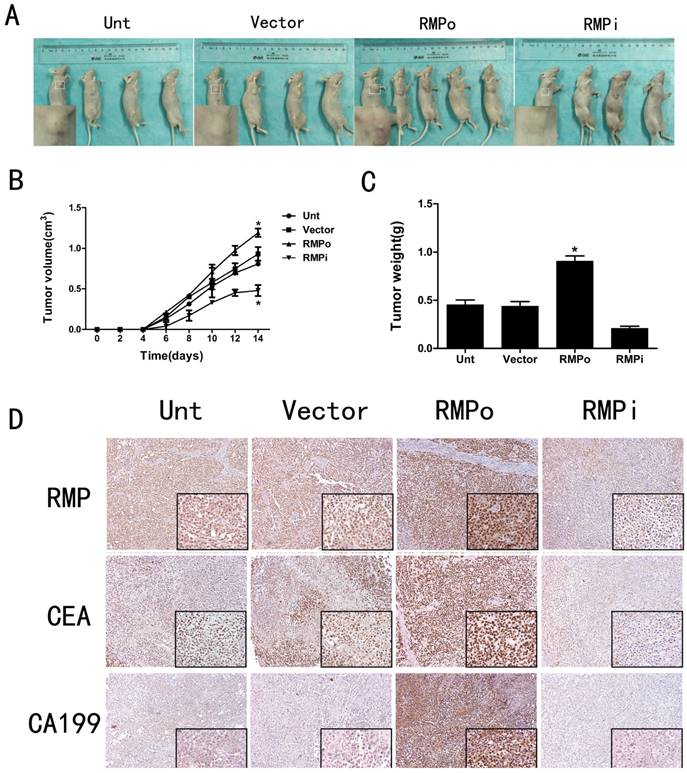
Effect of RMP on tumor progression in vivo
The CCK-8 assays have proved that RMP could promote cell proliferation in EC cells (Figure 3A and B). To further study the effects of RMP on tumor progression in vivo, we established four kinds of nude mouse models. The results showed that compared with the tumors in Unt or Vector groups, the volume and weight of tumors in RMPo group were obviously increased while distinctly decreased in RMPi group (Figure 5A, B and C). In addition, inmunohistochemical staining showed that the expression of RMP was enhanced in the xenograft tumors from RMPo cells, while decreased in tumors from RMPi cells, compared with control groups. The expression of EC marker CEA and CA199 were also improved in RMPo tumors, while decreased in RMPi tumors (Figure 5D). Thus, RMP might work as an oncogene of EC in vitro and vivo.
Discussion
Esophageal cancer is one of the common malignancies worldwide, especially in the East Asia, Africa and North America (16). ESCC is the most common clinical type of esophageal cancer in China (17). Limited clinical treatments, rare early-stage diagnosis and frequent recurrence lead to poor outcomes of EC (18). The 5-year survival rate of EC is only approximately 25% (19). As a result, a comprehensive understanding of the factors contributing to the development and progression of EC should aim to identify potential therapeutic targets in clinical management. RMP has been reported to serve as an oncogene in Hepatocellular Carcinoma (20), ovarian cancer and cervical cancer (12,13). However, the function of RMP in EC is still unknown. So it is urgent for us to clarify the molecular mechanisms underlying the development of EC.
RMP promotes EC cell growth in vitro by CCK-8 assays. (A) Cell growth of ECa-109 in Unt, Vector, RMPo and RMPi groups from 0h to 72h. (B) Cell growth of TE-1 in Unt, Vector, RMPo and RMPi groups from 0h to 72h.
In our study, the expression of RMP was much higher in EC tissues than in corresponding adjacent tissues. What's more, overexpression of RMP could reduce the susceptibility of EC cells to radiotherapy. At present, despite improvements in the management of esophageal cancer patients, the resistance of tumor to anticancer drugs such as cisplatin or irradiation remains a major obstacle in the non-surgical anticancer treatment of esophageal cancer (21-23). At the same time, understanding the molecular of the RMP associated with other genes is critical in using it as a therapeutic tool in EC. The integration of growth factor and nutrient signals is achieved through mammalian target of rapamycin (mTOR), which coordinately controls cell growth by modulating the activity of a series of effector proteins. Among these is S6 kinase 1 (S6K1), which promotes protein synthesis as well as cell survival (24,25). Functional analysis of URI in yeast revealed that it acts downstream of TOR to control nutrient-sensitive gene expression. In mammalian cells, the phosphorylation state of URI is influenced by signals that affect mTOR activity insofar as insulin or IGF1 treatment produces hyperphosphorylated URI, an effect blocked by rapamycin (6). The tight correlation between URI phosphorylation and mTOR activity implies a potential role for URI in mTOR signaling in mammalian cells. In its un(der)phosphorylated form, URI is bound to PP1γ and functions as its inhibitor (26-28). Dissociation of PP1γ, from bound URI is initiated by S6K1-mediated phosphorylation of URI at Ser-371, which when liberated, proceeds to dephosphorylate S6K1 and possibly BAD (29,30). Therefore, according to these theories, the relationships among RMP, mTOR, S6K1 and PPIγ are worth researching, which may make us understand the pathogenesis of EC well.
In conclusion, the effects of RMP on proliferation and radioresistance have been verified in the present study. As a result, a new biomarker in EC is helpful in the development of a better therapy strategy for EC patients.
Upregulation of RMP enhances the radioresistance of EC cells. (A) Cell apoptosis analysis of ECa-109 and TE-1 cells in Unt, Vector, RMPo and RMPi groups; (B) Expression of Bax mRNA and Bcl-2 mRNA in Unt, Vector, RMPo and RMPi groups through RT-qPCR analysis.
Effect of RMP on tumor progression in vivo. (A) Nude mouse models in Unt, Vector, RMPo and RMPi groups (B) Volume of tumors in Unt, Vector, RMPo and RMPi groups; (C) Weight of tumors in Unt, Vector, RMPo and RMPi groups; (D) Expression of RMP, CEA and CA199 in Unt, Vector, RMPo and RMPi groups by Inmunohistochemical staining.
Acknowledgements
This study was supported by project of Suzhou Science and Technology Development Plan (SYSD2015076), and the National Natural Science Foundation of China (No.81800279).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87-108
2. Yarhusseini A, Sharifzadeh L, Delpisheh A, Veisani Y, Sayehmiri F, Sayehmiri K. Survival rate of esophageal carcinoma in Iran - A systematic review and meta-analysis. Iran J Cancer Prev. 2014;7:61-65
3. Han B, Li W, Sun Y, Zhou L, Xu Y, Zhao X. A prolyl-hydroxylase inhibitor, ethyl-3, 4-dihydroxybenzoate, induces cell autophagy and apoptosis in esophageal squamous cell carcinoma cells via up-regulation of BNIP3 and N-myc downstream-regulated gene-1. PLoS One. 2014;9:e107204
4. Wang Z, Qiao Q, Chen M, Li X, Wang Z, Liu C. et al. miR-625 down-regulation promotes proliferation and invasion in esophageal cancer by targeting Sox2. FEBS Lett. 2014;588:915-921
5. Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N. et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol. 1998;18:7546-7555
6. Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M. et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2013;302:1208-1212
7. Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D. et al. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052-5060
8. Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. URI-1 is required for DNA stability in C. elegans. Development. 2006;33:621-629
9. Kirchner J, Vissi E, Gross S, Szoor B, Rudenko A, Alphey L. et al. Drosophila Uri, a PP1alpha binding protein, is essential for viability, maintenance of DNA integrity and normal transcriptional activity. BMC Mol Biol. 2008;9:36
10. Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N. et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol. 1998;18:7546-7555
11. Wei W, Gu JX, Zhu CQ, Sun FY, Dorjsuren D. et al. Interaction with general transcription factor IIF (TFIIF) is required for the suppression of activated transcription by RPB5-mediating protein (RMP). Cell Res. 2003;13:111-120
12. Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK. et al. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer cell. 2011;19:317-32
13. Gu J, Li X, Liang Y. et al. Upregulation of URI/RMP gene expression in cervical cancer by high-throughput tissue microarray analysis. Int J Clin Exp Pathol. 2013;6:669-677
14. Gu J, Liang Y, Qiao L. et al. Expression analysis of URI/RMP gene in endometrioid adenocarcinoma by tissue microarray immunohistochemistry. Int J Clin Exp Pathol. 2013;6:2396-2403
15. Fan JL, Zhang J, Dong LW. et al. URI regulates tumorigenicity and chemotherapeutic resistance of multiple myeloma by modulating IL-6 transcription. Cell Death Dis. 2014;13:5 e1126
16. Zhou W, Wang Q, Xu Y, Jiang J, Guo J, Yu H, Wei W. RMP promotes epithelial-mesenchymal transition through NF-κB/CSN2/Snail pathway in hepatocellular carcinoma. Oncotarget. 2017Jun20;8(25):40373-40388
17. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
18. Zeng H, Zheng R, Zhang S, Zuo T, Xia C, Zou X. et al. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thoracic Cancer. 2016;7:232-237
19. Allum WH, Blazeby JM, Griffin SM. et al. Guidelines for the management of oesophageal and gastric cancer. Gut. 2011;60:1449-1472
20. Herskovic A, Russell W, Liptay M, Fidler MJ, Al-Sarraf M. Esophageal carcinoma advances in treatment results for locally advanced disease: review. Ann Oncol. 2012;23(5):1095-1103
21. Yang S, Wang H, Guo Y. et al. RMP Plays Distinct Roles in the Proliferation of Hepatocellular Carcinoma Cells and Normal Hepatic Cells. Int J Biol Sci. 2013;9:637-648
22. Lindner K, Borchardt C, Schöpp M. et al. Proton pump inhibitors (PPIs) impact on tumour cell survival, metastatic potential and chemotherapy resistance,and affect expression of resistance-relevant miRNAs in esophageal cancer. J Exp Clin Cancer Res. 2014;33:73
23. Su H, Jin X, Zhang X. et al. Identification of microRNAs involved in the radioresistance of esophageal cancer cells. Cell Biol Int. 2014;38:318-25
24. Wang F, Xia J, Wang X. et al. miR-145 inhibits proliferation and invasion of esophageal squamous cell carcinoma in part by targeting c-Myc. Onkologie. 2013;36:754-8
25. Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729-734
26. Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471-484
27. Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R. et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell. 2007;28:28-40
28. Dhar R, Basu A. Constitutive activation of p70 S6 kinase is associated with intrinsic resistance to cisplatin. Int J Oncol. 2008;32:1133-1137
29. Freilinger A, Rosner M, Krupitza G, Nishino M, Lubec G, Korsmeyer SJ. et al. Tuberin activates the proapoptotic molecule BAD. Oncogene. 2006;25:6467-6479
30. Datta SR, Ranger AM, Lin MZ, Sturgill JF, Ma YC, Cowan CW. et al. Survival factormediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev Cell. 2002;3:631-643
31. Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. p70S6 kinase signals cell survival as well as growth, inactivating the proapoptotic molecule BAD. Proc Natl Acad Sci USA. 2001;98:9666-9670
Author contact
![]() Corresponding authors: Haitao Ma, Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215000, China. E-mail: mht7403com.
Corresponding authors: Haitao Ma, Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215000, China. E-mail: mht7403com.

 Global reach, higher impact
Global reach, higher impact