Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3728-3734. doi:10.7150/jca.32281 This issue Cite
Research Paper
Centromere Protein N Participates in Cellular Proliferation of Human Oral Cancer by Cell-Cycle Enhancement
1. Department of Oral Science, Graduate School of Medicine, Chiba University, Chiba, Japan
2. Department of Dentistry and Oral-Maxillofacial Surgery, Chiba University Hospital, Chiba, Japan
Received 2018-12-14; Accepted 2019-5-11; Published 2019-6-9
Abstract
Centromere protein N (CENP-N), an important member of the centromere protein family, is essential for kinetochore assembly and chromosome segregation; however, the relevance of CENP-N in cancers remains unknown. The aim of this study was to investigate CENP-N expression and its functional mechanisms in oral squamous cell carcinoma (OSCC). CENP-N expression was up-regulated significantly in vitro and in vivo in OSCCs. Overexpressed CENP-N was closely (p < 0.05) correlated with tumor growth using quantitative reverse transcriptase-polymerase chain reaction, immunoblot analysis, and immunohistochemistry. CENP-N knockdown (shCENP-N) cells showed depressed cellular proliferation by cell-cycle arrest at the G1 phase with up-regulation of p21Cip1 and p27Kip1 and down-regulation of cyclin D1, CDK2, and CDK4. Interestingly, we newly discovered that calcitriol (1, 25-dihydroxyvitamin D3) controlled the CENP-N expression level, leading to inhibition of tumor growth similar to shCENP-N cells. These results suggested that CENP-N plays a critical role in determining proliferation of OSCCs and that calcitriol might be a novel therapeutic drug for OSCCs by regulating CENP-N.
Keywords: Calcitriol, Cell-cycle arrest at G1 phase, Cellular proliferation, Centromere protein N, Oral squamous cell carcinoma
Introduction
A centromere, an intersection of the short and long arms of the chromosome, forms kinetochore on centromeric chromatin in mitosis. During cellular division, kinetochore proteins regulate DNA separation by spindle microtubules [1]. Therefore, the centromere is extremely important in cell-cycle regulation and chromosomal segregation [2]. Centromere proteins (CENPs), which comprise 18 subtypes, are related dynamical to association and dissociation during mitosis with microtubule regulation. Among the CENPs, CENP-A, a centromere-specific nucleosome, acts as a foundation for centromere assembly. CENP-A integrates centromeric chromatin instead of histone H3 for maintenance of centromeric chromatin [1,3].
CENP-N binds directly to the centromere-targeting domain of CENP-A [4,5]. CENP-N depletion causes down-regulation of several CENPs and is considered essential for making a new centromere [6]. Other functions of CENP-N, including in cancer research, are unclear, whereas centromere, mainly the CENP complex, is well elucidated.
The current study found that CENP-N is overexpressed in oral squamous cell carcinoma (OSCC) in vitro and in vivo. We also identified a novel drug for tumor growth suppression through CENP-N down-regulation.
Materials and methods
Ethics statement
The ethics committee of Chiba University approved the protocol of the current research project (protocol number, 680). All subjects provided written informed consent for inclusion in this study.
OSCC cells and OSCC tissue samples
We purchased 10 OSCC cell lines from RIKEN BioResource Center (Tsukuba, Ibaraki, Japan) and the Japanese Collection of Research Bioresources Cell Bank (Ibaraki, Osaka, Japan). We harvested human normal oral keratinocytes (HNOKs) from healthy patients as normal control cells and cultured them as described previously [7-9]. OSCC samples were resected at the Department of Oral-Maxillofacial Surgery, Chiba University Hospital. The pathologists at Department of Pathology of Chiba University Hospital diagnosed these samples as OSCC as mentioned previously [10-13].
Gene expression analysis
Quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) was performed as described previously [7,8]. The sequences of the gene-specific primers were as follows: CENP-N (5′-CAGACTCCGTACGCCTTCAC-3′ and 5′-GGTCCATTTTCACAATCTGATG-3′) and glyceraldehyde-3-phosphate dehydrogenase (5′-AGCCACATCGCTCAGACAC-3′ and 5′-GCCCAATACGACCAAATCC-3′) (Universal Probe-Library, Roche, Tokyo, Japan).
Immunoblot analysis
Protein extraction from OSCCs and HNOKs and immunoblot analysis were performed as described previously [13,14]. The antibodies were anti-CENP-N (rabbit, 16751-1-AP, Proteintech, Rosemont, IL, USA), anti-α-tubulin (mouse, sc-5286, Santa Cruz Biotechnology, Shanghai, China), anti-CDK2 (rabbit, #2546), anti-CDK4 (rabbit, #12790), anti-p21 Cip1 (rabbit, #2947), anti-p27Kip1 (rabbit, #3686), and anti-cyclin D1 (rabbit, #2978) (Cell Signaling Technology Japan, Tokyo, Japan).
Immunohistochemistry (IHC)
IHC of OSCCs and normal tissues (100 samples) was performed as described previously [13], with appropriate antibodies such as anti-CENP-N (Proteintech). The OSCCs and normal tissues were obtained from the primary OSCC patients from 2007 to 2016, excluding distant metastasis. The IHC scores were calculated from the staining intensity and proportion of the cells stained for CENP-N. Regarding the intensity of the stained cells, we classified four levels (0, 1, 2, 3) as described previously [14]. The median score of all OSCCs was 153.99. CENP-N was classified as positive and negative using the median score of all OSCCs as the cut-off points [15,16]. Cases with a score over the median score were considered CENP-N-positive.
Transfection
Transfection with shRNA vector was performed as described previously [10,11,17-19]. We transfected control shRNA (shMock) (sc-37007, SCB) and CENP-N shRNA (shCENP-N) (sc-76640, SCB) into Ca9-22 and SAS cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). RT-qPCR and immunoblot analysis were performed to investigate the efficiency of CENP-N knockdown.
Cellular proliferation
To investigate the effectiveness of CENP-N knockdown on cellular growth, we performed a cellular proliferation assay as mentioned previously [20-24]. For details, shCENP-N and shMock cells (Ca9-22 and SAS) were disseminated in six-well plates (1×104 viable cells/well). Every 24 h, the cells were trypsinized and counted in triplicate using a hemocytometer (up to 168 h).
Cell-cycle analysis
shCENP-N and shMock cells (Ca9-22 and SAS) were synchronized in the G2/M phase using 200 ng/ml nocodazole (Sigma-Aldrich Japan, Tokyo, Japan) for 16 h as reported previously [10,14,20-24]. After treatment, these cells were analyzed using the BD AccuriTM C6 Flow Cytometer (Becton-Dickinson, Tokyo, Japan) with the CycleTEST Plus DNA reagent kit (Becton-Dickinson).
Calcitriol treatment
To determine the optimal concentrations of calcitriol (1α, 25-dihydroxyvitamin D3) (Cayman Chemical, Ann Arbor, MI, USA) for CENP-N expression in OSCC cells, we performed RT-qPCR and immunoblot analysis after treatment with calcitriol. We then evaluated whether calcitriol affected the cellular proliferation and cell cycle by treating the OSCC cells with calcitriol for 72 h. In the cellular proliferation assay, the OSCC cells were cultured with calcitriol. Dimethylsulfoxide was used as the experimental control.
Statistical analysis
Fisher's exact test and Student's t-test were used to analyze the statistical differences. p < 0.05 indicates statistical significance. All data are expressed as the mean ± standard error of the mean.
Results
Increased CENP-N mRNA and protein expression levels in OSCCs
We evaluated the status of CENP-N expression by RT-qPCR and immunoblot analysis in 10 OSCC cell lines and HNOKs. Compared to the HNOKs, CENP-N mRNA expression status in the 10 OSCC cell lines was up-regulated significantly (Fig. 1A, p < 0.05). Immunoblot analysis showed that CENP-N protein increased in the 10 OSCC cell lines compared to the HNOKs (Fig. 2B).
Increased CENP-N mRNA and protein expression levels in OSCCs. (A) Significant (*p < 0.05, Student's t-test) up-regulation of CENP-N mRNA is seen in 10 OSCC cells compared with the HNOKs by RT-qPCR. The data are expressed as the mean ± standard error of the mean (SEM) from three assays. (B) The expression of CENP-N protein is up-regulated in OSCC cells compared with the HNOKs by immunoblot analysis. (C) Representative IHC results for CENP-N protein in normal oral tissue and primary OSCC tissue. Scale bars = 50 μm. (D) The status of CENP-N protein expression in primary OSCC (n = 100) and its normal counterparts by the IHC scoring system.
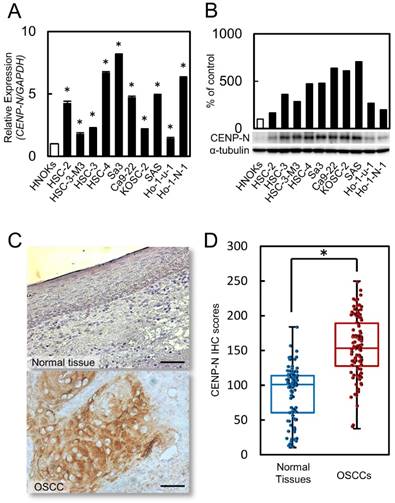
CENP-N expression status in clinical specimens of primary OSCCs and correlation in clinical classifications
We performed IHC to investigate the CENP-N expression in primary OSCCs. High expression of CENP-N was observed in the nucleus of the OSCC specimens. However, the normal oral mucosa was not stained by the CENP-N antibody. Fig. 1C shows representative CENP-N data in normal oral tissue and primary OSCC using IHC. These IHC scores in primary OSCCs were clearly greater in the mucosa than in normal oral tissues (Fig. 1D, p < 0.05). The scores in the normal oral tissues ranged from 10.4 to 184 (median, 101.1) and in primary OSCC tissues from 37.6 to 249.8 (median, 153.9). Table 1 shows the correlation between the CENP-N protein expression status and clinicopathological classification of the patients with OSCC. Among the clinical classifications, the CENP-N expression level was correlated significantly (p < 0.05) with the primary tumor size (Table 1).
Correlation between CENP-N expression and clinical classification in OSCCs.
| Clinical classification | Results of immunostaining No. patients | |||
|---|---|---|---|---|
| Total | CENP-N negative | CENP-N positive | p-value | |
| Age at surgery (years) | ||||
| < 70 | 59 | 28 | 31 | 0.685* |
| ≧70 | 41 | 22 | 19 | |
| Gender | ||||
| Male | 53 | 30 | 23 | 0.229* |
| Female | 47 | 20 | 27 | |
| T-primary tumor size | ||||
| T1+T2 | 57 | 35 | 22 | 0.015*† |
| T3+T4 | 43 | 15 | 28 | |
| N-regional lymph node metastasis | ||||
| Negative | 61 | 32 | 29 | 0.682* |
| Positive | 39 | 18 | 21 | |
| TNM stage | ||||
| I+ II | 43 | 25 | 18 | 0.225* |
| III+ IV | 57 | 25 | 32 | |
| Vascular invasion | ||||
| Negative | 64 | 36 | 28 | 0.144* |
| Positive | 36 | 14 | 22 | |
| Histopathologic type | ||||
| Well | 58 | 32 | 26 | 0.311* |
| Moderately+ poorly | 42 | 18 | 24 | |
*Fisher's exact test. †p < 0.05.
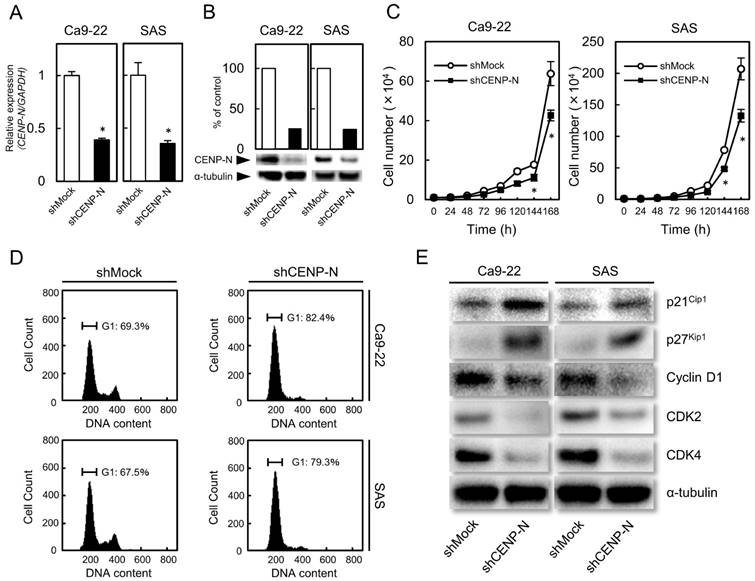
shRNA knockdown cells of CENP-N in OSCC
Since overexpression of CENP-N was observed in OSCCs (Fig. 1), we established the shCENP-N and shMock cells from the OSCC cell lines (Ca9-22 and SAS). To investigate the efficiency of the transfection, we performed RT-qPCR and immunoblot analysis. The CENP-N mRNA expression in the shCENP-N cells was significantly (p < 0.05) lower than that in the shMock cells (Fig. 2A). The CENP-N protein levels in the shCENP-N cells also decreased compared with the shMock cells (Fig. 2B).
Cellular growth of shCENP-N and shMock cells
To evaluate the effect of the shCENP-N on cellular growth, a cellular proliferation assay was performed. Compared with the shMock cells, we found a significant (p < 0.05) decrease of cellular proliferation in the shCENP-N cells (Fig. 2C).
Cell-cycle analysis of shCENP-N and shMock cells
Flow cytometric analysis showed that the proportion of the G1 phase in the shCENP-N cells was clearly (p < 0.05) higher than that in the shMock cells (Fig. 2D). We also evaluated the protein levels of p21Cip1, p27Kip1, cyclin D1, CDK2, and CDK4. As expected, p21Cip1 and p27Kip1 protein levels were up-regulated, and cyclin D1, CDK2, and CDK4 protein levels were down-regulated in the shCENP-N (Fig. 2E). These data indicated that the shCENP-N inhibited cellular proliferation by cell-cycle arrest at the G1 phase.
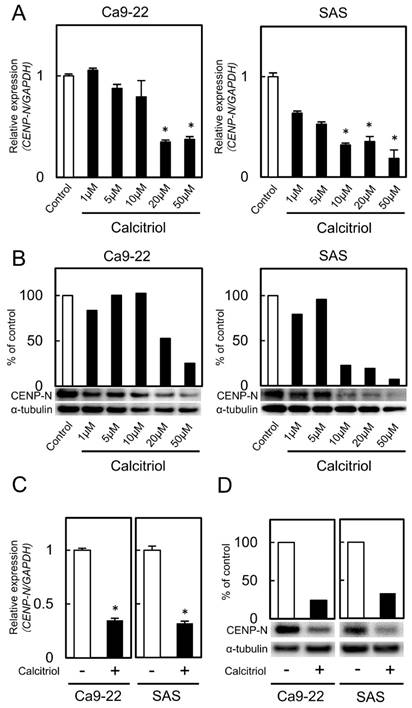
Effect of calcitriol treatment
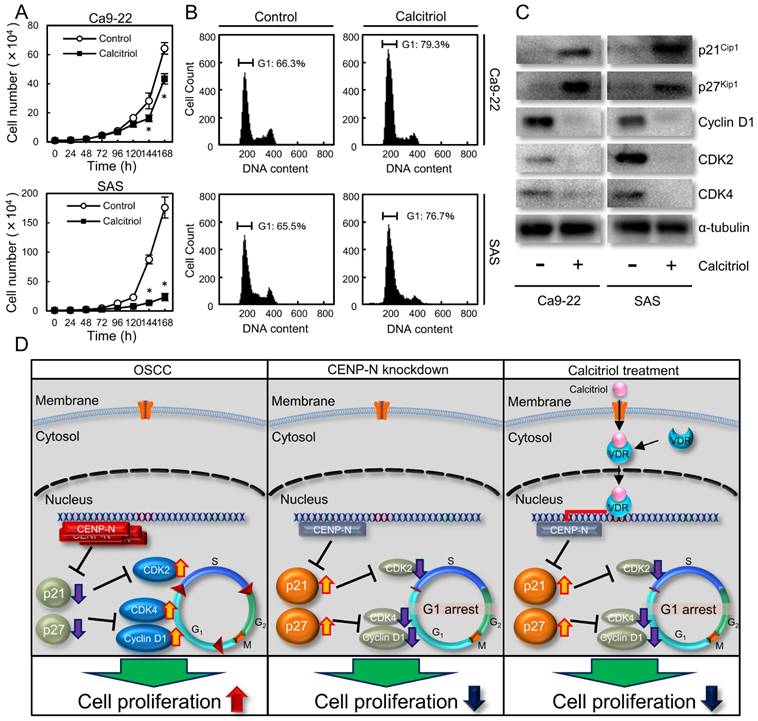
We treated the cells with calcitriol to determine the optimal concentrations for inhibiting CENP-N expression. Calcitriol inhibited CENP-N mRNA and protein expression levels in a dose-dependent manner (Fig. 3A, B; optimal concentrations, 20 μM with the Ca9-22 cell line and 10 μM with the SAS cell line). CENP-N mRNA and protein levels in the cells treated with an optimal concentration of calcitriol decreased significantly (p < 0.05) compared with control cells (Fig. 3C, D). To assess the effect of calcitriol on cellular growth, we performed a cellular proliferation assay. The cellular growth of the calcitriol-treated cells was significantly (p < 0.05) lower than that of control cells (Fig. 4A). In the cell-cycle analysis, the percentage of the G1 phase in the calcitriol-treated cells was clearly (p < 0.05) higher than that of control cells (Fig. 4B). We also evaluated the protein levels of p21Cip1, p27Kip1, cyclin D1, CDK2, and CDK4. As expected, the p21Cip1 and p27Kip1 protein levels were up-regulated, and the cyclin D1, CDK2, and CDK4 protein levels were down-regulated in the cells treated with calcitriol (Fig. 4C). These data indicated that calcitriol treatment inhibited cellular proliferation by cell-cycle arrest at the G1 phase similar to CENP-N knockdown.
shRNA knockdown cells of CENP-N in OSCC (Ca9-22 and SAS-derived transfectants). (A) CENP-N mRNA expression in shCENP-N cells is significantly (*p < 0.05, Student's t-test) lower than that in shMock cells. (B) Immunoblot analysis shows that the CENP-N protein levels in the shCENP-N cells are also markedly lower than that in shMock cells. (C) The shCENP-N cells and shMock cells were counted on 7 consecutive days. The results are expressed as the mean ± standard error of the mean (SEM) of values from three assays. Cellular growth is inhibited significantly (*p < 0.05, Student's t-test) after 144 h culture in the shCENP-N cells. (D) Flow cytometric analysis shows that the percentage of the G1 phase in the shCENP-N cells is increased compared with the shMock cells. (E) Immunoblot analysis shows up-regulation of p21Cip1 and p27Kip1 and down-regulation of cyclin D1, CDK2, and CDK4 in the shCENP-N cells compared with shMock cells.
CENP-N mRNA and protein levels in calcitriol-treated cells. (A, B) Calcitriol significantly inhibits CENP-N mRNA and protein expression levels in the Ca9-22 and SAS cell lines in a dose-dependent manner (*p < 0.05, Student's t-test). (C) The expression of CENP-N mRNA after treatment with calcitriol (optimal concentration, 20 µM in the Ca9-22 cell line and 10 µM in the SAS cell line). The expression of CENP-N mRNA in the calcitriol-treated cells is lower than in the control cells (*p < 0.05, Student's t-test). (D) The expression of CENP-N protein in the calcitriol-treated cells is lower than in the control cells.
Discussion
The current study showed that CENP-N was up-regulated in human OSCC and clinically positively correlated with tumor size. In the CENP-N knockdown models, cellular proliferation was regulated by arresting cell-cycle progression at the G1 phase, suggesting that CENP-N is highly correlated with the tumor size of the human OSCCs.
Of the CENPs, only CENP-A integrates centromeric chromatin instead of histone H3. CENP-A acts as an oncogene in the regulation of cellular growth in liver cancer [25]. CENP-A also is a reliable biomarker for tumor progression and prognosis in breast, ovarian, and lung cancers [26-30]. CENP-N binds the centromere-targeting domain of CENP-A; however, the functions of CENP-N in cancer research have not been reported to date.
Similar to the previous data, which showed that knockdown of cyclin D1 and CDK4 shows negative regulation of CENP-N by microarray [31], we found that CENP-N knockdown cells not only down-regulated cyclin D1 and CDK4 but also up-regulated p21cip1 and p27kip1. Thus, our date indicated for the first time that CENP-N plays a decisive role in tumor growth in OSCC by cell-cycle progression.
Calcitriol is the most biologically active metabolite of vitamin D with high affinity for the vitamin D receptor (VDR) and has been reported to have anticancer effects [32]. Calcitriol acts on bone and mineral homeostasis, inhibits proliferation, and induces differentiation of various normal and malignant cells [33]. However, the exact molecular mechanism behind these anticancer effects is unknown. Calcitriol prescribed for patients with hypocalcemia, osteoporosis, and prevention of corticosteroid-induced osteoporosis, affects the cell cycle with accumulation of cells at the G1 phase of the cell cycle. In addition, calcitriol inhibits its downstream genes, including CENP-A [34,35]. Since CENP-N is a critical binding partner of CENP-A, we speculated that calcitriol also controls CENP-N expression. Consistent with our hypothesis, CENP-N expression decreased after treatment with calcitriol. In addition to our CENP-N knockdown experiments, calcitriol also had an anticancer effect on OSCCs by cell-cycle arrest at the G1 phase with up-regulation of p21Cip1 and p27Kip1, and down-regulation of cyclin D1, CDK2, and CDK4 (Fig. 4D). Therefore, these data suggested that CENP-N is an important target for cancer treatment using calcitriol.
In conclusion, CENP-N is a useful biomarker involved in the proliferation of OSCCS, and calcitriol might be a novel therapeutic drug for OSCCs through regulation of CENP-N.
Abbreviations
CENP-N: Centromere protein N; OSCC: oral squamous cell carcinoma; CENP-A: centromere protein A; VDR: vitamin D receptor.
Effect of calcitriol treatment. (A) To assess the effect of calcitriol on cellular proliferation, the calcitriol-treated cells and control cells were counted on 7 consecutive days. The results are expressed as the mean ± standard error of the mean (SEM) of values from three assays. Cellular growth is inhibited significantly (*p < 0.05, Student's t-test) after 144 h culture in the calcitriol-treated cells. (B) Flow cytometric analysis shows the percentage of G1 phase in calcitriol-treated cells is increased compared with control cells. (C) Immunoblot analysis shows up-regulation of p21Cip1 and p27Kip1 and down-regulation of cyclin D1, CDK2, and CDK4 in the calcitriol-treated cells compared with the control cells. (D) Schematic representation of the cellular proliferation pathway by CENP-N. CENP-N silencing and calcitriol-treatment are involved in G1 phase-related genes, leading to delay of cellular proliferation.
Acknowledgements
We thank Lynda C. Charters for editing this manuscript. We declare that all authors have no conflicts of interest.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hoischen C, Yavas S, Wohland T, Diekmann S. CENP-C/H/I/K/M/T/W/N/L and hMis12 but not CENP-S/X participate in complex formation in the nucleoplasm of living human interphase cells outside centromeres. PLoS One. 2018;13:e0192572
2. Westhorpe FG, Straight AF. The Centromere: Epigenetic Control of Chromosome Segregation during Mitosis. Cold Spring Harb Perspect Biol. 2015;7:a015818
3. Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33-46
4. Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896-902
5. Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L. et al. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev. 2015;29:1058-73
6. Tian T, Li X, Liu Y, Wang C, Liu X, Bi G. et al. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 2018;28:374-8
7. Kasamatsu A, Uzawa K, Nakashima D, Koike H, Shiiba M, Bukawa H. et al. Galectin-9 as a regulator of cellular adhesion in human oral squamous cell carcinoma cell lines. Int J Mol Med. 2005;16:269-73
8. Endo Y, Uzawa K, Mochida Y, Shiiba M, Bukawa H, Yokoe H. et al. Sarcoendoplasmic reticulum Ca2+ ATPase type 2 downregulated in human oral squamous cell carcinoma. Int J Cancer. 2004;110:225-31
9. Kasamatsu A, Iyoda M, Usukura K, Sakamoto Y, Ogawara K, Shiiba M. et al. Gibberellic acid induces α-amylase expression in adipose-derived stem cells. Int J Mol Med. 2012;30:243-7
10. Koyama T, Ogawara K, Kasamatsu A, Okamoto A, Kasama H, Minakawa Y. et al. ANGPTL3 is a novel biomarker as it activates ERK/MAPK pathway in oral cancer. Cancer Med. 2015;4:759-69
11. Miyamoto I, Kasamatsu A, Yamatoji M, Nakashima D, Saito K, Higo M. et al. Kinesin family member 14 in human oral cancer: A potential biomarker for tumoral growth. Biochem Biophys Reports. 2015;3:26-31
12. Corson TW, Huang A, Tsao M-S, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741-53
13. Usukura K, Kasamatsu A, Okamoto A, Kouzu Y, Higo M, Koike H. et al. Tripeptidyl peptidase II in human oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2013;139:123-30
14. Saito T, Kasamatsu A, Ogawara K, Miyamoto I, Saito K, Iyoda M. et al. Semaphorin7A Promotion of Tumoral Growth and Metastasis in Human Oral Cancer by Regulation of G1 Cell Cycle and Matrix Metalloproteases: Possible Contribution to Tumoral Angiogenesis. PLoS One. 2015;10:e0137923
15. Ronchetti L, Melucci E, De Nicola F, Goeman F, Casini B, Sperati F. et al. DNA damage repair and survival outcomes in advanced gastric cancer patients treated with first-line chemotherapy. Int J Cancer. 2017;140:2587-95
16. Toeda Y, Kasamatsu A, Koike K, Endo-Sakamoto Y, Fushimi K, Kasama H. et al. FBLIM1 enhances oral cancer malignancy via modulation of the epidermal growth factor receptor pathway. Mol Carcinog. 2018;57:1690-97
17. Yamatoji M, Kasamatsu A, Kouzu Y, Koike H, Sakamoto Y, Ogawara K. et al. Dermatopontin: A potential predictor for metastasis of human oral cancer. Int J Cancer. 2012;130:2903-11
18. Saito Y, Kasamatsu A, Yamamoto A, Shimizu T, Yokoe H, Sakamoto Y. et al. ALY as a potential contributor to metastasis in human oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2013;139:585-94
19. Takahara T, Kasamatsu A, Yamatoji M, Iyoda M, Kasama H, Saito T. et al. SIPA1 promotes invasion and migration in human oral squamous cell carcinoma by ITGB1 and MMP7. Exp Cell Res. 2017;352:357-63
20. Ishida S, Kasamatsu A, Endo-Sakamoto Y, Nakashima D, Koide N, Takahara T. et al. Novel mechanism of aberrant ZIP4 expression with zinc supplementation in oral tumorigenesis. Biochem Biophys Res Commun. 2017;483:339-45
21. Iyoda M, Kasamatsu A, Ishigami T, Nakashima D, Endo-Sakamoto Y, Ogawara K. et al. Epithelial Cell Transforming Sequence 2 in Human Oral Cancer. PLoS One. 2010;5:e14082
22. Shimizu F, Shiiba M, Ogawara K, Kimura R, Minakawa Y, Baba T. et al. Overexpression of LIM and SH3 Protein 1 Leading to Accelerated G2/M Phase Transition Contributes to Enhanced Tumourigenesis in Oral Cancer. PLoS One. 2013;8:e83187
23. Ferrero M, Ferragud J, Orlando L, Valero L, Sánchez del Pino M, Farràs R. et al. Phosphorylation of AIB1 at Mitosis Is Regulated by CDK1/CYCLIN B. PLoS One. 2011;6:e28602
24. Díaz-Rodríguez E, Álvarez-Fernández S, Chen X, Paiva B, López-Pérez R, García-Hernández JL. et al. Deficient spindle assembly checkpoint in multiple myeloma. PLoS One. 2011;6:e27583
25. Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L. et al. ShRNA-Targeted Centromere Protein A Inhibits Hepatocellular Carcinoma Growth. PLoS One. 2011;6:e17794
26. McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72
27. Wu Q, Qian Y-M, Zhao X-L, Wang S-M, Feng X-J, Chen X-F. et al. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012;77:407-14
28. Qiu J-J, Guo J-J, Lv T-J, Jin H-Y, Ding J-X, Feng W-W. et al. Prognostic value of centromere protein-A expression in patients with epithelial ovarian cancer. Tumor Biol. 2013;34:2971-5
29. Wu Q, Chen Y-F, Fu J, You Q-H, Wang S-M, Huang X. et al. Short hairpin RNA-mediated down-regulation of CENP-A attenuates the aggressive phenotype of lung adenocarcinoma cells. Cell Oncol. 2014;37:399-407
30. Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J. et al. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin. 2015;8:2
31. Molenaar JJ, Ebus ME, Koster J, Sluis P v, van Noesel CJM, Versteeg R. et al. Cyclin D1 and CDK4 Activity Contribute to the Undifferentiated Phenotype in Neuroblastoma. Cancer Res. 2008;68:2599-609
32. Ma Y, Yu W-D, Hidalgo AA, Luo W, Delansorne R, Johnson CS. et al. Inecalcitol, an analog of 1,25D₃, displays enhanced antitumor activity through the induction of apoptosis in a squamous cell carcinoma model system. Cell Cycle. 2013;12:743-52
33. Haussler MR, Whitfield GK, Haussler CA, Hsieh J-C, Thompson PD, Selznick SH. et al. The Nuclear Vitamin D Receptor: Biological and Molecular Regulatory Properties Revealed. J Bone Miner Res. 1998;13:325-49
34. Wang QM, Luo X, Studzinski GP. Cyclin-dependent kinase 6 is the principal target of p27/Kip1 regulation of the G1-phase traverse in 1,25-dihydroxyvitamin D3-treated HL60 cells. Cancer Res. 1997;57:2851-5
35. Verlinden L, Eelen G, Beullens I, Van Camp M, Van Hummelen P, Engelen K. et al. Characterization of the Condensin Component Cnap1 and Protein Kinase Melk as Novel E2F Target Genes Down-regulated by 1,25-Dihydroxyvitamin D 3. J Biol Chem. 2005;280:37319-30
Author contact
![]() Corresponding authors: A. Kasamatsu, Department of Dentistry and Oral-Maxillofacial Surgery, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone/fax: +81-43-226-2300. K. Uzawa, Department of Oral Science, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone/fax: +81-43-226-2300. E-mail addresses: kasamatsuachiba-u.jp (A. Kasamatsu), uzawakchiba-u.jp (K. Uzawa).
Corresponding authors: A. Kasamatsu, Department of Dentistry and Oral-Maxillofacial Surgery, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone/fax: +81-43-226-2300. K. Uzawa, Department of Oral Science, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone/fax: +81-43-226-2300. E-mail addresses: kasamatsuachiba-u.jp (A. Kasamatsu), uzawakchiba-u.jp (K. Uzawa).

 Global reach, higher impact
Global reach, higher impact