Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3746-3756. doi:10.7150/jca.32052 This issue Cite
Research Paper
Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis
1. Department of Gastroenterology, Affiliated Hospital of Jiangsu University, Zhenjiang, Jiangsu 212001, China
2. Department of Gastroenterology, Songjiang Hospital Affiliated Shanghai First People's Hospital, Shanghai Jiao Tong University, Shanghai 201600, China
3. Department of General Surgery, Changhai Hospital, Second Military Medical University, Shanghai, 200433, China
4. Department of Gastroenterology, Changhai Hospital, Second Military Medical University, Shanghai, 200433, China
5. Department of Cell Biology, School of Medicine, Jiangsu University, Zhenjiang, Jiangsu 212013, China.
# These authors have contributed equally to this work.
Received 2018-12-6; Accepted 2019-5-18; Published 2019-6-9
Abstract
Background: Recent studies have shown that circulating long noncoding RNAs (lncRNAs) could be stably detectable in the blood of cancer patients and play important roles in the diagnosis of many different cancers. However, the value of lncRNAs in the diagnosis of pancreatic cancer (PC) has not been fully explored.
Methods: Eleven PC-related lncRNAs were selected by analyzing bioinformatics databases. The expression levels of the lncRNAs were further analyzed in a small set of plasma samples from a training group including 30 noncancer samples (15 healthy and 15 chronic pancreatitis (CP) subjects) and 15 PC samples. Then, the candidate lncRNAs were validated with data from 46 healthy controls, 97 CP patients and 114 PC patients. Receiver operating characteristic (ROC) curves were employed to evaluate the diagnostic performance of the identified lncRNAs.
Results: After selection and validation, three characteristic plasma candidate lncRNAs, ABHD11-AS1, LINC00176 and SNHG11, were identified, and their levels were significantly higher in PC patients than in normal controls. We found that among the three candidate lncRNAs, ABHD11-AS1 showed the best diagnostic performance for the detection of PC. Furthermore, ABHD11-AS1 had a higher area under the ROC curve (AUC) than CEA, CA199 and CA125 for early PC diagnosis, while the combination of ABHD11-AS1 and CA199 was more effective than ABHD11-AS1 alone.
Conclusions: Plasma ABHD11-AS1 could serve as a potential biomarker for detecting PC, and the combination of ABHD11-AS1 and CA199 was more efficient for the diagnosis of PC than ABHD11-AS1 alone, particularly for early tumor screening.
Keywords: lncRNA, ABHD11-AS1, plasma, biomarker, diagnosis, early pancreatic cancer
Background
Pancreatic cancer (PC) is the fourth leading cause of cancer death in Western societies, and despite recent research and therapeutic developments, its median 5-year survival remains less than 5% [1]. The lack of an effective early detection method is one of the most important reasons for the poor survival of patients with the disease. Most patients with PC remain asymptomatic until the disease reaches an advanced stage [2]. Current biomarkers, such as carbohydrate antigen (CA) 199 and carcinoembryonic antigen (CEA), are classic tumor markers commonly used in the management of patients with PC. However, these tumor markers have limited utility in the early detection of PC due to a lack of sufficiently high diagnostic sensitivity and specificity [3]. Therefore, the importance of exploring new biomarkers with high sensitivity and specificity for the early detection of PC should be emphasized.
Long noncoding RNAs (lncRNAs) are longer than 200 base-pairs and lack protein-coding capability [4]. Previous studies have indicated that lncRNAs play vital roles in numerous cellular processes ranging from embryonic stem cell pluripotency and cell-cycle regulation to diseases such as cancer [5]. Dysregulated lncRNAs play oncogenic roles in different types of tumors, including PC [6]. Moreover, aberrant expression levels of lncRNAs have been shown to serve as potential diagnostic or prognostic biomarkers for many human malignancies, such as lung [7], breast [8], colon [9], and liver [10] cancers. Higher HOTAIR expression levels have also been shown in pancreatic tumors than in nontumor tissue [11]. Although these lncRNAs have shown great promise as a new type of tumor marker, they cannot be used for clinical screening purposes because of the difficulty in obtaining tissue biopsies from patients suspected to have PC.
Circulating RNAs in the plasma or serum have been an emerging area of focus in noninvasive diagnostic applications [12]. More recently, studies have characterized several lncRNAs in human fluids as potential tumor markers. For example, POU3F3 was found to be significantly higher in the plasma of esophageal squamous cell carcinoma patients than in that of normal controls, and it was effective for detecting cancer at an early stage [13]. In addition, the plasma lncRNA HULC was identified as a novel marker for the detection of hepatocellular carcinoma [14]. However, there have been no systematic reports on the role of circulating lncRNAs in PC, and it is unclear whether serum lncRNAs have a clinicopathological influence on PC.
In this study, we selected 11 candidate lncRNAs closely related to PC from the Cancer Genome Atlas (TCGA) database. We investigated the expression of 11 lncRNAs in 30 noncancer individuals (15 healthy and 15 CP subjects) and 15 PC patients. ABHD11-AS1, LINC00176, and SNHG11 were selected as candidate lncRNAs to be validated in larger samples by a real-time PCR assay. The correlation between lncRNAs and the survival rate of PC patients was assessed to explore their prognostic potential. Furthermore, the specificity and sensitivity of ABHD11-AS1 were evaluated. Our results indicated that plasma ABHD11-AS1 levels might be a useful biomarker for diagnosing early pancreatic cancer (EPC).
Materials and Methods
Plasma samples and clinical information
All of the clinical plasma samples were obtained from Shanghai Second Military Medical University (2014 to 2016). Among the specimens, 46 were from healthy controls recruited from interested hospital visitors and employees who did not have a diagnosis of PC or chronic pancreatitis (CP); 114 were patients with PC, and 97 were CP patients. A diagnosis was established based on histological evidence of pancreatic adenocarcinoma from a biopsy obtained through surgery, endoscopic ultrasound (EUS), or interventional radiology. All patient diagnoses were confirmed by pathology or imaging. No patients received chemotherapy or radiotherapy before the samples were collected. Plasma samples and complete follow-up data were available for each patient (additional clinical characteristics are presented in Table 1). Venous blood samples were collected from each participant into an EDTA tube. Then, the samples were separated into plasma and cellular components by centrifugation at 1600 rcf for 10 min. The supernatants were collected, and the samples were centrifuged again at 16,000 rcf for 10 min to obtain the plasma. All plasma samples were stored at -80°C before processing. The data analysis was performed in a blinded manner. The clinical stage of cancer was based on the AJCC staging system [15].
Study design
To identify potential plasma lncRNA biomarkers for PC, a step-by-step discovery procedure was designed that included four parts: initial lncRNA profiling, screening phase, a validation phase, and an analysis of candidate lncRNAs in the database.
Part I: We selected differentially expressed genes in PC via a bioinformatics approach. We queried the pancreatic adenocarcinoma dataset of TCGA from the cBioportal (http://www.genenames.org/). The cancer data and genomic profiles were subjected to further analysis. Finally, the data were decoded, and the lncRNAs were sorted by expression level.
Part II: In the screening phase, using a set of plasma samples from 30 noncancer individuals (15 healthy and 15 CP subjects) and 15 PC patients. We assessed the expression of lncRNAs in the three groups. On the basis of the results, 3 candidate lncRNAs were selected for confirmation and analysis.
Part III: The expression of the three selected lncRNAs was validated in 46 healthy controls, 97 CP patients and 114 PC patients. The associations between ABHD11-AS1 expression level and clinicopathological factors were evaluated by the Mann-Whitney test, one-way analysis of variance (ANOVA), and chi-square test. To evaluate the predictive value of the selected lncRNAs for PC and CP, we generated a receiver operating characteristic (ROC) curve. A two-tailed p value of 0.05 or less was considered statistically significant.
Part IV: Correlations between PC patient survival and candidate lncRNA gene expression were determined through analysis of TCGA databases, which are available through Oncolnc (www.oncolnc.org).
RNA extraction, reverse transcription and quantitative PCR
Total RNA was extracted from 10 ml of plasma using RNAiso Plus (Takara, Shiga, Japan). Reverse transcription was performed using a RevertAid First Strand cDNA Synthesis Kit (Thermo, Waltham, MA, USA) according to the manufacturer's specifications. Each real-time PCR mixture (10 μl) contained 5 μl of 2×ChamQ SYBR Green qPCR Master Mix (High Rox Premixed) (Vazyme), 1 μl of primer (10 μM) and 1 μl of cDNA. The primer sequences are shown in Table 2. The cycling conditions consisted of an initial denaturation at 95°C for 5 min, followed by 40 cycles at 95°C, for 15 sec and 60°C for 30 sec. The last stage was 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. In addition, qPCR was performed on a StepOnePlusTM. 5S RNA was used as a control. The expression levels of the lncRNAs were calculated using the ΔCt method, where ΔCt = Cttarget - Ctreference, with a smaller ΔCt value indicating higher expression. Data processing was performed, and the real-time PCR data were analyzed by the comparative CT method [16]. To reduce errors, the experiments were performed in triplicate for every independent RNA sample. Statistical calculations were performed with SPSS 17.0, and p values less than 0.05 were considered statistically significant when an independent samples t-test was used.
Results
Clinical characteristics of the study population
We collected plasma samples from 46 healthy controls (29 males and 17 females, [mean age of 62.35 years, SD 8.5]), 97 CP patients (33 males and 64 females, [mean age of 51.64 years, SD 8.4]), and 114 PC patients (48 males and 66 females, [mean age of 55.25 years, SD 8.3]). As shown in Table 1, there were no significant differences in age (t-test) or sex (Pearson v2 test) among the three groups.
Table 1 summarizes the clinical characteristics of the PC cases except for the 15 PC cases used in Phase I. Among the 114 PC patients, the maximum tumor diameter was < 5 cm in 38 patients (33.3%). Seven (6.1%), 77 (67.5%) and 14 (12.3%) patients were in stages I, II, III and IV. Detectable levels of CEA (>5 µg/l), CA199 (>37 U/ml), and CA125 (>U/ml) were found in 66 (57.9%), 88 (77.2%), and 24 (21.1%) patients, respectively.
Clinicopathological features of Pancreatic carcinoma and cancer-free control samples
| Healthy controls (n=46) | Chronic pancreatitis (n=97) | Pancreatic cancer (n=114) | |
|---|---|---|---|
| Age, y | |||
| <60 | 25 (54.3%) | 56 (57.7%) | 45 (39.5%) |
| ≥60 | 21 (45.6%) | 41 (42.3%) | 69 (60.5%) |
| Sex | |||
| Women | 17 (37.0%) | 33 (34.0%) | 48 (42.1%) |
| Men | 29 (63.0%) | 64 (66.0%) | 66 (57.9%) |
| Tumor location | |||
| Pancreas head | 45 (39.5%) | ||
| Pancreas body/tail | 64 (56.1%) | ||
| Unknown | 5 (4.4%) | ||
| Tumor stage | |||
| I | 7 (6.1%) | ||
| II-IV | 91 (79.8%) | ||
| Unknown | 16 (14.1%) | ||
| Positive ratio | |||
| CEA | 66 (57.9%) | ||
| CA199 | 88 (77.2%) | ||
| CA125 | 24 (21.1%) |
Screening of PC-associated candidate lncRNAs
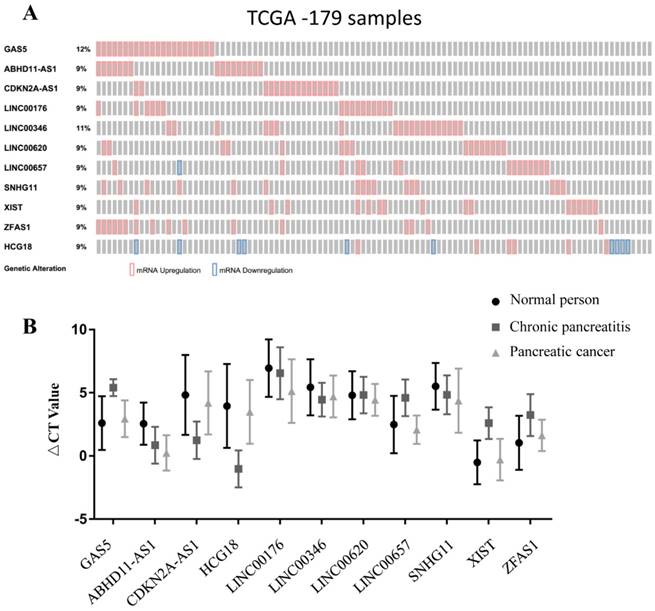
To select lncRNAs with differential expression, we screened the TCGA dataset in cBioportal. In this study, we selected eleven lncRNAs, namely, Gas5, ABHD11-AS1, CDKN2A-AS1, HCG18, LINC00176, LINC00346, LINC00620, LINC00657, SNHG11, Xist, and ZFAS1, with expression changes ranging from 9% to 12% (max) in pancreatic carcinoma (Fig. 1A). These eleven lncRNAs were then subjected to qPCR for further screening, which was performed with samples from 30 noncancer individuals and 15 PC patients. The expression levels of ABHD11-AS1, LINC00176 and SNHG11 were higher in PC patients than in normal controls and CP patients. The expression levels of GAS5, CDKN2A-AS1, HCG18, LINC00657, XIST and ZFAS1 were significantly higher in CP patients than in normal controls and PC patients. The expression of LINC00346 in PC patients and CP patients was similar, and LINC00620 expression was similar in normal subjects and CP patients. Therefore, the above eight lncRNAs were ruled out for subsequent study, and ABHD11-AS1, LINC00176 and SNHG11 were chosen (Fig. 1B).
Identification of predictive PC-associated lncRNAs in plasma
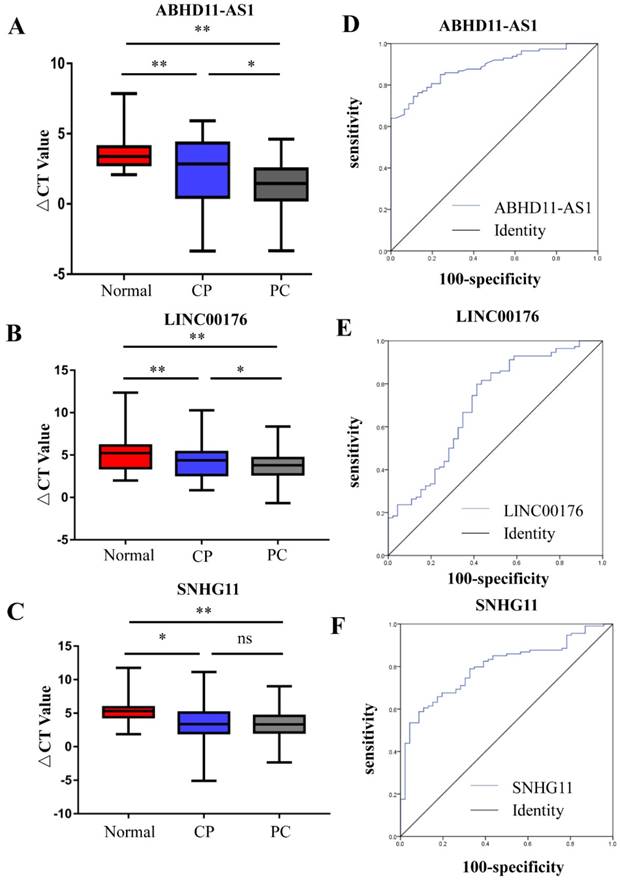
In the selection phase, 3 of 11 pancreatic cancer-related lncRNAs were chosen due to the difference in their plasma expression in PC patients and nontumorous subjects. The three lncRNAs were measured by RT-qPCR in another set of plasma samples from 114 PC patients, 97 PC patients and 46 healthy subjects. 5S RNA was the normalization control. The box plots and mean ΔCt values of the lncRNAs show that the plasma levels of ABHD11-AS1, LINC00176 and SNHG11 were higher in the patients with CP or PC than in the healthy subjects. Furthermore, the plasma levels of ABHD11-AS1 (Fig. 2A) and, LINCO00176 (Fig. 2B) relative to those of healthy controls were significantly higher in PC patients than in patients with CP, but SNHG11 did not exhibit the same pattern (Fig. 2C). We further evaluated the diagnostic value of the three selected lncRNAs (ABHD11-AS1, SNHG11, LINC00176) by ROC curves and area under the curve (AUC) values for all PC patients and healthy controls. The ROC curves are illustrated. When comparing the PC group with the normal group, the AUC value was 0.887 (95% confidence interval (CI): 0.839-0.936, p<0.0001) for ABHD11-AS1 (Fig. 2C), 0.707 (95% CI: 0.614-0.800, p<0.0001) for LINC00176 (Fig. 2D) and 0.79 (95% CI: 0.728-0.867, p<0.0001) for SNHG11 (Fig. 2E). Thus, all three lncRNAs were effective for distinguishing PC patients from healthy controls. ABHD11-AS1, in particular, had a high AUC value.
Biomarker selection and validation phase for PC-associated lncRNAs. (A) Selection of differentially expressed lncRNAs in PC from the TCGA database. (B) Validation of PC-related lncRNAs in human plasma.
Combination of candidate lncRNAs and tumor markers for PC diagnosis
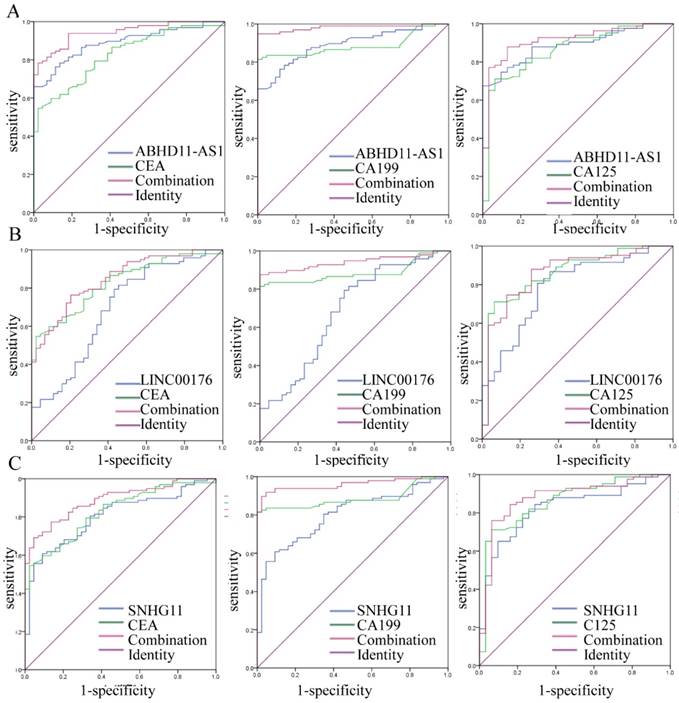
Increasing evidence has shown that the combination of several tumor markers could improve diagnostic accuracy. In this study, we combined ABHD11-AS1, LINC00176 and SNHG11, and the results showed that the combination yielded almost the same results as ABHD11-AS1 alone for PC detection (Fig. S1). However, combining the lncRNAs tumor markers such as CEA, CA199, and CA125 resulted in more effective PC detection. As shown in Figure 3, the combination of ABHD11-AS1 and CEA yielded an AUC of 0.943 (95% CI: 0.909-0.978, p<0.0001), the combination of ABHD11-AS1 and the combination of CA199 yielded an AUC of 0.982 (95% CI: 0.961-0.999, p<0.0001), and ABHD11-AS1 and CA125 yielded an AUC of 0.914 (95% CI: 0.857-0.970, p<0.0001); these AUCs were significantly better than the AUC for ABHD11-AS1 (AUC = 0.888) or CEA (AUC = 0.826), CA199 (AUC = 0.886), or CA125 (AUC = 0.861) alone (Fig. 3A). LINC00176 and CEA yielded an AUC of 0.847 (95% CI: 0.783-0.911, p<0.0001), LINC00176 and CA199 yielded an AUC of 0.941 (95% CI: 0.903-0.980, p<0.0001), and LINC00176 and CA125 yielded an AUC of 0.862 (95% CI: 0.784-0.940, p<0.0001), which were also better than the AUCs for LINC00176 (AUC = 0.692), CEA (AUC = 0.826), CA199 (AUC = 0.886), or CA125 (AUC = 0.861) alone (Fig. 3B). SNHG11 resulted in a similar effect (Fig. 3B). ROC analysis proved that the combination of plasma ABHD11-AS1, LINC00176, and SNHG11 levels was more accurate as a marker for detecting PC than the lncRNAs, CEA, CA199, or CA125 alone.
PC-related lncRNAs in the plasma of PC patients, CP patients, and normal controls were measured. RT-qPCR was used to analyze the plasma levels of ABHD11-AS1 (A), LINC00176 (B), and SNHG11 (C) in 114 PC patients, 97 PC patients and 46 healthy subjects. (D-E) ROC curves were used to evaluate the ability of the plasma lncRNAs ABHD11-AS1, LINC00176, and SNHG11 to detect PC. (ABHD11-AS1, 0.887, p<0.0001; LINC00176, 0.707, p<0.0001; SNHG11, 0.79, p<0.0001.)
Combination of the candidate lncRNAs and tumor markers for PC diagnosis. As the ROC curves show, for the diagnosis of PC, the accuracy of plasma ABHD11-AS1(A), LINC00176 (B), and SNHG11(C) combined with CEA, CA199 or CA125 was higher than the accuracy of the lncRNAs, CEA, CA199, and CA125 alone.
Candidate lncRNA expression in normal subjects and in patients with EPC or CP
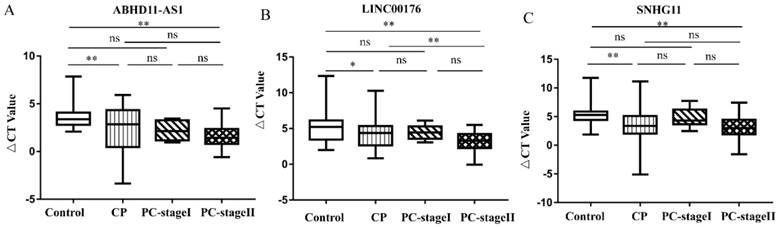
The above results demonstrate the ability of the candidate lncRNAs to distinguish patients with PC from healthy control individuals. We also tried to distinguish, in a minimally invasive and convenient manner, patients with EPC (stage I n=7 and stage II n=77) and patients with CP from control individuals. CP is a risk factor for the development of PC [17, 18]. We compared the expression of three lncRNAs in control individuals, CP patients and EPC patients. As Figure 4A shows, the plasma levels of ABHD11-AS1 increased progressively in the four groups, although some of the increases failed to reach significance. Compared with that in the healthy subjects, the expression of ABHD11-AS1 was increased in EPC patients, including stage I (P=0.206) and stage II (p <0.0001) patients. There was also a trend toward an increase in plasma ABHD11-AS1 expression levels among the patients with CP or EPC. ABHD11-AS1 expression was compared between the following categories: CP and PC-stage I (P=0.999), CP and PC-stage II (P=0.185), and PC-stage I, and PC-stage II (P =0.805). However, the box plot of LINC00176 (Fig. 4B) and SNHG11 (Fig. 4C) shows that there was no obvious distinction between CP and PC-stage I.
Predictive power of the candidate lncRNAs in EPC patients
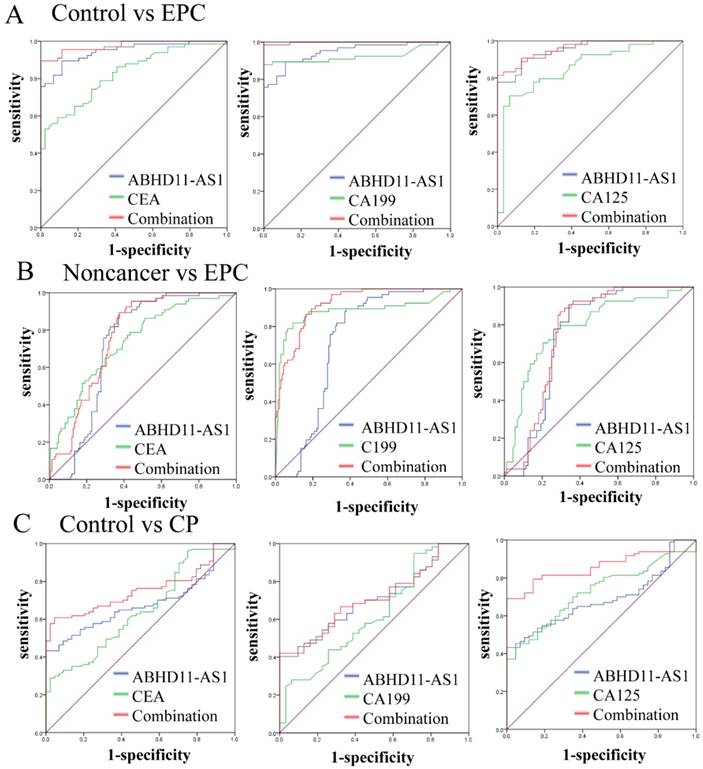
Subsequently, we evaluated the predictive power of ABHD11-AS1 in EPC patients. The results revealed that ABHD11-AS1 had a high AUC value (0.947, 95% CI: 0.908-0.985, p<0.001) for discriminating between patients with EPC and control individuals. We compared the AUC values of ABHD11-AS1 with those of CEA, CA199 and CA125, three blood-based biomarkers used for EPC detection. The AUC values of the ABHD11-AS1 signature were markedly higher than those of CEA (0.826 95% CI: 0.751-0.901, p < 0.001), CA199 (0.925 95% CI: 0.869-0.980, p < 0.001) and CA125 (0.855 95% CI: 0.771-0.939, p<0.0001) for discriminating EPC patients from normal controls. Furthermore, the AUC values of ABHD11-AS1 combined with the tumor markers were markedly increased. For discriminating EPC patients from controls, the AUC value of ABHD11-AS1 in combination with the other markers was as follows: 0.990 (95% CI: 0.978-0.999) with CEA, 0.998 (95% CI: 0.869-0.980) with CA199 and 0.980 (95% CI: 0.957-0.999) with CA125, all p < 0.001 (Fig. 5A). The results indicated that the plasma lncRNA ABHD11-AS1 level is a more accurate biomarker than CEA, CA199 or CA125 for EPC diagnosis and that the combination of ABHD11-AS1 and CA199 provided the greatest discrimination.
For the discrimination between patients with EPC and noncancer individuals, ABHD11-AS1 had an AUC of 0.722 (95% CI: 0.654-0.791), lower than that of CEA (0.736), CA199 (0.882), and CA125 (0.789). However, the AUC was increased when ABHD11-AS1 was combined with other tumor markers; in particular, its combination with CA199 (0.921; 95% CI: 0.886-0.956) provided the best performance (Fig. 5B). For the discrimination between patients with EPC and CP, ABHD11-AS1 had an AUC of 0.621 (95% CI: 0.534-0.708, p<0.009), which was lower than that of CEA (0.695), CA199 (0.863), and CA125 (0.751). However, the AUC was increased when ABHD11-AS1 was combined with the tumor markers; in particular, its combination with CA199 (0.895; 95% CI: 0.848-0.942) had the best performance (Fig. S2).
Lastly, for discriminating CP cases from normal cases, ABHD11-AS1 yielded an AUC of 0.686 (95% CI: 0.600-0.772, p<0.001), and its combination with other markers also yielded an AUC of 0.862 (95% CI: 0.803-0.921, p<0.001), which is higher than that of ABHD11-AS1 alone or its combination with CA199 and CA125 (Fig. 5C). The sensitivity, specificity, and Youden index of ABHD11-AS1, the tumor markers, and its combination with tumor markers for distinguishing EPC patients from healthy controls are summarized in Table 2. However, the AUC values of LINC00176 (Fig. S3 A) and SNHG11 (Fig. S3 B) were both lower than those of CEA, CA199 and CA125.
RNA nucleotide sequences
| primer | sequences | |
|---|---|---|
| Gas5-RT-5.2 | sense | CAGTGTGGCTCTGGATAGCA |
| Gas5-RT-3.2 | anti-sense | TTAAGCTGGTCCAGGCAAGT |
| ABHD11-AS1-5.1 | sense | ATGAAGCCATTGCCAAGAAG |
| ABHD11-AS1-3.1 | anti-sense | GCCTCTCTCTGCAGCTGATT |
| CDKN2A-AS1-5.1 | sense | ATCCCACTGTGGCAGAGAAC |
| CDKN2A-AS1-3.1 | anti-sense | CGAGGTTTGGGATATTTTGC |
| LINC00176-5.1 | sense | CCAGCTCAAATCGTTGGAAT |
| LINC00176-3.1 | anti-sense | AACTCCAAAAAGCTGCAGGA |
| LINC00346-5.1 | sense | ACAAAGGATGACGAGGGTTG |
| LINC00346-3.1 | anti-sense | GGGGCTTGTTCTTTTTCTCC |
| LINC00620-5.1 | sense | AGCAAAGATTGCTGCCTGTT |
| LINC00620-3.1 | anti-sense | CTTCCACGAGTTTTGGGAAA |
| LINC00657-RT-5.1 | sense | AAAGAGGTTGCCGACGTATG |
| LINC00657-RT-3.1 | anti-sense | CAGGTCTTCCAGCTCCATGT |
| SNHG11-5.1 | sense | CGACAAAAGGGGAAACTGAG |
| SNHG11-3.1 | anti-sense | TCCAAGGGAGAGCTGTCATC |
| Xist-5.1 | sense | TGGGGAATGGATTATTGGAA |
| Xist-3.1 | anti-sense | GGACAATGACGAAGCCACTT |
| ZFAS1-5.1 | sense | TTGCAGTCAGGCTTCATACG |
| ZFAS1-3.1 | anti-sense | CTACTTCCAACACCCGCATT |
| HCG18 | sense | CCACAGGTACTGGTGGTGTG |
| HCG18 | anti-sense | CAAGCTCCAAGGATCTCTGC |
| 5SRNA | sense | GTCTACGGCCATACCACCCTGAA |
| 5SRNA | anti-sense | AAGCCTACAGCACCCGGTATTCC |
Performance of ABHD11-AS1 for distinguishing EPC patients from healthy subjects
| Sensitivity | Specificity | Youden index | |
|---|---|---|---|
| ABHD11-AS1 | 0.894 | 0.886 | 0.780 |
| CEA | 0.530 | 0.977 | 0.508 |
| CA199 | 0.879 | 1.000 | 0.879 |
| CA125 | 0.704 | 0.935 | 0.639 |
| ABHD11-AS1+CEA | 0.894 | 1.000 | 0.894 |
| ABHD11-AS1+CA199 | 0.985 | 1.000 | 0.985 |
| ABHD11-AS1+CA125 | 0.815 | 1.000 | 0.815 |
Expression of the candidate lncRNAs in the E PC patients, CP patients and normal controls. (A-C) Comparison of the lncRNA expression levels of control, CP and EPC individuals.
Correlation between the expression of the three lncRNAs and the disease‑specific survival rate
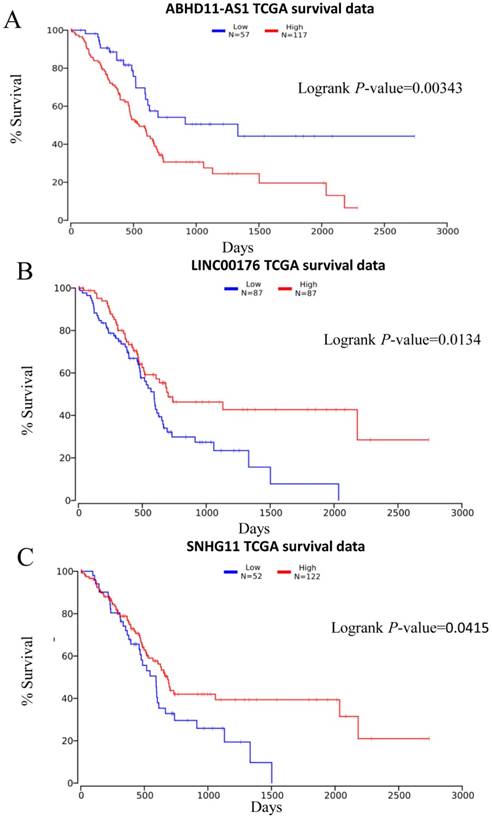
Subsequently, we obtained more information from the TCGA database (http://www.oncolnc.org/) and evaluated the correlation of the lncRNA ABHD11-AS1 with patient outcome. Survival analysis was carried out for 174 patients. The Kaplan-Meier survival analysis showed that patients with low expression of ABHD11-AS1 had a significantly lower disease-specific survival rate than those with high expression (p = 0.0045, Fig. 6A). This finding was consistent with our results and suggested that lncRNA ABHD11-AS1 acts as an oncogene and might be a prognostic marker for PC.
Evaluation of plasma ABHD11-AS1 for the detection of EPC. (A) ROC curves of ABHD11-AS1 combined with CEA, CA199 or CA125 for distinguishing EPC patients from controls. (B) ROC curves of ABHD11-AS1 combined with CEA, CA199 or CA125 to distinguish EPC patients from noncancer patients. (C) ROC curves of ABHD11-AS1 combined with CEA, CA199 or CA125 to distinguish CP patients from controls.
Discussion
The early detection and prevention of PC are important and have attracted considerable clinical and research interest [19]. The earlier this disease can be diagnosed, the earlier life-saving treatments can begin, thus greatly increasing the survival rate of patients with this cancer above the present 4-5% [20]. The low diagnostic rate is one factor contributing to the low survival rate. Previous studies have demonstrated that cell-free nucleic acids are detectable in the plasma and serum of cancer patients and therefore may be utilized as a tool for cancer diagnosis [21]. Patients at high risk are encouraged to enroll in screening clinical trials using magnetic resonance imaging or EUS.10 [22]. However, both of these techniques are expensive, and EUS is also an invasive diagnostic modality. Noninvasive and cost-effective diagnostic tests are urgently needed to screen the population at risk for developing PC and to detect these tumors at earlier stages that are amenable to more effective therapy. In previous studies, cancer-related lncRNA screening was performed based on the different expression profiles of tumor samples and matched normal samples. However, tumor tissues, especially PC, are difficult to acquire. Current diagnostic tools for PC, including the serological markers CEA and CA199, have low specificity and sensitivity. Numerous studies have focused on the lncRNAs in plasma or serum as potential tumor markers for cancer diagnosis and prognosis. Hence, to explore pancreatic carcinoma-associated lncRNAs, we analyzed tumor databases and sample data verified by RT-PCR. We found differences in the expression of the lncRNA ABHD11-AS1 among PC patients, CP patients, and healthy controls in our study, indicating that this lncRNA may be a predictive biomarker for cancer detection.
Prediction of PC patient survival rate by plasma lncRNA expression. Plasma levels of ABHD11-AS1 (A), LINC00176 (B), and SNHG11 (C) to evaluate PC patient' outcomes.
Previous studies have reported that increased plasma lncRNA POU3F3 levels can be used as an ideal tumor biomarker for esophageal squamous cell carcinoma (ESCC) detection and that the combination of POU3F3 and SCCA is better for detection, particularly at the early stage [13]. LncRNAuc003wbd and lncRNA-AF085935 are potential biomarkers for hepatocellular carcinoma (HCC) and hepatitis B virus (HBV) screening [23]. We aimed to find potential biomarkers for PC. In this study, we chose eleven PC-associated lncRNAs based on the TCGA database and examined the expression levels of these eleven lncRNAs in plasma samples from normal control, CP and PC subjects by RT-qPCR. ABHD11-AS1, LINC00176, and SNHG11 were chosen as PC-associated candidate lncRNAs for further study. Next, we validated that the plasma levels of the three lncRNAs were notably upregulated in patients with CP and PC, and that their expression was downregulated in healthy controls. We suggest that all three lncRNAs can distinguish PC samples from healthy samples. Furthermore, ROC curves for ABHD11-AS1 were constructed for differentiating EPC from CP and healthy controls. An AUC of 0.947 implied that ABHD11-AS1 was effective in distinguishing EPC subjects from normal persons and may serve as a potential biomarker for the detection of EPC. Consistent with our results, it has been reported that ABHD11-AS1 was higher in the tissues [24] and gastric juice [25] of subjects with gastric cancer than in those of normal controls.
CP is a progressive inflammatory disorder that is believed to arise from repeated overt or silent episodes of acute pancreatitis [26]. Compared with the general population, patients with CP have a markedly increased risk of PC [17]. In our study, when distinguishing CP patients from healthy subjects, the AUC of ABHD11-AS1 reached a low value of 0.680. For differentiating EPC patients and CP patients, the AUC was 0.621. Combining ABHD11-AS1 with CA199 led to greater effectiveness. These results suggest that evaluating a combination of biomarkers may provide more predictive value than assessing ABHD11-AS1 alone.
Conclusion
In conclusion, the lncRNA, ABHD11-AS1, was highly expressed in the plasma of CP patients and EPC patients. All of these findings indicate that ABHD11-AS1 might be a useful candidate biomarker for screening CP and EPC; the results also provided us with a new approach for studying the function of lncRNAs in cancer biology. Additionally, our study has the following limitations. The population of enrolled patients and controls was relatively small, so a larger sample should be used in the future to verify our results. In terms of ethnicity, the research was conducted in an Asian population. Thus, it remains unknown whether this lncRNA would have the same predictive value in Caucasian and African groups. Evaluating the associations between ABHD11-AS1 and other human cancers is recommended for future studies.
Abbreviations
PC, pancreatic cancer; EPC, early pancreatic cancer; lncRNA, long noncoding RNA; CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen; CA125, carbohydrate antigen 125; ROC curve, receiver operating characteristic curve; AUC, area under the curve.
Supplementary Material
Supplementary figures.
Acknowledgements
Ethics approval and consent to participate
The research was conducted in strict accordance with the protocol approved by the Jiangsu University Affiliated Hospital, and written informed consent was obtained from each participant prior to blood sample collection. All experiments were performed in accordance with the ethical standards established in the 1964 Declaration of Helsinki and its later amendments.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81672402), Provincial Natural Science Foundation of Jiangsu, China (BK20171305), Jiangsu Key R & D Program Social Development Project, China (BE2018689), Research Programs of Jiangsu Provincial Commission of Health and Family Planning, China (H201434), “Six one Project” Research Projects of High-level Medical Personnel of Jiangsu Province (LGY2016054), “Six Talents Peak” High-level Talent Selection and Training Project of Jiangsu Province (2014-WSW-038), Science and Technology Support Program Project of Zhenjiang, China (SH2014024) and Qing Lan Project Funding of Jiangsu Colleges and Universities.
Authors' contributions
L-YW, FW, H-JB, W-DW, Z-MT, ZG, XW and CW carried out the experiments and analyzed the data. L-WY, K-XY, and LL provided the clinical plasma samples. G-AH and XM contributed to the study design and, manuscript drafting and provided funds for this study. L-YW and FW wrote the first and final drafts of the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501
2. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet (London, England). 2011;378:607-20
3. Locker GY, Hamilton S, Harris J. et al. 2006 Update of ASCO Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. Journal of Oncology Practice. 2006;2:314-6
4. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ. et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circulation research. 2015;116:1143-56
5. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145-66
6. Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V. et al. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Molecular cancer. 2011;10:141
7. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180-9
8. Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R. et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748-62
9. Wang R, Du L, Yang X, Jiang X, Duan W, Yan S. et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. Journal of cancer research and clinical oncology. 2016;142:2291-301
10. Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G. et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology (Baltimore, Md). 2012;56:2231-41
11. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R. et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616-25
12. Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World journal of gastroenterology. 2015;21:8527-40
13. Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH. et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Molecular cancer. 2015;14:3
14. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed research international. 2013;2013:136106
15. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471-4
16. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101-8
17. Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F. et al. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849-52
18. Gukovsky I, Li N, Todoric J, Gukovskaya A, Karin M. Inflammation, autophagy, and obesity: common features in the pathogenesis of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1199-209.e4
19. Mohammed A, Janakiram NB, Li Q, Madka V, Ely M, Lightfoot S. et al. The epidermal growth factor receptor inhibitor gefitinib prevents the progression of pancreatic lesions to carcinoma in a conditional LSL-KrasG12D/+ transgenic mouse model. Cancer prevention research (Philadelphia, Pa). 2010;3:1417-26
20. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY). 2008;321:1801-6
21. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature reviews Cancer. 2011;11:426-37
22. Canto MI, Goggins M, Hruban RH, Petersen GM, Giardiello FM, Yeo C. et al. Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2006;4:766-81 quiz 665
23. Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:3231-6
24. Lin X, Yang M, Xia T, Guo J. Increased expression of long noncoding RNA ABHD11-AS1 in gastric cancer and its clinical significance. Medical oncology. 2014;31:42
25. Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X. et al. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:1183-8
26. Pinho AV, Chantrill L, Rooman I. Chronic pancreatitis: a path to pancreatic cancer. Cancer letters. 2014;345:203-9
Author contact
![]() Corresponding authors: Aihua Gong, Department of Cell Biology, School of Medicine, Jiangsu University, 301 Xuefu, Zhenjiang, China 212013. Tel: +86-13775369530; Email: ahg5ujs.edu.cn. Min Xu, Department of Gastroenterology, Affiliated Hospital of Jiangsu University, Jiangsu University, Zhenjiang, China 212001. Tel: +86-15862990603 Email: peterxu1974com
Corresponding authors: Aihua Gong, Department of Cell Biology, School of Medicine, Jiangsu University, 301 Xuefu, Zhenjiang, China 212013. Tel: +86-13775369530; Email: ahg5ujs.edu.cn. Min Xu, Department of Gastroenterology, Affiliated Hospital of Jiangsu University, Jiangsu University, Zhenjiang, China 212001. Tel: +86-15862990603 Email: peterxu1974com

 Global reach, higher impact
Global reach, higher impact