Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3789-3797. doi:10.7150/jca.31166 This issue Cite
Review
The role of Wnt signaling pathway in tumor metabolic reprogramming
1. NHC Key Laboratory of Carcinogenesis, Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, Hunan 410078, China
2. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute and School of Basic Medical Science, Central South University, Changsha 410078, China
3. Hunan Key Laboratory of Nonresolving Inflammation and Cancer, Disease Genome Research Center, The Third Xiangya Hospital, Central South University, Changsha, Hunan, China.
4. Department of Cancer Biology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio 44195, USA
# These authors contributed equally.
Received 2018-11-2; Accepted 2019-5-29; Published 2019-6-9
Abstract
The occurrence and development of tumors is a complex process involving long-term multi-factor participation. In this process, tumor cells from a set of abnormal metabolic patterns that are different from normal cells. This abnormal metabolic change is called metabolic reprogramming of tumors. Wnt signaling pathway is one of the critical signaling pathways regulating cell proliferation and differentiation. In recent years, it has been found that Wnt signaling participates in the occurrence and development of malignant tumors by affecting metabolic reprogramming. This paper reviews the role of Wnt signaling in tumor metabolic reprogramming to provide crucial theoretical guidance for targeted therapy and drug response of tumors.
Keywords: tumor metabolic reprogramming, Wnt signaling, targeted therapy
Introduction
The occurrence and development of tumors is a complex process involving long-term multi-factor participation [1-6]. In this process, tumor cells from a set of abnormal metabolic patterns different from normal cells, providing the energy and raw materials needed for their life activities. This abnormal metabolic change is called metabolic reprogramming of tumors [7-10]. In 1930, Warburg proposed the Warburg effect of abnormal glucose metabolism in tumors, that is, tumor cells obtain energy mainly through anaerobic glycolysis [11]. Subsequently, it was found that tumor cells also have changes in amino acid metabolism and fat metabolism, thereby providing energy and raw materials for biomacromolecule synthesis [12-14]. In recent years, it has been found that the metabolic reprogramming of tumor cells also includes pentose phosphate bypass, lipid and protein and anabolism associated with nucleic acid synthesis, and a large number of endogenous oxygen free radicals, which are involved in the development of tumors has played an important role [15-18].
Wnt signaling pathway is one of the critical signaling pathways regulating cell proliferation and differentiation and plays an essential role in normal physiological activities such as growth and development and pathological processes including malignant tumors [19-23]. The Wnt signaling pathway is composed of various signal molecules, ligands and receptors such as Wnt protein and β-catenin, and is very conservative in evolution [24]. It mainly consists of three pathways: the classical Wnt pathway, the Wnt/Ca2+ pathway, and the planar cell polarity (PCP) pathway. In recent years, a large number of studies have found that the Wnt signaling pathway can affect the occurrence of obesity and diabetes by affecting the metabolism of normal cells [25-28]. Further studies have shown that the Wnt pathway can also change the metabolism of tumor cells, thereby participating in the occurrence and development of malignant tumors by changing metabolic reprogramming [29-32].
1. Tumor metabolism reprogramming is one of the characteristics of malignant tumors
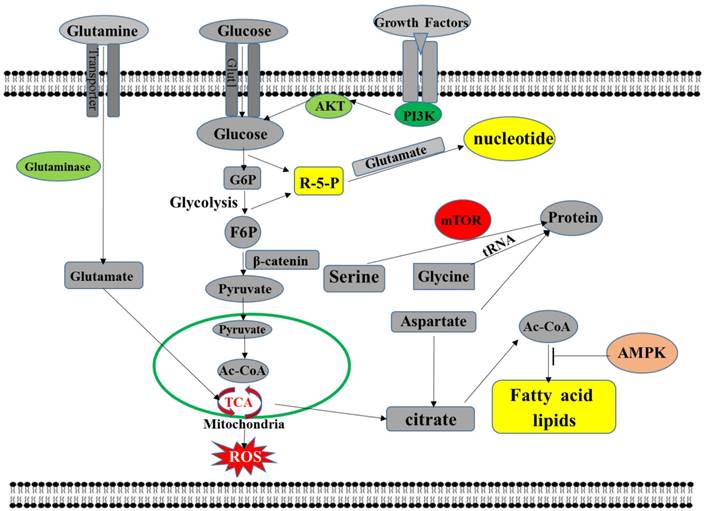
Metabolic reprogramming is one of the prominent features of tumor cells, including all metabolic changes in tumor cells [33-36]. Abnormal glucose metabolism is the earliest discovered metabolic reprogramming. Under aerobic conditions, tumor cells are also mainly powered by the glycolysis pathway. This metabolic abnormality is also known as the Warburg effect [37-39]. That is, when the mitochondrial function of the tumor cells is dysfunctional, the cells obtain energy mainly by enhancing anaerobic glycolysis [40-44]. That is, after glucose is metabolized to pyruvic acid, it does not enter the tricarboxylic acid cycle for aerobic oxidation but is converted to lactic acid by lactate dehydrogenase [45]. However, because tumor cells are energy-efficient through the glycolysis pathway, to maintain the energy required for their life activities, tumor cells, in addition to increased glucose consumption, also increase energy supply by increasing fat metabolism [46-49]. It is characterized by de novo synthesis of fatty acids and active beta-oxidation, thus providing an adequate supply of energy through increased fat metabolism [50]. In addition, due to the uncontrolled nature of tumor cell proliferation, anabolism of fatty acid synthesis and amino acid metabolism of tumor cells has also changed [51, 52]. In tumor cells, the pentose phosphate pathway, the hexose synthesis pathway, the serine/glycine synthesis pathway, and the glutamate-glutamine cycle are all increased, thereby providing the required raw materials for the synthesis of biological macromolecules such as ribonucleic acid and protein [53-59].Also, the number of oxygen free radicals (ROS) in tumor cells increased compared with normal cells, and increased ROS stimulated the proliferation of tumor cells [60-62]. The purpose of tumor metabolic reprogramming is to drive limited nutrients or intermediate metabolites to be more "effectively" utilized by tumor cells to support the vigorous metabolic demands of tumor cell development and progression [63] (Figure 1).
metabolic pathways in cancer cells. Highly proliferating cells promote glucose catabolism and glutamine catabolism by regulating key metabolic pathways, driving molecular synthesis and maintaining energy balance through molecular interactions. Cancer cells use aerobic glycolysis to produce ATP and promote pyruvate synthesis to promote glycolysis by increasing the expression of glycolytic enzymes. Tumor cells also produce macromolecules such as NADPH and 5-carbon sugar-driven nucleic acids via the pentose phosphate pathway. The citrate from the TCA cycle is exported to the cytosol and further converted to acetyl-CoA for the synthesis of lipid acids.
The network of Wnt signaling regulates tumor metabolism reprogramming. The activated Wnt signaling pathway promotes the up-regulation of MCT-1, CYC1 and ATP synthase by the downstream transcription factor TCF/LEF, resulting in the secretion of intracellular lactate and the occurrence of aerobic glycolysis. Wnt signaling pathway can also up-regulate the expression of GLUT-1, LDH, PKM2, SLC1A5 and other genes by c-Myc promoting glycolysis, nucleotide and fatty acid synthesis in tumor cells. The non-canonical Wnt signaling pathway promotes aerobic glycolysis by activating Akt-mTOR to stabilize the expression of mTORC1 and β-catenin. The activated mTOR pathway promotes glucose uptake by increasing glucose transporter expression. On the other hand, the mTOR pathway can also lead to an increase in fatty acid synthesis by up-regulating the expression of acetyl-CoA, resulting in an increase in fatty acid oxidative metabolism. In addition, activation of the mTOR pathway can also result in upregulation of glucose-6-phosphate dehydrogenase, resulting in enhanced pentose phosphate bypass and promotion of ribonucleic acid synthesis. ROS levels can directly affect the transcriptional activity of β-catenin. ROS can interact with TCF4, alter the binding of β-catenin to TCF, and interact with the transcriptional link factor of FOXO3a, thereby changing the gene expression of cells and promoting tumorigenesis.
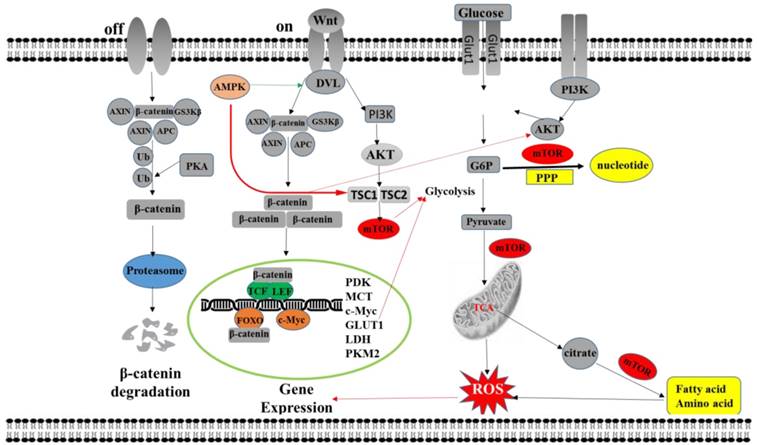
2. Wnt signaling regulates tumor metabolism reprogramming
As a highly conserved signaling pathway in evolution, the Wnt signaling pathway plays an essential role in many biological processes such as growth, development, metabolism, and stem cell maintenance, while the Wnt pathway is out of control and occurs in diseases such as cancer, obesity, and diabetes [64, 65]. The Wnt signaling pathway acts as a critical regulatory pathway, similar to that involved in metabolic alterations in normal cells, and is also engaged in swollen metabolic reprogramming in tumor cells. Current research indicates that the Wnt pathway can also mediate the regulation of tumor cell metabolism, thereby participating in tumor development and progression by affecting tumor cell metabolism reprogramming [66]. The current study found that the Wnt signaling pathway can participate in the metabolic reprogramming of tumors through the regulation of multiple downstream signaling pathways such as TCF/LEF, c-myc and Akt-mTOR pathway [67-69]. The Wnt signaling pathway can also indirectly affect metabolic pathways by directly regulating the expression of rate-limiting enzymes in metabolic pathways and by regulating other oncogenes, leading to metabolic changes in cells (Figure 2).
2.1 Wnt pathway participates in tumor metabolic reprogramming through TCF/LEF pathway
Current studies indicate that the metabolism of malignant tumor cells is mainly regulated by the canonical Wnt pathway , while the classical Wnt signaling pathway can also exert biological functions through the downstream transcription factor TCF/LEF [70-73]. When TCF/LEF is activated, the expression of MCT-1, CYC1, and ATP synthase is up-regulated, resulting in intracellular lactate secretion and aerobic glycolysis [74]. This not only leads to an increase in aerobic glycolysis but also provides essential energy for tumor cells, while a large amount of lactic acid secretion also induces secretion of factors such as VEGF, thereby promoting tumor angiogenesis [75-77]. In addition to affecting tumor cells themselves, the Wnt pathway is also involved in the formation of the tumors microenvironment [78-80]. Although alterations in fatty acid (FA) metabolism in cancer cells have received less attention compared to other metabolic alterations such as glucose or glutamine metabolism, recent studies have uncovered the importance of lipid metabolic reprogramming in carcinogenesis [81]. By activating TCF/LEF to inhibit the expression of PPARγ and C/EBPα in adipocytes around the tumor, it promotes the fat degradation of fat cells, thus providing the necessary conditions for the fat supply of tumor cells [82, 83].
2.2 Wnt pathway participates in tumor metabolic reprogramming through the c-mycpathway
Metabolic alterations attributable to Myc are due to quantitative and not qualitative differences in its behavior, thus making it somewhat easier to understand its role in normal metabolic processes. In metabolic reprogramming, the classical Wnt signaling pathway can be involved in tumor metabolic reprogramming by over-activation of c-Myc [84, 85], c-Myc's overexpression in cancer can most likely be attributed to the fact that it is a major transcriptional integrator of most, if not all, normal and oncogenic growth factor pathways, c-Mycis activated as a transcription factor by binding to a specific gene sequence (CACGTG), the most prominent transcript families under Myc's control tend to encode proteins that supervise energy production, anabolic pathways, protein synthesis, thereby up-regulating the expression of genes such as GLUT-1, PDH, PFK1, HK, LDH, PKM2, and SLC1A5[86-90], thereby correspondingly causing cells. Increased internal glucose uptake, accelerated glycolysis, and increased glutamine-glutamate cycling. Thereby promoting the glycolysis, nucleotide and fatty acid synthesis in tumor cells, providing tumor cells with the substances and energy required for proliferation [91-94]. Also, c-myc can also up-regulate the expression of FOXOs, thereby increasing the production of reactive oxygen species and inducing autophagy in tumor cells, thereby recycling intracellular proteins and lipids through autophagy to other growth and substances required for proliferation[95-101].
2.3 Wnt pathway involved in tumor metabolic reprogramming through the Akt-mTOR pathway
Cells grow and proliferate when nutrients, growth factors and the cellular energy status trigger carbohydrate catabolism and the synthesis of essential building blocks such as proteins, nucleotides and lipids. The evolutionarily conserved Ser/Thr protein kinase target of rapamycin mTORC1 is activated by other pathways such as Wnt signalling. The Wnt signaling pathway is involved in tumor metabolic reprogramming mainly through activation of Akt -mTOR. Wnt signalling stimulate mTORC1 through the PI3K-phosphoinositide-dependent kinase 1 (PDK1)-AKT pathway. Activated AKT phosphorylates tuberous sclerosis complex 2 (TSC2; also known as tuberin) to activate the metabolic pathways that ultimately drive cell growth [102, 103]. In comparison to mTORC1 regulation, mTORC2 regulation is poorly understood, mTORC2actived by the hydrophobic motif in a subset of AGC family kinases such as PKA, PKG, PKC , including AKT Glycogen synthase kinase 3β (GSK3β) Wnt signalling inhibits GSK3β and the TSC complex, and thus activates mTORC1 and mTORC2 [104-107]. The activated mTOR pathway promotes glucose uptake by cells by increasing the expression of glucose transporters [108]. On the other hand, the mTOR pathway can also lead to an increase in fatty acid synthesis by up-regulating the expression of acetyl-CoA, resulting in the increased oxidative metabolism of fatty acids [109-111]. Activation of the mTOR pathway can also lead to up-regulation of glucose-6-phosphate dehydrogenase, leading to enhanced pentose phosphate bypass and more feedstock for ribonucleic acid synthesis [112-116].
2.4 Wnt pathway participates in tumor metabolic reprogramming by regulating the expression of rate-limiting enzymes in metabolic pathways
Regulation of the Wnt signaling pathway also includes control of metabolic enzymes, and β-catenin-mediated c-Myc expression leads to up-regulation of several rate-limiting glycolytic genes, including those for glucose transporter 1 (GLUT-1) ), LDH and pyruvate kinase, the last step in catalyzing glycolysis produces ATP and pyruvate, which promotes aerobic glycolysis in cancer cells [117-120]. When the mitochondrial function of tumor cells is dysfunctional, the cells obtain energy mainly by enhancing anaerobic glycolysis. After glucose is metabolized to pyruvic acid, it does not enter the tricarboxylic acid cycle for aerobic oxidation but is converted to lactic acid by lactate dehydrogenase [121-124]. In ovarian cancer, a large number of metabolic genes have been found to be targets of transcription, including participation in glutamine metabolism and fatty acid metabolism [125-127]. In breast cancer, Wnt/β-catenin increases the aerobic glycolysis process by reducing the expression of the cytochrome c oxidase that inhibits mitochondrial respiration [128, 129]. Also, recent studies have demonstrated that Wnt5b regulates the expression of oxidative phosphorylation-related genes, such as cytochrome C1 and ATP synthase, through the canonical Wnt pathway [130]. In colorectal cancer cells, aerobic glycolysis is promoted, pyruvate synthesis is increased, and pyruvate dehydrogenase (PDH) activity in mitochondria is inhibited to reduce pyruvate oxidation, resulting in more conversion of pyruvate to lactic acid. The glycolytic enzyme PKM2 is a pleopoietic protein that acts as a transcriptional coactivator. In the signaling of various cancer cell types, PKM2 interacts with β-catenin to cause β-catenin transcriptional changes, resulting in up-regulation of c-myc expression [131, 132].
In addition to the above regulation, the Wnt signaling pathway is also regulated by the expression of reactive oxygen species (ROS) [133]. The phenomenon of elevated reactive oxygen species is common in tumor cell, which is mainly caused by accelerated metabolic activity, mitochondrial dysfunction, cell receptor signal enhancement, oncogene, oxidase and cyclooxygenase activity [134-136]. Low levels of ROS, by reversibly oxidizing protein sulfhydryl groups, altering protein structure and function involved in the transduction of cellular signaling pathways, also can cause DNA instability and mutations, leading to genomic instability and, ultimately, canceration of cells [137]. Mitochondria are an important source of ROS. In tumor cells, ROS levels can directly affect the transcriptional activity of β-catenin. ROS can replace the interaction between β-catenin and TCF4, and change the binding of β-catenin to TCF to transcriptional catenin with FOXO3a. The role of factors, which in turn changes the gene expression of cells, regulates the occurrence and development of tumors [138, 139]. The oxidative stress process also activates DVL levels upstream of the canonical Wnt signal, which interacts with thioredoxin family proteins to activate independent Wnt signals outside the classical Wnt signaling pathway [140-142]. Thus, oxidative stress in tumor cells regulates the canonical Wnt signaling pathway to alter gene transcriptional changes, promoting tumor cell autophagy via the mTORC1-independent pathway [143, 144]. Under nutrient pressure or other stress conditions, tumor cells can recycle cellular proteins and lipids and convert them into other substances needed for survival, helping tumor cells to pass through the harsh cellular microenvironment [145-148].
3. Prospect
Metabolic reprogramming of cells is one of the important features of tumors. And with the deepening of research, people's understanding of tumor metabolism reprogramming is no longer limited to changes in glucose metabolism such as glycolysis and tricarboxylic acid cycle, but also includes many metabolic pathways such as fatty acid metabolism, cholesterol metabolism, and amino acid metabolism change [149-152]. As one of the important signaling pathways regulating cell metabolism reprogramming, Wnt signaling pathway has been found to regulate the metabolism of tumor cells through downstream signaling pathways such as mTOR and c-myc.In addition to the pathways mentioned above, recent studies have shown that the Wnt pathway can also interact with hippo pathway regulating tumor metabolism. Wnt pathway mediates TAZ expression, upregulates IRS1 and stimulates Akt- and Glut4-mediated glucose metabolism [153].In addition Wnt pathway a key scaffolding protein Dishevelled (DVL), is responsible for nuclear export of phosphorylated YAP, DVL is also required for YAP intracellular trafficking induced metabolic stress. Note that the p53/LATS2 and LKB1/AMPK tumor suppressor axes regulating tumor metabolism reprogram [154]. Stearoyl-CoA desaturase 1 (SCD1)a central enzymatic in the conversion of saturated fatty acids, enhances the production of lipid-modified Wnt proteins that activate the canonical Wnt pathway. Activation of the Wnt pathway leads to the release of both β-catenin and YAP/TAZ from the destruction complex. This enables β-catenin and YAP/TAZ to translocate to the nucleus where, upon interaction with their transcriptional partners, they mediate the cancer metabolism reprogramming [155]. In addition, studies have shown that non-coding RNAs such as miRNA and lncRNA as well as circular RNA can regulate tumor metabolism by regulating the Wnt pathway [156-169].Also, current studies have found that abnormal changes in cellular metabolism precede tumorigenesis. Therefore, molecular markers related to tumor metabolism have important clinical value, which can provide new ideas for the development of new molecular markers and early diagnosis of tumors [170-172]. The relationship between abnormal activation of the Wnt signaling pathway and tumors and multi-level target anticancer therapy of the Wnt signaling pathway has become a new research hotspot in tumor molecular biology. These findings provide direct evidence that metabolic changes can promote tumorigenesis, and the key nodes of its regulation are also becoming potential targets in tumor diagnosis and treatment and will provide important theoretical guidance for targeted therapy and precision medicine of tumors. The tumor microenvironment is closely related to the occurrence and development of cancer [173-177]. Nutritional competition between cells can significantly affect cell growth, survival, and function. Glucose restriction caused by tumors alters the metabolism of T cells, which in turn affects their function. Studies have shown that, tumor glucose consumption metabolically limits T cells, inhibiting their mTOR activity, glycolysis, and IFN-γ production, allowing tumors to develop further. This mechanism can help people develop more effective cancer immunotherapy, better liberate the immune system, and achieve long-lasting anti-cancer effects [178-182]. Wnt pathway regulates PDL1 to regulate tumor immune escape by regulating the expression of ALDH [183]. Tumor cells can also derived Wnt ligands stimulate M2-like polarization of TAMs through c-Myc via canonical Wnt/β-catenin signaling, which results in tumor immunosuppression [184]. In conclusion, through in-depth study of Wnt pathway and tumor metabolic reprogramming, it will further expand people's understanding of the etiology of malignant tumors and will provide an important theoretical basis for humans to overcome malignant tumors finally.
Acknowledgements
This study was supported by grants from The National Natural Science Foundation of China (81572787, 81672683, 81672993, 81672688, 81702907, 81772901, 81772928, 81803025 and 81872278), the Overseas Expertise Introduction Project for Discipline Innovation (111 Project, No. 111-2-12), and the Natural Science Foundation of Hunan Province (2016JC2035, 2017SK2105, 2018JJ3704, 2018JJ3815, 2019JJ50872, 2019JJ50354, 2019JJ50780, 2018SK21210 and 2018SK21211).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wei F, Wu Y, Tang L. et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60(11):1271-1275
2. Tu C, Zeng Z, Qi P. et al. Genome-Wide Analysis of 18 Epstein-Barr Viruses Isolated from Primary Nasopharyngeal Carcinoma Biopsy Specimens. J Virol. 2017;91(17):e00301-00317
3. Tu C, Zeng Z, Qi P. et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole-genome sequencing. Carcinogenesis. 2018;39(12):1517-1528
4. Fan C, Tang Y, Wang J. et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9(16):2852-2864
5. Xiong F, Deng S, Huang HB. et al. Effects and mechanisms of innate immune molecules on inhibiting nasopharyngeal carcinoma. Chin Med J (Engl). 2019;132(6):749-752
6. Wu C, Li M, Meng H. et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62(5):640-647
7. Chang CH, Qiu J, O'Sullivan D. et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162(6):1229-1241
8. Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23(1):27-47
9. Jiang X, Wang J, Deng X. et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18(1):10
10. Wang YA, Li XL, Mo YZ. et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17(1):168
11. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309-314
12. Bhutia YD, Babu E, Ramachandran S. et al. Amino Acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75(9):1782-1788
13. Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610-2623
14. Nakagawa H, Hayata Y, Kawamura S. et al. Lipid Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers (Basel). 2018;10(11):447
15. Tang L, Wei F, Wu Y. et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37(1):87-102
16. Venneti S, Thompson CB. Metabolic Reprogramming in Brain Tumors. Annu Rev Pathol. 2017;12:515-545
17. Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol. 2017;48:47-53
18. Jia D, Park JH, Jung KH. et al. Elucidating the Metabolic Plasticity of Cancer: Mitochondrial Reprogramming and Hybrid Metabolic States. Cells. 2018;7(3):1-19
19. Pate KT, Stringari C, Sprowl-Tanio S. et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33(13):1454-1473
20. Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13(7):513-532
21. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461-1473
22. Tammela T, Sanchez-Rivera FJ, Cetinbas NM. et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545(7654):355-359
23. Zeng ZY, Zhou YH, Zhang WL. et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol. 2007;38(1):120-133
24. Lee SY, Jeon HM, Ju MK. et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012;72(14):3607-3617
25. Liu H, Fergusson MM, Wu JJ. et al. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4(158):ra6
26. Karner CM, Long F. Wnt signaling and cellular metabolism in osteoblasts. Cell Mol Life Sci. 2017;74(9):1649-1657
27. Fuster JJ, Zuriaga MA, Ngo DT. et al. Noncanonical Wnt signaling promotes obesity-induced adipose tissue inflammation and metabolic dysfunction independent of adipose tissue expansion. Diabetes. 2015;64(4):1235-1248
28. Wei F, Jing YZ, He Y. et al. Cloning and characterization of the putative AFAP1-AS1 promoter region. J Cancer. 2019;10(5):1145-1153
29. Rada P, Rojo AI, Offergeld A. et al. WNT-3A regulates an Axin1/NRF2 complex that regulates antioxidant metabolism in hepatocytes. Antioxid Redox Signal. 2015;22(7):555-571
30. Sherwood V WNT signaling. an emerging mediator of cancer cell metabolism? Mol Cell Biol. 2015;35(1):2-10
31. Tao H, Yang JJ, Shi KH. et al. Wnt signaling pathway in cardiac fibrosis: New insights and directions. Metabolism. 2016;65(2):30-40
32. Adebayo Michael AO, Ko S, Tao J. et al. Inhibiting Glutamine-Dependent mTORC1 Activation Ameliorates Liver Cancers Driven by beta-Catenin Mutations. Cell Metab. 2019;29(5):1135-1150
33. Hensley CT, Faubert B, Yuan Q. et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164(4):681-694
34. Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168(4):657-669
35. Andrejeva G, Rathmell JC. Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors. Cell Metab. 2017;26(1):49-70
36. Vernieri C, Casola S, Foiani M. et al. Targeting Cancer Metabolism: Dietary and Pharmacologic Interventions. Cancer Discov. 2016;6(12):1315-1333
37. Chocarro-Calvo A, Garcia-Martinez JM, Ardila-Gonzalez S. et al. Glucose-induced beta-catenin acetylation enhances Wnt signaling in cancer. Mol Cell. 2013;49(3):474-486
38. Cantor JR and Sabatini DM Cancer cell metabolism. one hallmark, many faces. Cancer Discov. 2012;2(10):881-898
39. Granger A, Mott R, Emambokus N. Hacking Cancer Metabolism. Cell Metab. 2016;24(5):643-644
40. Buck MD, O'Sullivan D, Klein Geltink RI. et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 2016;166(1):63-76
41. Bogdanovic E. IDH1, lipid metabolism and cancer: Shedding new light on old ideas. Biochim Biophys Acta. 2015;1850(9):1781-1785
42. Valcarcel-Jimenez L, Gaude E, Torrano V. et al. Mitochondrial Metabolism: Yin and Yang for Tumor Progression. Trends Endocrinol Metab. 2017;28(10):748-757
43. Viale A, Corti D, Draetta GF Tumors, mitochondrial respiration. a neglected connection. Cancer Res. 2015;75(18):3685-3686
44. Li X, Han G, Li X. et al. Mitochondrial pyruvate carrier function determines cell stemness and metabolic reprogramming in cancer cells. Oncotarget. 2017;8(28):46363-46380
45. Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017;26(1):39-48
46. Davidson SM, Papagiannakopoulos T, Olenchock BA. et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23(3):517-528
47. DeBerardinis RJ, Lum JJ, Hatzivassiliou G. et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11-20
48. Hansen M, Flatt T, Aguilaniu H Reproduction, fat metabolism, life span. what is the connection? Cell Metab. 2013;17(1):10-19
49. Ringstad N A Controlled Burn. Sensing Oxygen to Tune Fat Metabolism. Cell Rep. 2016;14(7):1569-1570
50. Yi M, Li J, Chen S. et al. Emerging role of lipid metabolism alterations in Cancer stem cells. J Exp Clin Cancer Res. 2018;37(1):118
51. Ho PC, Bihuniak JD, Macintyre AN. et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162(6):1217-1228
52. Zechner R, Zimmermann R, Eichmann TO. et al. FAT SIGNALS-lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15(3):279-291
53. Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39(8):347-354
54. Stincone A, Prigione A, Cramer T. et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc. 2015;90(3):927-963
55. Kuehne A, Emmert H, Soehle J. et al. Acute Activation of Oxidative Pentose Phosphate Pathway as First-Line Response to Oxidative Stress in Human Skin Cells. Mol Cell. 2015;59(3):359-371
56. Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17(4):351-359
57. Hackmann TJ, Ngugi DK, Firkins JL. et al. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ Microbiol. 2017;19(11):4670-4683
58. Amelio I, Cutruzzola F, Antonov A. et al. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39(4):191-198
59. Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572-583
60. Li L, Tan J, Miao Y. et al. ROS and Autophagy: Interactions and Molecular Regulatory Mechanisms. Cell Mol Neurobiol. 2015;35(5):615-621
61. Schieber MS, Chandel NS. ROS links glucose metabolism to breast cancer stem cell and EMT phenotype. Cancer Cell. 2013;23(3):265-267
62. Panieri E and Santoro MM ROS homeostasis and metabolism. a dangerous liason in cancer cells. Cell Death Dis. 2016;7(6):e2253
63. Mondal S, Bhattacharya K, Mandal C. Nutritional stress reprograms dedifferention in glioblastoma multiforme driven by PTEN/Wnt/Hedgehog axis: a stochastic model of cancer stem cells. Cell Death Discov. 2018;4:110
64. Mzoughi S, Zhang J, Hequet D. et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat Genet. 2017;49(9):1354-1363
65. Langton PF, Kakugawa S, Vincent JP. Making, Exporting, and Modulating Wnts. Trends Cell Biol. 2016;26(10):756-765
66. Lu W, Pelicano H, Huang P Cancer metabolism. is glutamine sweeter than glucose? Cancer Cell. 2010;18(3):199-200
67. Wong N, Ojo D, Yan J. et al. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356(2 Pt A):184-191
68. Poli V, Fagnocchi L, Fasciani A. et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun. 2018;9(1):1024
69. Sun H, Yang W, Tian Y. et al. An inflammatory-CCRK circuitry drives mTORC1-dependent metabolic and immunosuppressive reprogramming in obesity-associated hepatocellular carcinoma. Nat Commun. 2018;9(1):5214
70. Yang W, Zheng Y, Xia Y. et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295-1304
71. Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4(11):a007906
72. Lu FI, Sun YH, Wei CY. et al. Tissue-specific derepression of TCF/LEF controls the activity of the Wnt/beta-catenin pathway. Nat Commun. 2014;5:5368
73. Doumpas N, Lampart F, Robinson MD. et al. TCF/LEF dependent and independent transcriptional regulation of Wnt/beta-catenin target genes. EMBO J. 2019;38(2):e98873
74. Wu Z, Wei D, Gao W. et al. TPO-Induced Metabolic Reprogramming Drives Liver Metastasis of Colorectal Cancer CD110+ Tumor-Initiating Cells. Cell Stem Cell. 2015;17(1):47-59
75. Jiang L, Yin M, Wei X. et al. Bach1 Represses Wnt/beta-Catenin Signaling and Angiogenesis. Circ Res. 2015;117(4):364-375
76. Zhang L, Wang H, Li C. et al. VEGF-A/Neuropilin 1 Pathway Confers Cancer Stemness via Activating Wnt/beta-Catenin Axis in Breast Cancer Cells. Cell Physiol Biochem. 2017;44(3):1251-1262
77. Wu C, Chen J, Chen C. et al. Wnt/beta-catenin coupled with HIF-1alpha/VEGF signaling pathways involved in galangin neurovascular unit protection from focal cerebral ischemia. Sci Rep. 2015;5:16151
78. Zhang Q, Lou Y, Zhang J. et al. Hypoxia-inducible factor-2alpha promotes tumor progression and has crosstalk with Wnt/beta-catenin signaling in pancreatic cancer. Mol Cancer. 2017;16(1):119
79. Chirumbolo S, Bjorklund G. Can Wnt5a and Wnt non-canonical pathways really mediate adipocyte de-differentiation in a tumour microenvironment? Eur J Cancer. 2016;64:96-100
80. Meyer IS, Jungmann A, Dieterich C. et al. The cardiac microenvironment uses non-canonical WNT signaling to activate monocytes after myocardial infarction. EMBO Mol Med. 2017;9(9):1279-1293
81. Nakagawa H, Hayata Y, Kawamura S. et al. Lipid Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers (Basel). 2018;10(11):pii E447
82. Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12(4):364-371
83. Sun Y, Campisi J, Higano C. et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18(9):1359-1368
84. Cowling VH, D'Cruz CM, Chodosh LA. et al. c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol Cell Biol. 2007;27(14):5135-5146
85. Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3(8):a014217
86. Stine ZE, Walton ZE, Altman BJ. et al. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5(10):1024-1039
87. Dejure FR and Eilers M MYC and tumor metabolism. chicken and egg. EMBO J. 2017;36(23):3409-3420
88. Togel L, Nightingale R, Chueh AC. et al. Dual Targeting of Bromodomain and Extraterminal Domain Proteins, and WNT or MAPK Signaling, Inhibits c-MYC Expression and Proliferation of Colorectal Cancer Cells. Mol Cancer Ther. 2016;15(6):1217-1226
89. Luan W, Wang Y, Chen X. et al. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6(15):13006-13018
90. Liang F, Li Q, Li X. et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49(3):1046-1056
91. Tateishi K, Iafrate AJ, Ho Q. et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin Cancer Res. 2016;22(17):4452-4465
92. Goetzman ES, Prochownik EV. The Role for Myc in Coordinating Glycolysis, Oxidative Phosphorylation, Glutaminolysis, and Fatty Acid Metabolism in Normal and Neoplastic Tissues. Front Endocrinol (Lausanne). 2018;9:129
93. Gong Z, Yang Q, Zeng Z. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):3683-3695
94. Xu K, Xiong W, Zhou M. et al. Integrating ChIP-sequencing and digital gene expression profiling to identify BRD7 downstream genes and construct their regulating network. Mol Cell Biochem. 2016;411(1-2):57-71
95. Dong C, Yuan T, Wu Y. et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23(3):316-331
96. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417(1):1-13
97. Masui K, Tanaka K, Akhavan D. et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18(5):726-739
98. Cianfanelli V, Fuoco C, Lorente M. et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17(1):20-30
99. Wei J, Long L, Yang K. et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. 2016;17(3):277-285
100. Song Y, Li X, Zeng Z. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7(24):36783-36799
101. Yan Q, Zeng Z, Gong Z. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6(39):41766-41782
102. Li Q, Chen P, Zeng Z. et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37(9):12503-12512
103. Xiao L, Wei F, Liang F. et al. TSC22D2 identified as a candidate susceptibility gene of multi-cancer pedigree using genome-wide linkage analysis and whole exome sequencing. Carcinogenesis. 2019 pii: bgz095. doi: 10.1093/carcin/bgz095. [Epub ahead of print]
104. Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40(2):310-322
105. Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169(6):985-999
106. Arques O, Chicote I, Puig I. et al. Tankyrase Inhibition Blocks Wnt/beta-Catenin Pathway and Reverts Resistance to PI3K and AKT Inhibitors in the Treatment of Colorectal Cancer. Clin Cancer Res. 2016;22(3):644-656
107. Huang J, Nguyen-McCarty M, Hexner EO. et al. Maintenance of hematopoietic stem cells through regulation of Wnt and mTOR pathways. Nat Med. 2012;18(12):1778-1785
108. Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15(3):155-162
109. Li J, Huang Q, Long X. et al. CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARalpha pathways. J Hepatol. 2015;63(6):1378-1389
110. Mo Y, Wang Y, Xiong F. et al. Proteomic analysis of the molecular mechanism of lovastatin inhibiting the growth of nasopharyngeal carcinoma cells. J Cancer. 2019;10(10):2342-2349
111. Lee G, Zheng Y, Cho S. et al. Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell. 2017;171(7):1545-1558
112. Zoncu R, Efeyan A, Sabatini DM mTOR. from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21-35
113. Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25(9):545-555
114. Cheng SC, Quintin J, Cramer RA. et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345(6204):1250684
115. Wolfson RL, Chantranupong L, Wyant GA. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543(7645):438-442
116. Mullins GR, Wang L, Raje V. et al. Catecholamine-induced lipolysis causes mTOR complex dissociation and inhibits glucose uptake in adipocytes. Proc Natl Acad Sci U S A. 2014;111(49):17450-17455
117. Hackett SR, Zanotelli VR, Xu W. et al. Systems-level analysis of mechanisms regulating yeast metabolic flux. Science. 2016;354(6311):2786-2803
118. Sweetlove LJ, Fernie AR. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat Commun. 2018;9(1):2136
119. Olson DG, Horl M, Fuhrer T. et al. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab Eng. 2017;39:169-180
120. Zaidi N, Swinnen JV, Smans K ATP-citrate lyase. a key player in cancer metabolism. Cancer Res. 2012;72(15):3709-3714
121. Sehrawat A, Roy R, Pore SK. et al. Mitochondrial dysfunction in cancer chemoprevention by phytochemicals from dietary and medicinal plants. Semin Cancer Biol. 2017;47:147-153
122. Schapira AH. Mitochondrial diseases. Lancet. 2012;379(9828):1825-1834
123. Nunnari J and Suomalainen A Mitochondria. in sickness and in health. Cell. 2012;148(6):1145-1159
124. Xia M, Zhang Y, Jin K. et al. Communication between mitochondria and other organelles: a brand-new perspective on mitochondria in cancer. Cell Biosci. 2019;9:27
125. Matassa DS, Amoroso MR, Lu H. et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016;23(9):1542-1554
126. Rosano L, Cianfrocca R, Masi S. et al. Beta-arrestin links endothelin A receptor to beta-catenin signaling to induce ovarian cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2009;106(8):2806-2811
127. Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52(4):590-614
128. Zhuang X, Zhang H, Li X. et al. Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nat Cell Biol. 2017;19(10):1274-1285
129. Srinivasan S, Guha M, Dong DW. et al. Disruption of cytochrome c oxidase function induces the Warburg effect and metabolic reprogramming. Oncogene. 2016;35(12):1585-1595
130. Harada T, Yamamoto H, Kishida S. et al. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci. 2017;108(1):42-52
131. Mendez-Lucas A, Li X, Hu J. et al. Glucose Catabolism in Liver Tumors Induced by c-MYC Can Be Sustained by Various PKM1/PKM2 Ratios and Pyruvate Kinase Activities. Cancer Res. 2017;77(16):4355-4364
132. Yang W, Xia Y, Ji H. et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118-122
133. Myant KB, Cammareri P, McGhee EJ. et al. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12(6):761-773
134. Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94(3):909-950
135. Cheung EC, Lee P, Ceteci F. et al. Opposing effects of TIGAR- and RAC1-derived ROS on Wnt-driven proliferation in the mouse intestine. Genes Dev. 2016;30(1):52-63
136. Muhlemann JK, Younts TLB, Muday GK. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc Natl Acad Sci USA. 2018;115(47):E11188-E11197
137. Tapeinos C, Pandit A. Physical, Chemical, and Biological Structures based on ROS-Sensitive Moieties that are Able to Respond to Oxidative Microenvironments. Adv Mater. 2016;28(27):5553-5585
138. Ferber EC, Peck B, Delpuech O. et al. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012;19(6):968-979
139. Cammareri P, Vincent DF, Hodder MC. et al. TGFbeta pathway limits dedifferentiation following WNT and MAPK pathway activation to suppress intestinal tumourigenesis. Cell Death Differ. 2017;24(10):1681-1693
140. Wang W, Li X, Lee M. et al. FOXKs promote Wnt/beta-catenin signaling by translocating DVL into the nucleus. Dev Cell. 2015;32(6):707-718
141. Jiang X, Charlat O, Zamponi R. et al. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol Cell. 2015;58(3):522-533
142. Xie Z, Khair M, Shaukat I. et al. Non-canonical Wnt induces chondrocyte de-differentiation through Frizzled 6 and DVL-2/B-raf/CaMKIIalpha/syndecan 4 axis. Cell Death Differ. 2018;25(8):1442-1456
143. Zeng H, Lu B, Zamponi R. et al. mTORC1 signaling suppresses Wnt/beta-catenin signaling through DVL-dependent regulation of Wnt receptor FZD level. Proc Natl Acad Sci U S A. 2018;115(44):E10362-E10369
144. Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377-388
145. Semenza GL Hypoxia-inducible factors. coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 2017;36(3):252-259
146. Katheder NS, Khezri R, O'Farrell F. et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541(7637):417-420
147. Galluzzi L, Bravo-San Pedro JM, Demaria S. et al. Activating autophagy to potentiate immunogenic chemotherapy and radiation therapy. Nat Rev Clin Oncol. 2017;14(4):247-258
148. Yu T, Guo F, Yu Y. et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170(3):548-563 e516
149. Liang JQ, Teoh N, Xu L. et al. Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat Commun. 2018;9(1):4490
150. Silvente-Poirot S and Poirot M Cancer. Cholesterol and cancer, in the balance. Science. 2014;343(6178):1445-1446
151. Marz W and Ritsch A Cholesterol Efflux Capacity. Choke Point of Reverse Cholesterol Traffic? J Am Coll Cardiol. 2016;67(21):2488-2491
152. Almhjell PJ, Boville CE, Arnold FH. Engineering enzymes for noncanonical amino acid synthesis. Chem Soc Rev. 2018;47(24):8980-8997
153. Hwang JH, Kim AR, Kim KM. et al. TAZ couples Hippo/Wnt signalling and insulin sensitivity through Irs1 expression. Nat Commun. 2019;10(1):421
154. Kim NH, Lee Y, Yook JI. Dishevelling Wnt and Hippo. BMB Rep. 2018;51(9):425-426
155. Mancini R, Noto A, Pisanu ME. et al. Metabolic features of cancer stem cells: the emerging role of lipid metabolism. Oncogene. 2018;37(18):2367-2378
156. Fan C, Tang Y, Wang J. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16(1):130
157. Wang Y, Mo Y, Gong Z. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25
158. Zhong Y, Du Y, Yang X. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79
159. Wang P, Xu J, Wang Y. et al. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358(6366):1051-1055
160. Zhou R, Wu Y, Wang W. et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134-142
161. Bo H, Fan L, Gong Z. et al. Upregulation and hypomethylation of lncRNA AFAP1-AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncology Reports. 2019;41:2431-2439
162. Fan C, Wang J, Tang Y. et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2018;40(2):380-391
163. Lian Y, Xiong F, Yang L. et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253
164. Tang Y, He Y, Zhang P. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17(1):77
165. He Y, Jing Y, Wei F. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9(2):235
166. He R, Liu P, Xie X. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145
167. Tang Y, Wang J, Lian Y. et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol Cancer. 2017;16(1):42
168. Bo H, Fan L, Li J. et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer. 2018;9(24):4677-4683
169. Fan CM, Wang JP, Tang YY. et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019 doi: 10.1111/cas.14034. [Epub ahead of print]
170. Zhao H, Yang L, Baddour J. et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250
171. Wei F, Wu Y, Tang L. et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br J Cancer. 2018;118(2):233-247
172. Zhang Y, Xia M, Jin K. et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17(1):45
173. Wang M, Zhao J, Zhang L. et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8(5):761-773
174. Kaur A, Webster MR, Marchbank K. et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532(7598):250-254
175. Deng X, Xiong F, Li X. et al. Application of atomic force microscopy in cancer research. J Nanobiotechnology. 2018;16(1):102
176. Duan S, Guo W, Xu Z. et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol Cancer. 2019;18(1):29-43
177. Wang H, Zhou Y, Oyang L. et al. LPLUNC1 stabilises PHB1 by counteracting TRIM21-mediated ubiquitination to inhibit NF-kappaB activity in nasopharyngeal carcinoma. Oncogene. 2019 doi: 10.1038/s41388-019-0778-6. [Epub ahead of print]
178. Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18(3):363-374
179. Phan AT, Goldrath AW, Glass CK. Metabolic and Epigenetic Coordination of T Cell and Macrophage Immunity. Immunity. 2017;46(5):714-729
180. Moshfegh CM, Collins CW, Gunda V. et al. Mitochondrial superoxide disrupts the metabolic and epigenetic landscape of CD4(+) and CD8(+) T-lymphocytes. Redox Biol. 2019:101141
181. Kishton RJ, Sukumar M, Restifo NP. Metabolic Regulation of T Cell Longevity and Function in Tumor Immunotherapy. Cell Metab. 2017;26(1):94-109
182. Ma EH, Bantug G, Griss T. et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017;25(2):345-357
183. Castagnoli L, Cancila V, Cordoba-Romero SL. et al. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene. 2019;38(21):4047-4060
184. Yang Y, Ye YC, Chen Y. et al. Crosstalk between hepatic tumor cells and macrophages via Wnt/beta-catenin signaling promotes M2-like macrophage polarization and reinforces tumor malignant behaviors. Cell Death Dis. 2018;9(8):793
Author contact
![]() Corresponding authors: guocdeedu.cn or xiongfedu.cn
Corresponding authors: guocdeedu.cn or xiongfedu.cn

 Global reach, higher impact
Global reach, higher impact