Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3842-3850. doi:10.7150/jca.33989 This issue Cite
Research Paper
RAG1 high expression associated with IKZF1 dysfunction in adult B-cell acute lymphoblastic leukemia
1. Department of Hematology, Zhongda Hospital, Medical School of Southeast University, Institute of Hematology Southeast University,Nanjing 210009, China
2. International Cooperative Leukemia Group and International Cooperative Laboratory of Hematology, Zhongda Hospital, Medical School of Southeast University, Nanjing 210009, China
3. Department of Pediatrics, Pennsylvania State University Medical College, Hershey, PA17033, USA
4. Genome Sciences and Bioinformatics Core Facility, Institute for Personalized Medicine, Penn State College of Medicine, Hershey, PA17033, USA
Received 2019-2-11; Accepted 2019-5-30; Published 2019-6-9
Abstract
The recombination mediated by recombination activating gene (RAG) is not only the dominant mutational process but also the predominant driver of oncogenic genomic rearrangement in acute lymphoblastic leukemia (ALL). It is further responsible for leukemic clonal evolution. In this study, significant RAG1 increase is observed in the subsets of B-ALL patients, and high expression of RAG1 is observed to be correlated with high proliferation markers. IKZF1-encoded protein, IKAROS, directly binds to the RAG1 promoter and regulates RAG1 expression in leukemic cells. CK2 inhibitor by increasing IKAROS activity significantly suppresses RAG1 expression in ALL in an IKAROS-dependent manner. Patients with IKZF1 deletion have significantly higher expression of RAG1 compared to that without IKZF1 deletion. CK2 inhibitor treatment also results in an increase in IKZF1 binding to the RAG1 promoter and suppression of RAG1 expression in primary ALL cells. Taken together, these results demonstrate that RAG1 high expression is associated with high proliferation markers in B-ALL. Our data for the first time proved that RAG1 expression is directly suppressed by IKAROS. Our results also reveal drive oncogenesis of B-ALL is driven by high expression of RAG1 with IKAROS dysfunction together, which have significance in an integrated prognostic model for adult ALL.
Keywords: RAG1, IKZF1, adult, acute lymphoblastic leukemia
Introduction
The recombination-activating gene 1 (RAG1) is the member of a recombination-activating gene family which play an important role in the rearrangement and recombination of the genes of immunoglobulin and T cell receptor molecules and is critical for the generation of T and B cells. However, the abnormalities in RAG family members are observed quite commonly in lymphoid malignancies [1-7]. The RAG-mediated gene deletions and translocations have been reported in acute lymphoblastic leukemia (ALL) [8-13]. RAG and activation-induced cytidine deaminase (AID) together drive leukemic clonal evolution due to chronic exposure to inflammatory stimuli, parallel to infections noted in children during their childhood [13]. It is also reported that a higher RAG1 mRNA level predicts shorter relapse free survival and overall survival in patients with ALL [13]. These reports suggest RAG1 is involved in the oncogenesis and relapse of ALL.
IKAROS, a kruppel-like zinc finger protein is encoded by the IKZF1 gene, and it plays an essential role for normal hematopoiesis and acts as a tumor suppressor in ALL. The genetic defects of IKZF1 in a single allele are associated with the development of human acute lymphoblastic leukemia, which is characterized by an increased risk of relapses and poorer prognosis. Recently, we reported the global binding profiling of IKAROS in ALL cells and found that IKAROS regulates the expression of gene targets through chromatin remodeling in ALL [14-20]. Our data from chromatin immunoprecipitation followed by sequencing (ChIP-seq) showed IKAROS binding peaks in the promoter region of the genes in multioncogenic pathways. We also observed that CK2 inhibitors increased the tumor suppressor activity of IKAROS and act as a functional activator of IKAROS [14-20]. However, it still remains unclear whether IKAROS regulates RAG1 expression.
Here we examined RAG1 expression in an adult patient with B-ALL and analyzed the correlation of RAG1 expression with clinical features. We observed that high expression of RAG1 correlates with high proliferation markers in adult ALL. We also identified RAG1 as a direct target of IKZF1-encoded protein, IKAROS. Deletion of IKZF1 was significantly associated with increased expression of RAG1 in adult B-ALL. Our findings reveal that IKZF1 deletion may be responsible for the high expression of RAG1 in B-ALL. High expression of RAG1 may work in conjunction with IKZF1 deletion to drive oncogenesis and relapse in B-ALL.
Materials and Methods
Patients and samples
Subjects with newly-diagnosed B-ALL (age12-77 years old) were recruited between June 2008 and June 2016. There were 131 patients who were recruited at the First Affiliated Hospital of Nanjing Medical University and Zhongda Hospital Southeast University. The patients' diagnosis was based on the WHO Diagnosis and Classification of ALL (2008). The approval for this study was received from the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University and Zhongda Hospital Southeast University, Nanjing, China.
Cytogenetic and molecular analyses
Cytogenetics and detection of most common IKZF1 deletion, Ik6 [27], CRLF2 rearrangements [28-30], BCR-ABL fusion gene/Ph chromosome [25, 31], PAX5 mutations [26] and SH2B3 mutations [17] etc. were performed as previously described. The StepOne Plus Real-time PCR system (Applied Biosystem-Thermofisher, Foster, CA, USA) was used for qPCR. Expression values of genes of interest (GOI) were calculated in each sample by a formula derived from a scatter graph of Ct values from serial dilutions of a template standard as described [16, 18, 20, 25]. Expression levels of GOIs were expressed as gene expression value of GOI/18s rRNA by normalized to the housekeeping genes. Subjects were allocated into high or low RAG1expression cohorts (3rd-4th quartiles vs. 1st-2nd quartiles) with a cut-off value determined by SPSS 20[16, 20].
The qPCR for RAG1 expression was similarly performed as above in Nalm6 cells. The results were n presented as fold induction over vector controls after normalized to those obtained with 18sRNA. Primers: 18s RNA, Sense: 5'-GTAACCCGTTGAACCCCATT-3', Antisense: 5'-CCATCCAATCGGTAGTAGCG-3'; RAG1 Sense: 5'-CGAGGAACGTGACCATGGA-3', Anti-sense: 5'-GGCAGTGTTGCAGATGTCACA -3'.
Plasmid construction, retroviral gene transfer,and cell culture reagents
The Nalm6 [32] has been previously described. RPMI 1640 medium (Cellgro, USA) supplemented with 10% fetal bovine serum (Hyclone, USA) and DMEM (Cellgro) supplemented with 10% fetal calf serum and 1% L-glutamine (Cellgro) were used for culture of Nalm6 and HEK293 cell, respectively, incubated at 37°C in a humidified atmosphere of 5% CO2. Primary human B-ALL were cultured in RPMI 1640 medium (Cellgro) supplemented with 10% fetal bovine serum (Hyclone). CX4945 was obtained from Sigma (St. Louis, MO, USA). Cells were cultured with or without CX4945 and collected for total RNA isolation. Human IKZF1 retroviral construct and retroviral production were described previously [14, 15, 19, 20].
Quantitative Chromatin Immunoprecipitation (qChIP)
For the qChIP assays, the chromatin was incubated with antibodies against IKAROS [14, 15, 19] or normal rabbit IgG (Abcam) as a control [14, 15, 19, 20]. The enrichment of the ChIP sample was evaluated over input with the qPCR data obtained in three or more replicates, using specific primers in the promoter region of RAG1 (forward: 5'-GCAACCAACCAACTCTGTTCAC-3', reverse: 5'-ACTGGTTTGTTTTGGACTTTCCTT-3'). The relative concentration of the qPCR product was presented as the fold change of the level of DNA-IKAROS samples in comparison to controls.
Luciferase reproter Assay
The promoter of RAG1 was cloned into the pGL4.15 vector (Promega, WI, USA), and transiently expressed in HEK293 cells for luciferase reporter assay. The promega luciferase assay reagents were used to measure the result on the luminometer following the manufacture's instruction [14, 16]. The firefly luciferase activities were expressed as a percentage of pcDNA3.1-IKZF1 transfection-induced luciferase activity versus that of the pcDNA3.1 vector. All transfection for reporter assays were performed independently, in triplicate, at least three times.
IKZF1 shRNA knockdown
The humanIKZF1 shRNA constructs in the GFP vector (pGFP-v-RS) (Origene) was transiently transfected into Nalm6 cells using the Neon Transfection System (Invitrogen, USA). The scrambled 29-mer shRNA cassette in the pGFP-v-RS vector was used as a control. IKZF1 Knockdown efficiency was confirmed by measurement of IKZF1 mRNA level with qPCR [14, 15, 19, 20]. Primers: IKZF1-F: 5'-GGCGCGGTGCTCCTCCT-3', IKZF1-R: 5'-TCCGACACGCCCTACGACA-3'.
Statistical analysis
The Mann-Whitney U-test was used for evaluation of median differences between the cohorts. Univariate and multivariate Cox models were performed for frequency differences with SPSS version 20.0. Mean values with bars representing the standard error of the mean (SEM) was used for data presentation. The shown data was the representative of 3 independent experiments. Statistical significance was determined by Student t-test for comparison of two groups or analysis of variance (ANOVA) for comparing more than two groups.
Results
RAG1 is highly expressed and associated with high proliferation in adult B-ALL
Firstly, we detected the RAG1 mRNA expression in 131 newly diagnosed adult B-ALL patients. We found that the RAG1 mRNA levels were significantly higher in patients with B- ALL compared to that in the normal controls (Fig 1). We also examined the RAG1 expression in microarray cohorts and found that RAG1 is highly expressed in the cohort studies of B-ALL patients (Supplemental Fig 1).
RAG1 expression in adult B-ALL. Comparison of RAG1 mRNA level in B-ALL with normal BM control by qPCR.
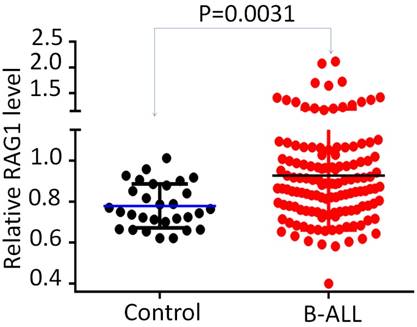
We further divided the patients into two subgroups based on the high (Quartile 1-2) or low (Quartile 3-4) RAG1 mRNA levels. The relationships between the RAG1 expression and clinical characteristics in this cohort of B-ALL were analyzed (Fig 2 and Supplemental Table 1). RAG1 high expression group demonstrated a significantly higher percentage of white blood cell (WBC) ≥30x109/L (67.7% vs. 38.5%, P<0.001) and higher median WBC (47.9×109/L vs. 17.0×109/L, P=0.025), which are markers of poorer prognosis in B-ALL (Fig 2A and Supplemental Table 1). In addition, patients with high RAG1 expression had a higher rate of blasts in peripheral blood (PB) (75.0% vs. 61.0%, P=0.026) (Fig 2B and Supplemental Table 1), indicating the higher proliferation of leukemic cells in patients with high RAG1 expression.
We did not observe the significant association of mutations in PAX5 and SH2B3 with RAG1 expression (Fig 2C, Supplemental Table 1); however, the percentage of CRLF2 mutations, which includes R186S in exon5, F232F in exon6 and A11A in exon 1 is significantly higher in the patients with RAG1 high expression compared to that of low expression (Fig 2C). Also, the rate of IK6, the most common deletion form of IKZF1 is significantly higher in patients with RAG1 high expression (Fig 2C). These data suggest RAG1 high expression associates with CRLF2 mutations and IKZF1 deletion.
IKAROS binds to the promoter of RAG1 and regulates its expression in B-ALL
To further address the potential link between the high RAG1 expression and high proliferation of B-ALL cells, we analyzed the motifs of transcription factors in the promoter region of RAG1. As we expected, certain core IKAROS binding motifs (GGGA or GGAA) were identified in the promoter region of RAG1 (Fig 3A). Notably, our chromatin immunoprecipitation followed by sequencing (ChIP-seq) data showed the strong binding peaks for IKZF1 in the RAG1 promoter region in B-ALL cell lines and primary cells from the B-ALL patients (Fig 3B and Supplemental Fig 2)[14, 15]. To further confirm, quantitative ChIP (qChIP) assays were performed and showed significant binding of IKAROS at the RAG1 promoter region in both B-ALL cell line Nalm6 (Fig 3C) and primary cells (Fig 3D).
Correlation of RAG1 expression with clinical features in adult B-ALL. (A-B) Correlation of RAG1 mRNA level with WBC count and blast in peripheral blood in B-ALL. (C) RAG1 expression with deletion and mutations in adult B-ALL.*P<0.05, ***P<0.001.
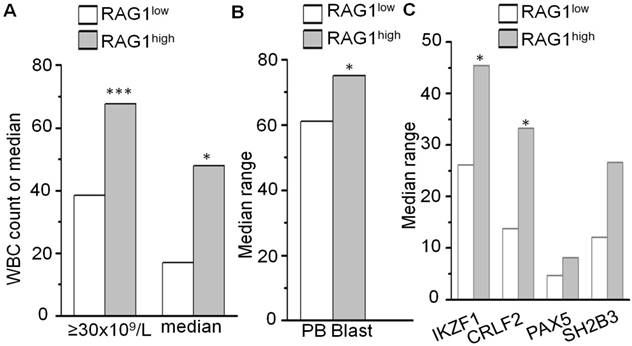
IKAROS binds the promoters of RAG1. (A) RAG1 promoter region with conserved IKZF1 core binding motifs (GGGA or GGAA, red). (B) IKAROS binding peaks in the RAG1promoter region identified by ChIP-seq. (B) qChIP data for IKAROS binding on RAG1 promoter in Nalm6 B-ALL (B) and primary B-ALL cells (C).
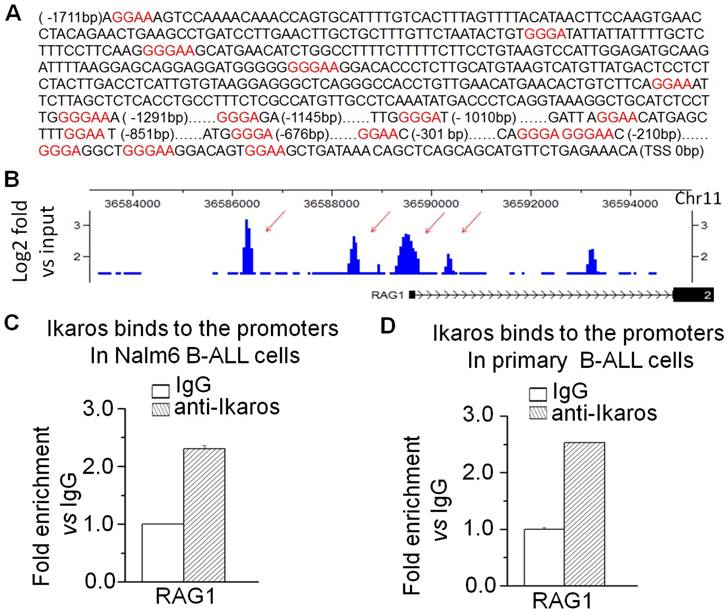
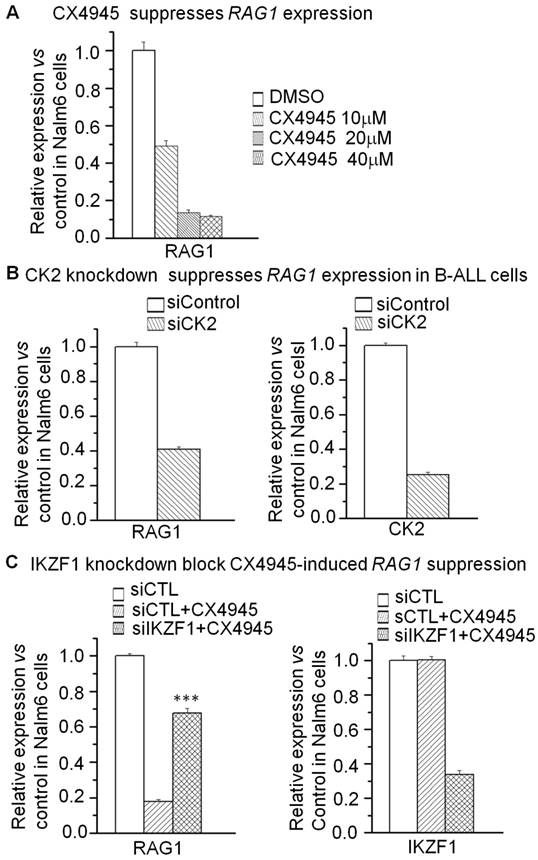
Moreover, we addressed whether RAG1 is the direct target of IKAROS and how IKAROS regulates RAG1 expression in B-ALL cells. We observed that IKAROS suppresses the promoter activity of RAG1 by luciferase reporter assay (Fig 4A). By over-expression of IKAROS in Nalm6 B-ALL cells, we observed that IKAROS could suppress RAG1 mRNA levels (Fig 4B). Conversely, efficient IKZF1 knockdown increased RAG1 expression in Nalm6 cells (Fig 4C).To further demonstrate that IKAROS negatively regulates RAG1 expression, Nalm6 B-ALL cells were treated with CK2 inhibitor (CX4945) which functioned as IKAROS activator. Significantly, CX4945 could suppress RAG1 expression in a dose-dependent manner (Fig 5A). Similarly, CK2 knockdown with shRNA could reduce RAG1 expression (Fig 5B). It worth noting that the CX4945-induced decrease of RAG1 expression could be blocked by IKZF1 knockdown with shRNA in Nalm6 cells (Fig 5C). These data indicate that RAG1 is the direct target of IKAROS and IKAROS suppresses RAG1 expression in B-ALL cells.
IKAROS suppresses RAG1 expression. (A) The promoter activity of RAG1 promoters measured by luciferase reporter assay following transfection with IKAROS or control vector in HEK293 cells. (B) Effect of expression of IKAROS in Nalm6 cells. The cells were transduced with a vector containing IKAROS as compared to the control vector. (C) qPCR of RAG1 and IKZF1 expression in the Nalm6 cells following IKZF1 shRNA treatment as compared to scramble shRNA cells. Gene expression is evaluated by RT-qPCR using total RNA isolated from the cells transfected with scramble shRNA (siControl) or IKZF1shRNA (siIKZF1) for 2 days.
CK2 inhibitor CX4945 suppresses the expression of RAG1 in an IKAROS-dependent manner. (A) Effect of CK2 inhibitor CX4945, which functions as IKAROS activator, on the expression of RAG1 mRNA level in Nalm6 cells with CX4945 treatment for 2 days. (B) Effect of CK2 knockdown on the expression of RAG1 in Nalm6 cells. (C) IKZF1 knockdown rescues the CX4945-induced change of RAG1 in Nalm6 B-ALL cells. Cells were treated with 10mM CX4945 for 2 days.***P<0.001.
Correlation of IKZF1 deletion with high RAG1 expression in patients with B-ALL
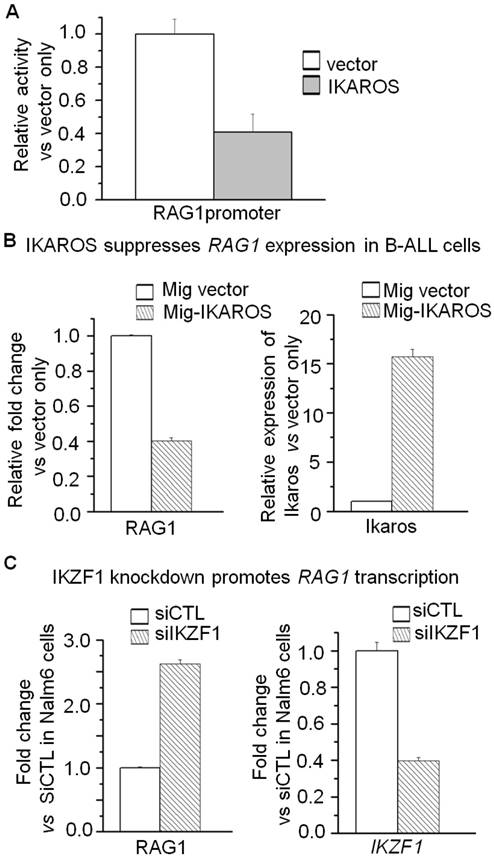
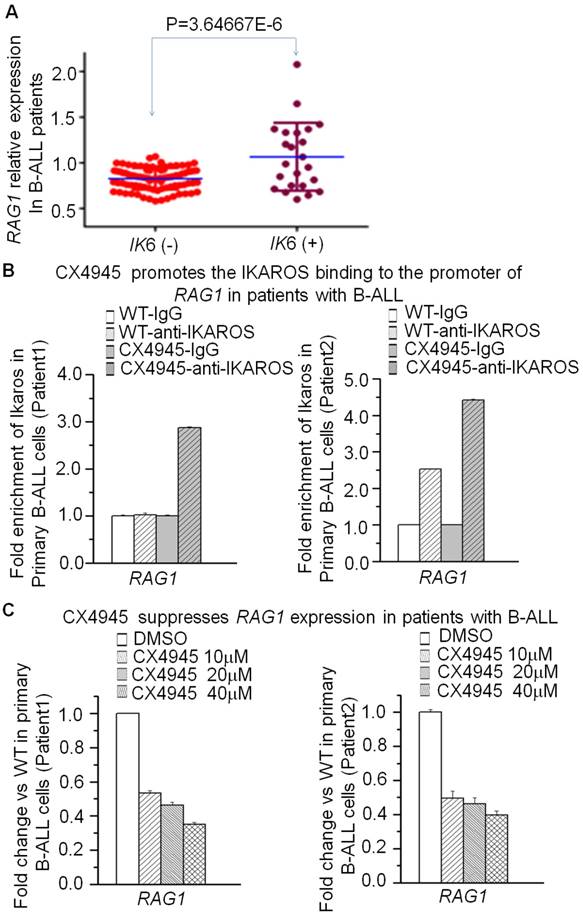
To explore the clinical association of IKAROS and RAG1 expression, we observed the significantly negative correlation of RAG1 mRNA level with IKZF1 expression in the cohort of B-ALL and B lymphoma patients (Supplemental Fig 3). More importantly, we analyzed the correlation of IKZF1 deletion with RAG1 expression in patients with B-ALL. A higher incidence of IK6, the most common protein produced by IKZF1 deletion was observed in B-ALL patients with the high RAG1 expression group (45.5% vs 26.2%, P=0.021) (Supplemental Table 1); and RAG1 mRNA level was significantly higher in patients with IKZF1 deletion than those without IKZF1 deletion (Fig 6A). These data indicate that IKZF1 deletion is associated with upregulation of RAG1 expression in primary B-ALL cells.
We have shown that CK2 inhibitor (CX4945) suppressed RAG1 expression in Nalm6 cells. To further demonstrate the potential clinical significance, we treated the primary cells from the patients with B-ALL. CX4945 could increase the binding of IKAROS on the RAG1 promoter region in primary B-ALL as compared to untreated controls (Fig 6B). In addition, CX4945 treatment inhibited expression of RAG1 mRNA in a dose-dependent manner in primary B-ALL cells (Fig 6C). These results indicate that IKZF1 binds to the promoter of RAG1 and that treatment with CK2 inhibitors, which can restore IKZF1 tumor suppressor activity, results in suppression of RAG1 expression in primary cells from B-ALL patients.
Correlation of RAG1 high expression with IKZF1 deletion in primary ALL cells. (A)Comparison of RAG1 expression in patients with or without IKZF1 deletion presented as RAG1/18s rRNA. The detection method for Ik6 (the most common IKAROS deletion) was done as our previously reported [23]. (B) CK2 inhibitor-CX4945 increased the IKZF1 binding to the promoters of RAG1 in primary B-ALL cells. (C) Effect of CK2 inhibitor-CX4945 functioning as IKZF1 activator on the expression of RAG1 in primary B-ALL cells with CX4945 treatment for 2 days.
Discussion
We observed that RAG1 is significantly higher in patients with B-ALL. Higher expression of RAG1 is associated with proliferation markers in B-ALL. IKAROS is crucial for the development of the hematopoietic system and its genetic deletion/mutations have been linked to the development of lymphoid leukemia. In particular, IKAROS functions as a major tumor suppressor involved in human B-cell acute lymphoblastic leukemia. In addition to a genetic defect, we also found that CK2 mediated-phosphorylation of IKAROS is another key reason leading to IKAROS dysfunction in ALL. Here, we found IKAROS directly suppresses RAG1 expression in B-ALL cells; CK2 inhibitor-CX4945 could suppress RAG1 expression by restoring IKAROS function in B-ALL cells. IKZF1 deletion is significantly associated with RAG1 high expression in B-ALL patients. These data reveal that RAG1 high expression together with IKAROS dysfunction may be responsible for the oncogenesis in B-ALL.
RAG1 is the key component of RAG complex which is the predominant driver of oncogenic genomic deletion and translocation, and also the cause of the clonal expansion in ALL.RAG complexes are constitutively expressed in pro- and pre-B cells and target recombination signal sequences (RSSs) in both Ig and non-Ig genes. Motif analyses show that deletion breakpoints in the tumor suppressor genes such as IKZF1, etc. have RSSs motif and RAG complex target to the RSS motifs [12, 13, 23]. The patients with IKZF1 deletion and mutations have increased risk of relapses and poor prognosis. Here we observed that RAG1 high expression is associated with IKZF1 deletion and patients with RAG1 high expression have a significantly higher detective rate of Ik6, the most common isoform of IKZF1 deletion. These data suggest RAG1 high expression may also affect IKZF1 deletion in adult B-ALL. Therefore, both RAG1 high expression and IKZF1 deletion may work together to amplify the oncogenic signaling responsible for oncogenesis in B-ALL (Supplemental Fig 4).
It is also reported that RAG-mediated recombination is the predominant driver of oncogenic rearrangement ETV6-RUNX1 in ALL [12] and plays critical roles underlying clonal expansion in pediatric ALL [12, 13]. Particularly RAG complex drives leukemic clonal evolution with repeated exposure to inflammatory stimuli, paralleling chronic infections in childhood under the same conditions [13]. We did observe the RAG1 high expression is associated with a high frequency of CRLF2 mutations in addition to IKZF1 deletion (Supplemental Table 1, Fig 2C). It is reported that both IKZF1 deletion and CRLF2 rearrangement are the drivers of high-risk ALL particularly the Ph-like ALL [17, 23-26]. Thus, these data suggested that high RAG1 expression may be also through oncogenic CRLF2 rearrangement to drive oncogenesis in B-ALL, although the association and underlying mechanisms of RAG activity with CRLF2 rearrangement need to be further determined.
Constitutive Rag expression is observed in a large portion of B-ALL cases [33]. Transcription factors such as FOXO1, FOXP1, NF-κB, PAX5β, E2A and Ikaros are reported to regulate Rag1 expression [34,35]. Here our data showed the Rag1 high expression in subsets of B-ALL and RAG1 high expression is associated with the cell proliferation markers in B-ALL, and also Rag1 expression is directly regulated by Ikaros in B-ALL. The RAG1 high expression may result in the Ikaros deletion and CRLF2 rearrangements, and then Ikaros dysfunction and may result in the Ikaros suppression on RAG1 expression and further induce the RAG1 high expression. This might be a positive feedback loop to amplify the oncogenic signaling in B-ALL (Supplemental Fig 4). Restoring IKAROS function by CK2 inhibitor can suppress the RAG1 expression and block the oncogenic amplification loop.
In summary, we for the first time reported RAG1 is a direct target of tumor suppressor gene IKZF1. We found that IKZF1 deletion (characterized by IK6 expression) is significantly correlated with high RAG1 expression. RAG1 is highly expressed and significantly correlated with high proliferation markers in a cohort of adult B-ALL patients. Our data suggested RAG1 high expression work together with IKZF1 deletion may drive the oncogenesis in B-ALL.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported in part by Milstein Medical Asian American Partnership (MMAAP) Foundation Research Project Award in Hematology (2017); The National Natural Science Foundation of China (81770172, 81270613); The Key Research & Technology Projects in Jiangsu Province (BE2017747); The Fundamental Research Funds for the Central Universities (2242017K40271, 2242016K40143); Jiangsu Provincial Key Medical Discipline (Zdxkb2016020); Jiangsu Province Key Medical Talents (RC2011077); The Scientific Research Foundation for the Returned Overseas Chinese Scholars; State Education Ministry (39th); China Postdoctoral Science Foundation (20090461134); Special grade of the financial support from China Postdoctoral Science Foundation (201003598); The Six Great Talent Peak Plan of Jiangsu (2010-WS-024) (to ZG). This work has also been partially supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) grants (R01CA209829, R01CA213912), Hyundai Hope on Wheels Scholar Grant, the Four Diamonds Fund of the Pennsylvania State University College of Medicine (to SD and CS); Bear Necessities Pediatric Cancer Foundation, Alex's Lemonade Stand Foundation, and the John Wawrynovic Leukemia Research Scholar Endowment (to SD).
Ethics approval and consent to participate
The written informed consent from all the patients was obaitned in accordance with the Declaration of Helsinki before enrollment in the study. The Institutional Review Board of the Nanjing Medical University and Zhongda Hospital Southeast University, Nanjing, China approved this study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Haines BB, Ryu CJ, Chen J. Recombination activating genes (RAG) in lymphoma development. Cell Cycle. 2006;5(9):913-916
2. Lee BS, Dekker JD, Lee BK. et al. The BCL11A transcription factor directly activates RAG gene expression and V(D)J recombination. Mol Cell Biol. 2013;33(9):1768-1781
3. Chen Z, Xiao Y, Zhang J. et al. Transcription factors E2A, FOXO1 and FOXP1 regulate recombination activating gene expression in cancer cells. PLoS One. 2011;6(5):e20475
4. Mijuskovic M, Chou YF, Gigi V. et al. Off-Target V(D)J Recombination Drives Lymphomagenesis and Is Escalated by Loss of the Rag2 C Terminus. Cell Rep. 2015;12(11):1842-1852
5. Deriano L, Chaumeil J, Coussens M. et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471(7336):119-123
6. Qiu JX, Kale SB, Yarnell Schultz H. et al. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7(1):77-87
7. Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13(23):3059-3069
8. Puiggros A, Venturas M, Salido M. et al. Interstitial 13q14 deletions detected in the karyotype and translocations with concomitant deletion at 13q14 in chronic lymphocytic leukemia: different genetic mechanisms but equivalent poorer clinical outcome. Genes Chromosomes Cancer. 2014;53(9):788-797
9. Wiemels J. Chromosomal translocations in childhood leukemia: natural history, mechanisms, and epidemiology. J Natl Cancer Inst Monogr. 2008;2008(39):87-90
10. Nickoloff JA, De Haro LP, Wray J. et al. Mechanisms of leukemia translocations. Curr Opin Hematol. 2008;15(4):338-345
11. Gollin SM. Mechanisms leading to nonrandom, nonhomologous chromosomal translocations in leukemia. Semin Cancer Biol. 2007;17(1):74-79
12. Papaemmanuil E, Rapado I, Li Y. et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116-125
13. Swaminathan S, Klemm L, Park E. et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16(7):766-774
14. Song C, Gowda C, Pan X. et al. Targeting casein kinase II restores IKAROS tumor suppressor activity and demonstrates therapeutic efficacy in high-risk leukemia. Blood. 2015;126(15):1813-1822
15. Song C, Pan X, Ge Z. et al. Epigenetic regulation of gene expression by IKAROS, HDAC1 and Casein Kinase II in leukemia. Leukemia. 2016;30(6):1436-1440
16. Ge Z, Guo X, Li J. et al. Clinical significance of high c-MYC and low MYCBP2 expression and their association with IKAROS dysfunction in adult acute lymphoblastic leukemia. Oncotarget. 2015;6(39):42300-42311
17. Ge Z, Gu Y, Xiao L. et al. Co-existence of IL7R high and SH2B3 low expression distinguishes a novel high-risk acute lymphoblastic leukemia with IKAROS dysfunction. Oncotarget. 2016;7(29):46014-46027
18. Ge Z, Zhou X, Gu Y. et al. IKAROS regulation of the BCL6/BACH2 axis and its clinical relevance in acute lymphoblastic leukemia. Oncotarget. 2017;8(5):8022-8034
19. Wang H, Song C, Ding Y. et al. Transcriptional Regulation of JARID1B/KDM5B Histone Demethylase by IKAROS, Histone Deacetylase 1 (HDAC1), and Casein Kinase 2 (CK2) in B-cell Acute Lymphoblastic Leukemia. J Biol Chem. 2016;291(8):4004-4018
20. Ge Z, Gu Y, Han Q. et al. Targeting High Dynamin-2 (DNM2) Expression by Restoring IKAROS Function in Acute Lymphoblastic Leukemia. Sci Rep. 2016;6:38004
21. Schwickert TA, Tagoh H, Gultekin S. et al. Stage-specific control of early B cell development by the transcription factor IKAROS. Nat Immunol. 2014;15(3):283-293
22. Iacobucci I, Storlazzi CT, Cilloni D. et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell'Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood. 2009;114(10):2159-2167
23. Mullighan CG, Miller CB, Radtke I. et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of IKAROS. Nature. 2008;453(7191):110-114
24. Churchman ML, Mullighan CG. IKAROS: Exploiting and targeting the hematopoietic stem cell niche in B-progenitor acute lymphoblastic leukemia. Exp Hematol. 2017;46:1-8
25. Ge Z, Gu Y, Zhao G. et al. High CRLF2 expression associates with IKZF1 dysfunction in adult acute lymphoblastic leukemia without CRLF2 rearrangement. Oncotarget. 2016;7(31):49722-49732
26. Lin ZK, Zhang R, Ge Z. et al. [Mutation and expression of PAX5 gene in adult acute lymphoblastic leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22(5):1206-1211
27. Liu P, Ge Z, Lin ZK. et al. [Expression characteristics of isoforms of IKAROS and Helios in patients with leukemia and their mechanism]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2012;20(4):812-817
28. Harvey RC, Mullighan CG, Chen IM. et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312-5321
29. Mullighan CG, Collins-Underwood JR, Phillips LA. et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41(11):1243-1246
30. Yoda A, Yoda Y, Chiaretti S. et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2010;107(1):252-257
31. Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125(26):3977-3987
32. Campana D, Janossy G, Bofill M. et al. Human B cell development. I. Phenotypic differences of B lymphocytes in the bone marrow and peripheral lymphoid tissue. J Immunol. 1985;134(3):1524-1530
33. Steenbergen EJ, Verhagen OJ, Van Leeuwen EF. et al. Distinct ongoing Ig heavy chain rearrangement processes in childhood B-precursor aute lymphoblatic leukemia. Blood. 1993;82(2):581-9
34. Ochodnicka-Mackovicova K, Bahjat M, Maas C. et al. The DNA Damage response regulates RAG1/2 expression in pre-B cells through ATM-FOXO1 Signaling. J Immunol. 2016;197(7):2918-2929
35. Chen Z, Xiao Y, xhang J. et al. Transcription factors E2A, FOXO1 and FOXP1 regualte recombination activating gene expression in cancer cells. PloS One. 2011;6(5):e20475
Author contact
![]() Corresponding author: Zheng Ge, M.D., Ph.D., Zhongda Hospital, Medical School of Southeast University, Department of Hematology, Institute of Hematology Southeast University, Nanjing 210009, China. Telephone: 86-25-83262468; FAX: 86-25-83262471; E-mail: Janege879com
Corresponding author: Zheng Ge, M.D., Ph.D., Zhongda Hospital, Medical School of Southeast University, Department of Hematology, Institute of Hematology Southeast University, Nanjing 210009, China. Telephone: 86-25-83262468; FAX: 86-25-83262471; E-mail: Janege879com

 Global reach, higher impact
Global reach, higher impact