Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3871-3882. doi:10.7150/jca.31266 This issue Cite
Research Paper
SIRT5 Promotes Hepatocellular Carcinoma Progression by Regulating Mitochondrial Apoptosis
1. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, The Second Hospital of Dalian Medical University, NO.467, Zhongshan Road, Dalian, Liaoning 116023, China.
2. Department of Vascular Surgery, The Second Hospital of Dalian Medical University, NO.467, Zhongshan Road, Dalian, Liaoning 116023, China.
3. CAS Key Lab of Separation Sciences for Analytical Chemistry, National Chromatographic Research and Analysis Center, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, NO.457, Zhongshan Road, Dalian, Liaoning 116023, China.
*These authors have contributed equally to this work.
Received 2018-11-6; Accepted 2019-5-12; Published 2019-6-10
Abstract
SIRT5 belongs to a family of NAD+-dependent lysine deacetylases called sirtuins. Although accumulating evidence indicates SIRT5 upregulation in cancers, including liver cancer, the detailed roles and mechanisms remain to be revealed. Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related deaths among men worldwide, and finding effective targets for HCC treatment and prevention is urgently needed. In the present study, we confirmed that mitochondrial sirtuins, particularly SIRT5, are more highly expressed in HCC cell lines than in normal liver cell lines. Moreover, SIRT5 knockdown suppresses HCC cell proliferation and SIRT5 overexpression promotes HCC cell proliferation. Furthermore, we verified that SIRT5 knockdown increases HCC cell apoptosis via the mitochondrial pathway. By co-IP and western blotting, we illustrated that SIRT5 deacetylates cytochrome c thus regulating HCC cell apoptosis. Taken together, our findings suggest that SIRT5 may function as a prognostic factor and drug target for HCC treatment.
Keywords: SIRT5, HCC, Cytochrome c, Acetylation
Introduction
Sirtuins (SIRTs) are a family of nicotinamide adenine dinucleotide (NAD+)-dependent lysine deacetylases. SIRTs are characterized by a highly conserved SIRT catalytic core domain [1]. SITRs are mammalian orthologues of the silent information regulator (SIR) 2 protein which was the first reported sirtuin gene from Saccharomyces cerevisiae [2, 3]. Seven SIRTs (SIRT1-7) exist in humans, and these SIRTs have different locations. SIRT1, SIRT6 and SIRT7 are present primarily in the nucleus. SIRT2 exists in the cytoplasm, while SIRT3, SIRT4 and SIRT5 are localized in mitochondria [4]. SIRTs participate in diverse cellular processes, such as cell fate, DNA stress repair, ageing and metabolism [5, 6] and play important roles in various diseases, such as neurodegenerative diseases [7, 8], cardiovascular diseases [9] and type Ⅱ diabetes [10]. SIRT5, a member of the SIRT family, is localized mainly in the mitochondria. SIRT5 has diverse catalytic activities of desuccinylation, deglutarylation and demalonylation, as well as roles in mitochondrial metabolism [11]. Currently, the most important roles of SIRT5 has been reported in the urea cycle in mitochondria. SIRT5 can deacetylate and stimulate carbamoyl phosphate synthetase (CPS1), leading to ammonia detoxification [12]. SIRT5 could regulate fatty acid metabolism through desuccinylase hydroxyl-coenzyme A dehydrogenase (HADH) which is an essential enzyme in fatty acid metabolism [13]. In mitochondrial oxidative phosphorylation, SIRT5 suppresses the biochemical activity of the important TCA cycle enzyme succinate dehydrogenase (SDH) and reduces the mitochondrial respiration driven by SDH [13]. Hala et al. reported that SIRT5 deletion enhanced mitochondrial DRP1 accumulation, leading to mitochondrial fragmentation and degradation during autophagy [14]. Excess mitochondrial reactive oxygen species (ROS) increase cellular oxidative stress, and SIRT5 could enhance cellular antioxidant defense by deglutarylating glucose-6-phosphate dehydrogenase (G6PD) and desuccinylating isocitrate dehydrogenase 2 (IDH2) [15]. SIRT5 could also eliminate ROS through desuccinylating and activating Cu/Zn superoxide dismutase 1 (SOD1) [16]. Furthermore, SIRT5 was also reported to be a regulator of mitochondrial energy metabolism, and SIRT5 overexpression increased ATP synthesis and oxygen consumption in HCC cell line HepG2 [17].
Researches on the relation between SIRT and cancers have been continuous since SIRTs were identified [18, 19]. However, correlations between SIRT5 and cancers have rarely been reported. The roles of SIRT5 in cancers were studied and revealed only recently. Existing evidence suggests that SIRT5 functions as an oncogene. In non-small cell lung cancer (NSCLC) SIRT5 could promote cell growth and enhance drug resistance [20]. Based on TCGA data, Gong et al revealed that SIRT5 was highly expressed in squamous cell carcinoma and that high SIRT5 expression was associated with poor overall survival in NSCLC patients [21]. In colorectal cancer (CRC), SIRT5 also promotes colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner [22]. Moreover, SIRT5 could promote CRC cell respiration, proliferation and tumour growth by deacetylating lactate dehydrogenase B (LDHB), a glycolytic enzyme that catalysis the conversion of lactate and NAD+ to pyruvate [23]. In triple-negative breast cancer (TNBC), SIRT5 is upregulated markedly and high SIRT5 expression has been associated with poor clinical prognosis [24]. SIRT5 could also control ammonia production by regulating glutaminase and glutamine metabolism, resulting in ammonia-induced autophagy and mitophagy in breast carcinoma and mouse myoblastoma cells [25]. SIRT5 has been reported to increase SHMT2 activity and enhance serine catabolism, leading to tumor cell proliferation [26]. In HEK293T cells, SIRT5 could bind to and desuccinylate pyruvate kinase M2 (PKM2), thus stimulating in cell proliferation and tumor growth [27]. On the other hand, SIRT5 has also reported to suppress the tumor progression. In gastric cancer, SIRT5 inhibits gastric cancer cell proliferation by suppressing aerobic glycolysis [28]. Furthermore, SIRT5 could facilitate S100A10 degradation, thus inhibiting the role of S100A10 in promoting gastric cancer invasion and migration [29]. Consequently, additional studies should be performed to explore the positive or negative roles of SIRT5 in more cancers.
Primary liver cancer is the fifth most prevalent malignant tumor and the second leading cause of cancer-related deaths among men worldwide, particularly in less developing countries, primary liver cancer accounts for approximately 50% of the total number of cases and deaths occurring in China alone [30]. According to Chinese statistical data, liver cancer was estimated as the first killer and most common cancer of men younger than 60 years old in 2015 [31]. Although the incidence of liver cancer has decreased gradually, the estimated deaths from liver cancer still ranked fifth among all cancers in males in the United States [32]. Hepatocellular carcinoma (HCC) is the most common and accounts for 85-90% of total primary liver cancer cases [33]. Surgical resection remains the most common therapeutic strategy for resectable HCC patients; however, many advanced HCC patients are not suitable for surgery [34] and the recurrence rate after surgical resection for HCC remains approximately 50-70% [35]. Liver transplantation, embolization, ablation, radiation therapy and chemotherapy have provided partial curative interventions. However, only approximately 30% of HCC patients meet the criteria for all the above therapies [36]; furthermore, despite so many therapies, the HCC five-year survival rate is as low as 17% [37]. Therefore, identifying new treatment targets or developing novel strategies for HCC management is urgently needed.
The role of SIRT5 in cancers has not been identified completely. Before 2013, few studies reported the effects of SIRT5 in HCC. Recently, an increasing number of studies have gradually revealed the role of SIRT5 in HCC. In fact, high-levels of gene amplification and expression have been reported for SIRT5 in liver cancer [38]. SIRT5 expression is upregulated in HCC tissues and cell lines, and its overexpression is positively correlated with tumor size, lymph node metastasis and TNM stage. SIRT5 upregulation also indicates poor clinical prognosis in HCC patients. SIRT5 directly targets and activates E2F transcription factor 1 (E2F1) thus promoting HCC cell proliferation and invasion [39]. However, the mechanisms of SIRT5 regulation in HCC need to be studied further to achieve complete understanding.
In the present study, we selected two HCC cell lines (Bel-7402, Huh7) to confirm the effects of SIRT5 in HCC. We found that SIRT5 could facilitate HCC progression by inhibiting HCC cell apoptosis. Furthermore, we revealed that SIRT5 could deacetylate cytochrome c (cyt c) and regulate cyt c distribution in the cytoplasm and mitochondria to suppress mitochondrial apoptosis. Our results suggest that SIRT5 is a potential marker and drug target for HCC diagnosis and treatment.
Materials and Methods
Reagents and antibodies
A Cell Cycle Detection Kit, Annexin V-APC/propidium iodide (PI) Apoptosis Detection Kit, JC-1 Apoptosis Detection Kit, and BCA Protein Quantitation Assay Kit were purchased from KeyGen Biotech Co., Ltd. (Nanjing, China). A Cell Counting Kit-8 assay was obtained from Bimake (Houston, TX, USA).
The following primary antibodies were used for western blot analysis: cleaved-caspase3 (cl-cas3), cleaved-PARP (cl-PARP), Bax, Bcl-2, cyt c, Cox IV, pan acetylation, β-actin (Proteintech, IL, USA), SIRT5 (Abcam, MA, USA). Antibodies against cyt c for the Co-IP assay were purchased from Cell Signaling Technology (Danvers, MA, USA). Peroxidase-conjugated polyclonal goat anti-rabbit and polyclonal goat anti-mouse IgG antibodies were obtained from Thermo-Pierce (IL, USA).
Cell culture
The human normal liver cell LO2 and hepatocellular carcinoma cell lines SK-Hep-1, 97-L, 97-H, Huh7, Bel-7402, Bel-7402/Fu were obtained from the tumor cell bank of the Chinese Academy of Medical Science. Cells were cultured in RPMI-1640 medium (Gibco, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA), 100units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, USA) in an incubator with 5% CO2 at 37℃.
Transfection
Cells (2 x 105) were seeded in 6-well plates, cultured for 24 hours and then transfected with 100 pmol siRNA using Lipofectamine 2000 (Invitrogen, USA) in serum-free medium according to the manufacturer's protocol. After 4-6 hours, the medium was replaced with complete culture medium. After 2 days, the transfected cells were harvested for protein extraction or used for other experiments. All siRNAs were obtained from GenePharma (Shanghai, China). The 3 SIRT5 siRNA sequences were as follows: human SIRT5 siRNA#1, 5'-GCCAAGUUCAAGUAUGGCATT-3'; siRNA#2, 5'-GCUGGUGUUAGUGCAGAAATT-3'; siRNA#3, 5'-GCAUCCCAGUUGAGAAACU-3'. The SIRT5 overexpressing lentivirus and the control lentivirus were purchased from GenePharma (Shanghai, China). 106 lentiviruses were added to each single well with gentle mixing and puromycin was used to select the SIRT5 overexpressing cell line.
Cell viability assay
Cell viability was determined by CCK-8 assay according to the manufacturer's instructions. Approximately 5000 cells/well were seeded into 96-well plates. After 24 hours, the cells were transfected with siRNA. The cells were cultured for another 24, 48 and 72 hours; medium was exchanged for 100 µl of RPMI-1640 and 10 µl of CCK-8 reagent. Then cells were incubated at 37℃ for 2 hours. Finally, light absorbance was measured at 450 nm using a microplate reader.
Colony formation assay
Cells were seeded in triplicate into a 6-well culture dish at 2000 cells/well. Cells were transfected with siRNA after seeding for 24 hours. Then, cells were cultured for 2 weeks, washed three times with PBS, fixed in methanol for 15 minutes and stained with a crystal violet solution for 15 minutes. Visible colonies were counted according to the cell numbers in each colony.
Cell apoptosis analysis
An Annexin V-APC/PI Apoptosis Detection Kit was used to quantify the percentage of apoptotic cells. After seeding in 6-well plates for 24 hours, cells were transfected with the siRNAs. After another 48 hours cells were harvested and stained with 5 µl of Annexin V-APC and 5 µl of PI in 500 µl of an apoptosis reaction solution in the dark at room temperature for 30 minutes. The stained cells were analysed using a BD FACSCantoTMII flow cytometer.
Cell cycle analysis
Cells were seeded in 6-well plates for 24 hours and then transfected with the siRNAs and cultured for 48 hours. The cells were harvested and resuspended in 2 ml of ice-cold 70% ethanol at 4℃ overnight. The fixed cells were washed with PBS and incubated with 100 µl of RNase A for 30 minutes at 37℃, stained with 400 µl of PI and placed in the dark at temperature for 30 minutes. The stained cells were analysed using a BD FACSCantoTM II flow cytometer.
Mitochondrial membrane potential detection
Mitochondrial membrane potential change were detected with JC-1 apoptosis detection kit. Harvested cells were washed with PBS twice and resuspended in 500 µl of incubation buffer containing 1 µl of JC-1. The cells were incubated with 5% CO2 at 37℃ for 20 minutes, washed twice and resuspended in incubation buffer. The stained cells were then analysed by a BD FACSCantoTM II flow cytometer.
Mitochondria isolation
A Mitochondria Isolation Kit (KeyGen Biotech, Shanghai, China) was used to isolate mitochondria according to the manufacturer's instructions. A total of 5×107 cells were harvested and washed with PBS at 4℃. The cells were resuspended in 0.8 ml of pre-cooling lysis buffer and ground 40 times. The cell lysates were washed with 0.2 ml of medium and centrifuged at 1200×g and 4℃ for 5 minutes. The supernatants were further centrifuged at 7000×g and 4℃ for 10 minutes. The precipitates were suspended with 0.3 ml of suspension buffer and centrifuged at 9500×g and 4℃ for 10 minutes. The precipitates were mitochondria.
Cellular and mitochondrial protein extraction
Harvested cells or mitochondria were lysed with RIPA lysis buffer (KeyGen Biotech, Shanghai, China) containing 1 mmol/l phosphatase inhibitor cocktail (KeyGen Biotech, Shanghai, China) and 1 mmol/l phenylmethylsulphonyl fluoride (KeyGen Biotech, Shanghai, China). The mixture was centrifuged for 10 minutes at 12000 x g, and the supernatant containing the proteins was saved.
Western blot analysis
Protein concentrations were determined using a BCA Protein Assay Kit and diluted with RIPA and loading buffer (KeyGen Biotech, Shanghai, China). Equal amounts of protein samples (30 µg) were separated by 8% or 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes (Bio-Rad, USA). The membranes were blocked with 5% skim milk for 2 hours at room temperature and incubated with primary antibodies (1:1000) and secondary antibodies (1:5000). Finally, the bands were detected by chemiluminescence.
Co-immunoprecipitation (co-IP) assay
The co-immunoprecipitated assay used Pierce Protein A/G Magnetic Beads (Rockford, IL, USA) according to the manufacturer's protocol. Cell lysates were combined with 10 µg of antibody and incubated for 1 hour with rotation overnight at 4℃. A 25 µl protein A/G magnetic beads slurry was added to the cell lysates and mixed with rotation at 4℃ for 30 minutes. The cell lysates were placed into a magnetic stand to collect the beads against the side of the tube, and the precipitates were saved. The beads were washed twice. Next, 100 µl of low-pH elution buffer was added and mixed for 15 minutes. The beads were separated magnetically and the supernatants were saved for next analysis.
Statistical analysis
SPSS 17.0 software was used for statistical analysis (IBM, USA). All experiments were repeated 3 times. The values are presented as the means ± standard deviation. Statistical analyses were performed using Student's t-test, and one-way ANOVA was used to analyse variance. A p-value < 0.05 represented statistical significance.
Results
SIRT5 is overexpressed in HCC cell lines
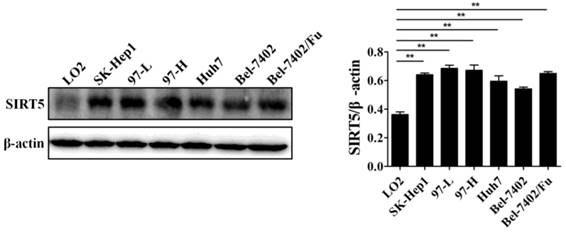
Previous work has demonstrated the role of SIRT5 in cancers including HCC. SIRT5 is overexpressed in HCC tissues and cells, and higher SIRT5 expression is associated lower overall survival in HCC patients [39]. We detected SIRT5 expression in a normal liver cell line (LO2) and different HCC cell lines (SK-Hep1, 97-L, 97-H, Huh7,Bel-7402, and Bel-7402/Fu which is resistant to 5-fluorouracil), and confirmed that SIRT5 is indeed upregulated in HCC cell lines compared to SIRT5 expression in the normal liver cells (Figure 1). In this study, we selected the Huh7 and Bel-7402 (Bel) cell lines in which SIRT5 was modestly higher than that in the LO2 normal liver cell line, to investigate SIRT5 roles and related mechanisms in HCC.
SIRT5 is overexpressed in most HCC cell lines. SIRT5 expression in different HCC cell lines and normal liver cell line was measured by western blotting. **P < 0.01.
SIRT5 knockdown suppresses HCC cell proliferation. A and B. Bel cells were transfected with siRNAs, the expression of SIRT5 was detected by western blotting. C and D. After transfection with control siRNA, siRNA#1 and siRNA#2, relative Bel and Huh7 cell viability was evaluated using a CCK-8 kit at 0 h, 24 h, 48 h, 72 h. E, F and G. After the Bel and Huh7 cells were transfected with control siRNA, siRNA#1 and siRNA#2, the cells were cultured for 2 weeks, and the colony numbers were counted. **P < 0.01.
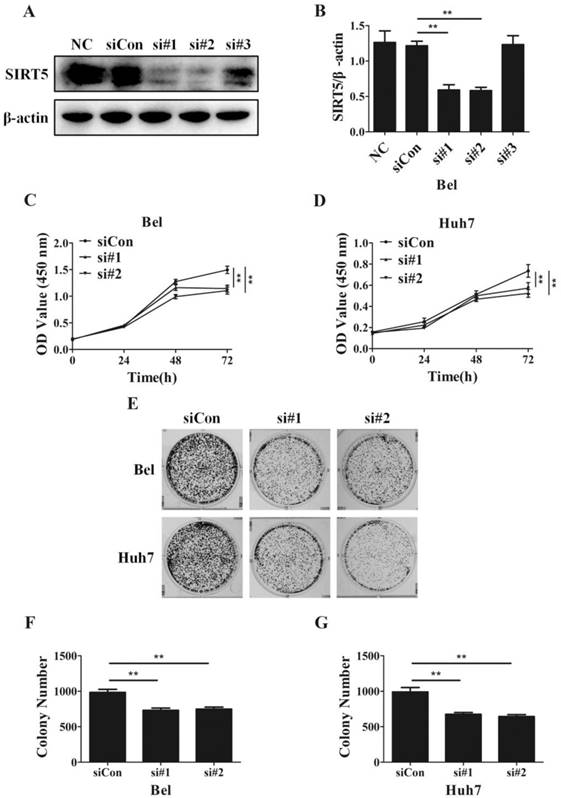
SIRT5 promotes the proliferation of HCC cells
Despite SIRT5 overexpression in HCC cells and tissues, SIRT5 specific roles in HCC still need to be determined completely. To investigate the effects of SIRT5 on HCC, we measured cell proliferation in HCC cells with different levels of SIRT5 expression. We introduced 3 different SIRT5-siRNAs (si#1-3) and a control-siRNA (siCon) into Bel cell lines for 48 hours. We measured SIRT5 expression by western blotting and selected si#1 and si#2 siRNAs, which possessed better interfering efficiency, to silence SIRT5 (Figure 2A, B). Using a CCK-8 cell viability assay, we found SIRT5 knockdown by si#1 and si#2 markedly decreased the cell proliferation rate (Figure 2C, D). We also conducted cell colony formation assays to determine whether SIRT5 knockdown inhibited proliferation. The number of colonies formed was 732.0 ± 31.7 for Bel-si#1 cells, 748.7 ± 27.8 for Bel-si#2 cells, 675.3 ± 24.8 for Huh7-si#1 cells and 644.3 ± 26.8 for Huh7-si#2 cells, while the number was 984.3 ± 42.4 and 990.7 ± 62.8 for Bel-siCon and Huh7-siCon cells respectively (Figure 2E-G). The results showed SIRT5 knockdown weakened HCC cell viability and proliferation ability.
Overexpression of SIRT5 promotes HCC cell proliferation. A, B and C. Bel and Huh7 cells were transfected with vector and SIRT5 lentivirus plasmids, the expression of SIRT5 was detected by western blot. D and E. After transfecttion with vector and SIRT5 lentivirus plasmids, relative Bel and Huh7 cell viability was measured using a CCK-8 kit at different time points. E, F and G. After Bel and Huh7 cells were transfected with vector and SIRT5 lentivirus plasmids, and the cells were cultured for 2 weeks, and the colony numbers were counted. **P < 0.01.
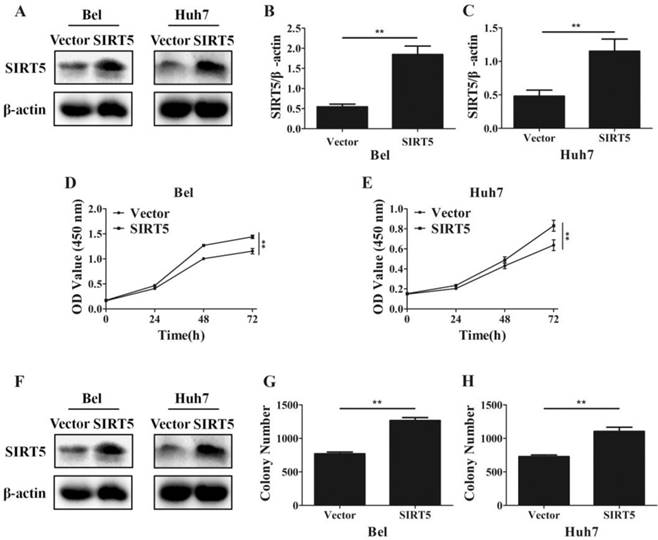
Furthermore, we established SIRT5 stable expression Bel (Bel-SIRT5) and Huh7 (Huh7-SIRT5) cells to verify that SIRT5 facilitated HCC cell proliferation. Western blotting showed SIRT5 was overexpressed in Bel-SIRT5 and Huh7-SIRT5 cells compared to that in Bel-vector and Huh7-vector cells respectively (Figure 3A-C). CCK-8 assays showed that the cell viability was significantly higher in SIRT5 stable expressed cells than in vector cells (Figure 3D, E). Cell colony formation assays also revealed a similar phenomenon, the number of colonies formed was 1267.0 ± 43.8 for Bel-SIRT5 cells, 1106.2 ± 61.1 for Huh7-SIRT5 cells, while the number was 770.0 ± 26.3 and 729.7 ± 23.6 for Bel-vector and Huh7-vector cells respectively (Figure 3F-H). Our results illustrated that SIRT5 could indeed promote HCC cells proliferation leading to HCC progression.
SIRT5 plays an important role in HCC cells apoptosis
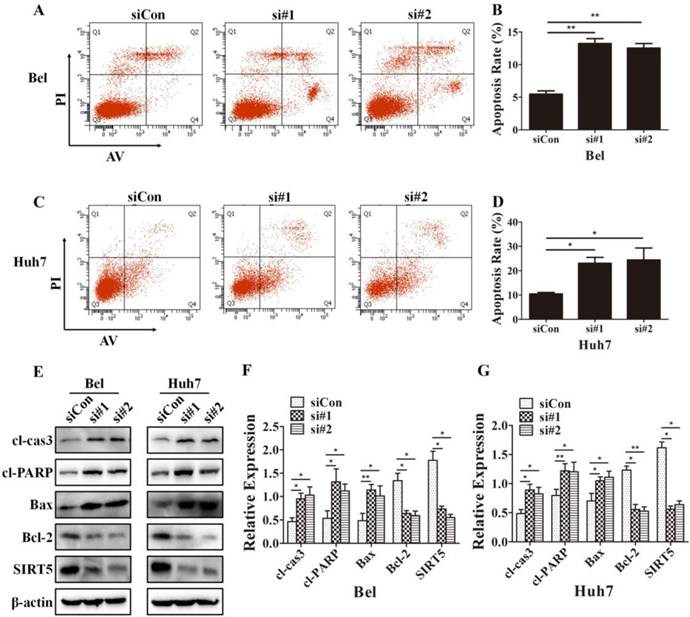
Because cells with different SIRT5 expression levels showed different proliferation rates, we further investigated whether SIRT5 affected cell apoptosis. We used Annexin V-APC/Propidium iodide (PI) staining-based fluorescence activated cell sorting (FACS) analysis to evaluate cell apoptosis. We observed the percentage of apoptotic cells was dramatically increased after SIRT5 knockdown. The percentages of apoptotic cells were 5.5 ± 0.5%, 13.2 ± 0.8% and 12.5 ± 0.7% in Bel-siCon, Bel-si#1, Bel-si#2 respectively (Figure 4A, B). Similar results were observed in Huh7 cells, the apoptotic cells percentages were 10.4 ± 0.6%, 23.0 ± 2.4% and 24.4 ± 4.9% in Huh7-siCon, Huh7-si#1, Huh7-si#2 in turn (Figure 4C, D). We subsequently compared the apoptosis-related proteins expression in cells with different SIRT5 expression levels. The pro-apoptotic proteins cl-cas3, cl-PARP, and Bax were upregulated, while the apoptosis suppressor Bcl-2 was downregulated (Figure 4E-G). These results revealed that SIRT5 could inhibit apoptosis in HCC cells.
Cell cycle dysregulation is a hallmark of cancers, we next determined the percentages of cell cycle distribution percentages by PI staining-based FACS analysis to explore whether SIRT5 could regulate the cell cycle. However, we did not observe differences between siCon and SIRT5 knockdown HCC cells (Supplementary material). The flow cytometry data indicated SIRT5 had no impact on HCC cell cycle.
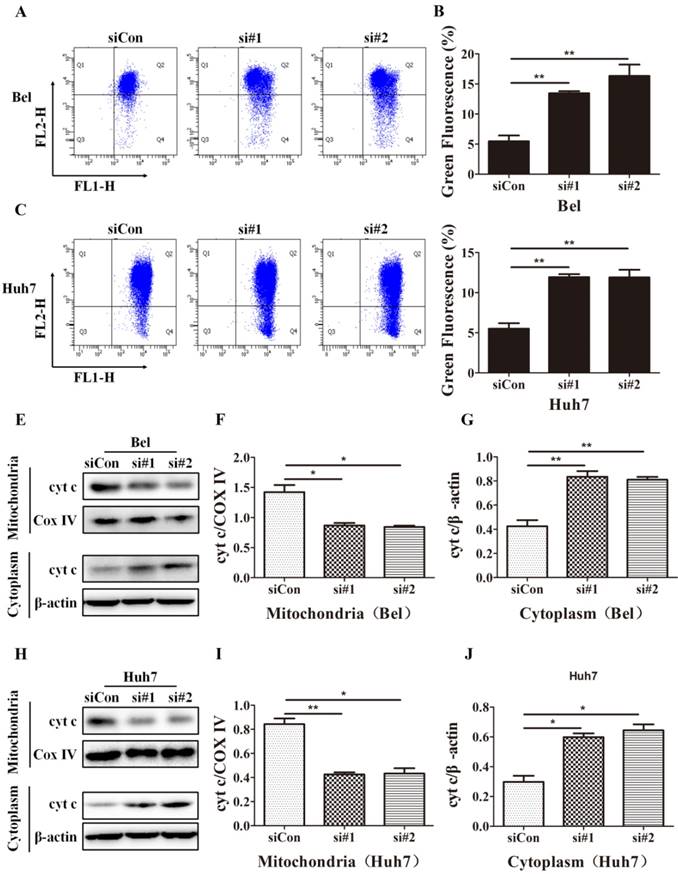
SIRT5 knockdown increases HCC cell apoptosis. A-D. After transfection with control siRNA, siRNA#1 and siRNA#2 in Bel and Huh7 cells, the apoptotic cells were stained with Annexin V-PI and detected by flow cytometry. E, F and G. The protein levels of cl-cas3, cl-PARP, Bax, Bcl-2 and SIRT5 were detected by western blotting. *P < 0.05, **P < 0.01.
SIRT5 regulates apoptosis through the mitochondrial pathway
Mitochondrial apoptosis is an important cell apoptosis pathway. As SIRT5 is located in mitochondria, we hypothesized that SIRT5 suppressed the mitochondrial pathway, thus inhibiting on apoptosis in HCC cells. In mitochondrial apoptosis, mitochondrial membrane permeability changes, lead to mitochondrial membrane potential disruption, this is a hallmark of mitochondrial apoptosis [40, 41]. To determine whether mitochondrial apoptosis is the underlying mechanism through which SIRT5 inhibits HCC cell apoptosis, we examined alterations in mitochondrial membrane potential with JC-1 dye, which is highly sensitive at detecting mitochondrial membrane potential alterations. Using flow cytometric analysis, we found that SIRT5 knockdown prominently decreased the mitochondrial membrane potential (Figure 5A-D).
Cyt c released from mitochondria to cytoplasm is another marker of mitochondrial apoptosis; this change is caused by increases in the mitochondrial outer membrane permeability [42]. To investigate the effect of SIRT5 on cyt c release, we extracted mitochondrial and cytoplasmic proteins and evaluated the cyt c distribution in cytoplasm and mitochondria. Western blotting results showed that SIRT5 knockdown significantly decreased cyt c distribution in mitochondria and increased cyt c distribution in cytoplasm (Figure 5E-J). These data demonstrated that SIRT5 regulated mitochondrial apoptosis, thus suppressing apoptosis in HCC cells.
SIRT5 deacetylates cyt c to induce mitochondrial apoptosis
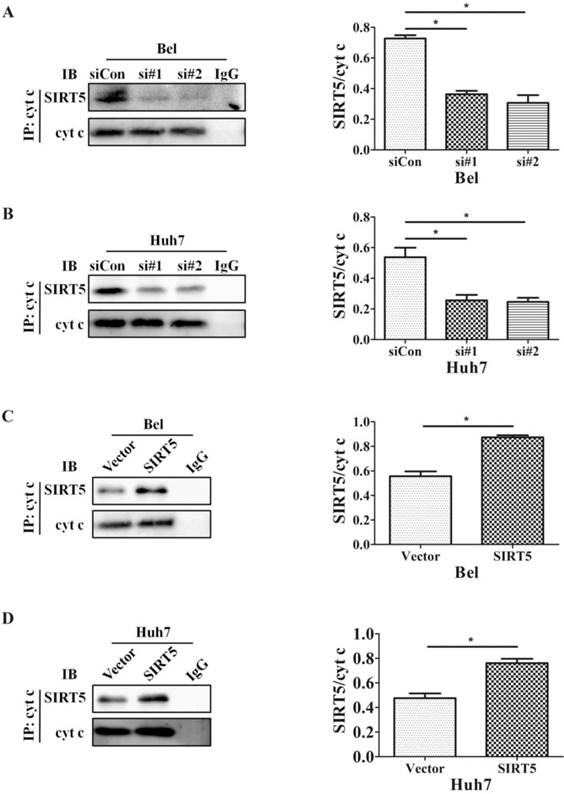
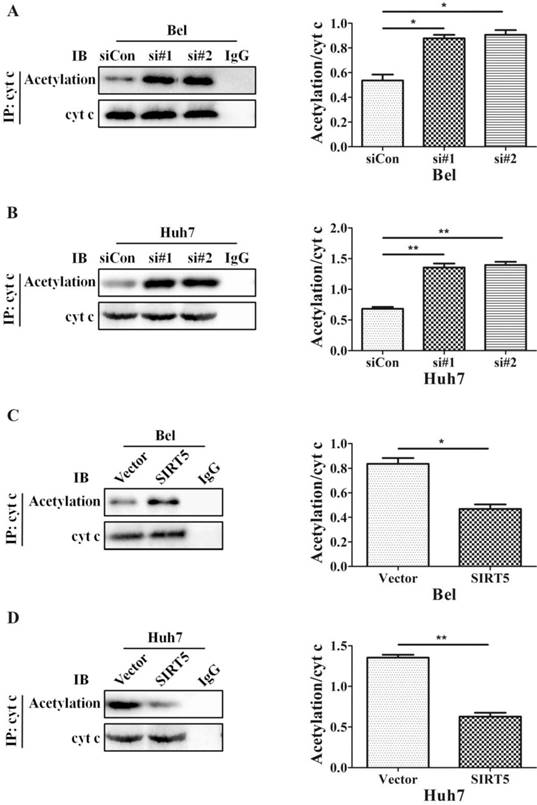
The SIRT family possesses potent deacetylase activity and can regulate cancer progression by deacetylating certain proteins. Previous studies showed SIRT1/SIRT5 play a vital role in modulating cancer cell survival by regulating promyelocytic leukemia protein deacetylation [43]. As a member of the SIRT family, SIRT5 is co-located with cyt c. Therefore, we hypothesized SIRT5 could regulate cyt c distribution by deacetylating cyt c. We used an anti-cyt c antibody to immunoprecipitate SIRT5 and found the association of SIRT5 and cyt c decreased considerably in SIRT5 knockdown cells (Figure 6A, B), while in SIRT5 stable expressed cells, the association of SIRT5 and cyt c was increased markedly (Figure 6C, D). These results indicated SIRT5 can combine with cyt c. We further detected cyt c acetylation levels by immunoprecipitation and western blotting. The results showed cyt c acetylation was higher in SIRT5 knockdown cells than that in siCon cells (Figure 7A, B), while cyt c acetylation was decreased significantly in SIRT5 overexpressing cells (Figure 7C, D). These data indicated SIRT5 could interact with cyt c and SIRT5-mediated cyt c deacetylation decreased cyt c release from mitochondria.
SIRT5 knockdown induces mitochondrial apoptosis in HCC cells. A-D. After control siRNA, siRNA#1 and siRNA#2 transfection, JC-1 dye was used to stain Bel and Huh7 cells, and alterations in mitochondrial membrane potential were detected by flow cytometric analysis. E-J. Cyt c in cytoplasm and mitochondria of Bel and Huh7 cells was evaluated by western blot. *P < 0.05, **P < 0.01.
SIRT5 interacts with cytochrome c. A and B. Silencing SIRT5 decreases the interaction between SIRT5 and cyt c in Bel and Huh7 cells. C and D. Overexpression of SIRT5 increases the interaction of SIRT5 and cyt c. *P < 0.05, **P < 0.01.
Discussion
Humans have made great efforts to conquer hepatocellular cancer, but liver cancer incidence and mortality have long remained high. Although more therapeutic strategies, such as surgical resection, liver transplantation, embolization, ablation, radiation therapy and chemotherapy have been utilized in the clinical treatment of liver cancer, the surgical recurrence and five-year survival rates remain unsatisfactory [34, 35, 37]. Thus, the development of new biomarkers or therapeutic targets for improving liver cancer diagnosis and outcomes is critical. Exploration studies for new markers or targets have focused on DNA, RNA, and proteins [44-46]. SIRTs play important roles in the progression of a wide variety of diseases, including cancers [46, 47]. As SIRTs are an NAD+-dependent lysine deacetylase family, they participate in cancer progression by regulating metabolism [48, 49]. As a member of SIRT family member, SIRT5 could also participate in cancer progression. SIRT5 has anti-apoptotic and anti-oxidative functions in neuroblastoma cells [50]. Similar functions were verified in other cancers including HCC [20, 22, 39]. However, the specific roles and mechanisms of SIRT5 in HCC have not been described in detail. In the present study, we confirmed SIRT5 was overexpressed in HCC cell lines which were consistent with previous studies, in which SIRT5 was overexpressed in liver and NSCLC and it could be an indicator of patient prognosis [20, 39].
We further explored the effects of SIRT5 on HCC cells. First, we showed that SIRT5 overexpression promoted HCC cell (Huh7 and Bel) proliferation, while SIRT5 knockdown could suppress HCC cell proliferation greatly. We also demonstrated that SIRT5 knockdown significantly inhibited HCC cell apoptosis. Additionally, we did not find positive or negative effects of SIRT5 on cell cycle. Although previous work has revealed several molecular pathways (Nrf2 and E2F1) involved in the positive effects of SIRT5 in cancers, the underlying mechanisms remain unknown.
SIRT5 regulates the acetylation of cytochrome c. A and B. Silencing SIRT5 decreases the acetylation of cyt c in Bel and Huh7 cells. C and D. SIRT5 Overexpression increases cyt c acetylation. *P < 0.05, **P < 0.01.
Apoptosis is a crucial feature of a variety of liver diseases, especially liver cancer. In liver cancer, cell apoptosis inhibition is often the cytological basis of liver tumor occurrence. Therefore, improvements in liver cancer diagnosis and treatment could be made from identifying liver cancer cell apoptosis mechanisms and promoting liver cancer cell apoptosis. SIRT3 is a tumor suppressor in HCC and could regulate mitochondrial apoptosis [51]. Thus, the anti-apoptotic role of SIRT5 may be one of the important underlying pathways; clarifying the mechanisms of apoptosis may help to illustrate the effects of SIRT5 in HCC. However, the anti-apoptotic mechanisms of SIRT5 have not been revealed. Similar to SIRT5 and SIRT3 localization, we confirmed that SIRT5 could inhibit HCC cell apoptosis by controlling mitochondrial apoptosis. In mitochondrial apoptosis, pro-apoptotic factors activate signaling and the pro-apoptotic protein Bax translocates to mitochondrial membranes thus changing mitochondrial membrane permeabilization. Then, the mitochondrial membrane is disrupted, and pro-apoptotic factors (such as cyt c and apoptosis protease-activating factor 1) in mitochondria are released into cytoplasm. In cytosol, cyt c combines with apoptosis protease-activating factor 1 (Apaf-1) and caspase-9 to constitute a large complex called the apoptosome. Subsequently, downstream caspases such as caspase-3 are activated by apoptosome leading to cell apoptosis [42]. Our present study showed that SIRT5 silencing induced mitochondrial membrane potential disruption, cyt c release from mitochondria to cytoplasm, and cl-cas3 upregulation, which indicated SIRT5 knockdown facilitated HCC cell mitochondrial apoptosis. We illustrated SIRT5 could suppress HCC cell mitochondrial apoptosis by decreasing cyt c release from mitochondria.
The most important role of SIRT5 is to induce the deacetylation of CPS1, which is the rate-limiting enzyme for urea synthesis in urea cycle. In normal liver cells, SIRT5 acts as a deacetylase to mediate ammonia detoxification by regulating urea cycle [12]. Acetylation is a reversible post-translational protein modification that plays vital roles in cancer carcinogenesis and development. Previous studies have reported that SIRT5 is able to co-localize with cyt c in the IMS and SIRT5 might regulate acetylation of cyt c [52, 53]. In our study, we further explored the role of SIRT5 in cyt c acetylation. Silencing SIRT5 increased acetylation of cyt c, while overexpressing SIRT5 decreased acetylation of cyt c. We demonstrated the important effect of acetylation on cyt c distribution in cells, and SIRT5 deacetylation of cyt c reduced cyt c release to cytoplasm, thus decreasing mitochondrial apoptosis in HCC cells.
In addition, we reported SIRT5 has positive roles in HCC cell proliferation, which indicated SIRT5 promoted liver tumorigenesis and liver cancer progression by mechanisms other than suppressing on HCC cell mitochondrial apoptosis. Furthermore, SIRT5 also has the activities of desuccinylation, deglytarylation and demalonylation, and many reports have indicated SIRT5 had same or stronger activity as a desuccinylase compared to that as a deacetylase in physiology and pathology [16, 54]. These results indicate that SIRT5 may function as a desuccinylase to facilitate liver carcinogenesis and liver cancer development, and encourage us to explore more effects and mechanisms of SIRT5 in HCC.
In summary, our findings identified the positive role of SIRT5 in HCC cell proliferation and apoptosis inhibition. This study illustrates a novel mechanism by which SIRT5 could suppress HCC cell mitochondrial apoptosis through deacetylation of cyt c. Overall, this work supports that SIRT5 could be a potential marker and target in clinical HCC diagnosis and therapy.
Supplementary Material
Supplementary figure.
Acknowledgements
This research was supported by National Natural Science Foundation of China (No. 81272368 and 81471755) and Clinical Capability Construction Project for Liaoning Provincial Hospitals (LNCCC-B03-2014).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1-13
2. Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913-21
3. Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793-8
4. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623-35
5. Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587-91
6. Merksamer PI, Liu Y, He W, Hirschey MD, Chen D, Verdin E. The sirtuins, oxidative stress and aging: an emerging link. Aging. 2013;5:144-50
7. Albani D, Polito L, Forloni G. Sirtuins as novel targets for Alzheimer's disease and other neurodegenerative disorders: experimental and genetic evidence. J Alzheimers Dis. 2010;19:11-26
8. Esteves AR, Lu J, Rodova M, Onyango I, Lezi E, Dubinsky R. et al. Mitochondrial respiration and respiration-associated proteins in cell lines created through Parkinson's subject mitochondrial transfer. J Neurochem. 2010;113:674-82
9. Ota H, Eto M, Ogawa S, Iijima K, Akishita M, Ouchi Y. SIRT1/eNOS axis as a potential target against vascular senescence, dysfunction and atherosclerosis. Journal of Atherosclerosis and Thrombosis. 2010;17:431-5
10. Avogaro A, de Kreutzenberg SV, Fadini GP. Insulin signaling and life span. Pflugers Arch. 2010;459:301-14
11. Roessler C, Tuting C, Meleshin M, Steegborn C, Schutkowski M. A novel continuous assay for the deacylase Sirtuin 5 and other deacetylases. J Med Chem. 2015;58:7217-23
12. Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560-70
13. Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919-30
14. Guedouari H, Daigle T, Scorrano L, Hebert-Chatelain E. Sirtuin 5 protects mitochondria from fragmentation and degradation during starvation. Biochim Biophys Acta Mol Cell Res. 2017;1864:169-76
15. Zhou L, Wang F, Sun R, Chen X, Zhang M, Xu Q. et al. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep. 2016;17:811-22
16. Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB. et al. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. 2013;441:191-5
17. Buler M, Aatsinki SM, Izzi V, Uusimaa J, Hakkola J. SIRT5 is under the control of PGC-1alpha and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J. 2014;28:3225-37
18. Voelter-Mahlknecht S, Mahlknecht U. The sirtuins in the pathogenesis of cancer. Clin Epigenetics. 2010;1:71-83
19. Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015;15:608-24
20. Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol. 2014;35:10699-705
21. Gong J, Wang H, Lou W, Wang G, Tao H, Wen H. et al. Associations of sirtuins with clinicopathological parameters and prognosis in non-small cell lung cancer. Cancer Manag Res. 2018;10:3341-56
22. Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL. et al. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun. 2018;9:545
23. Shi L, Yan H, An S, Shen M, Jia W, Zhang R. et al. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol Oncol. 2019;13:358-75
24. Xu L, Che X, Wu Y, Song N, Shi S, Wang S. et al. SIRT5 as a biomarker for response to anthracycline-taxane-based neoadjuvant chemotherapy in triple-negative breast cancer. Oncol Rep. 2018;39:2315-23
25. Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S. et al. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11:253-70
26. Yang X, Wang Z, Li X, Liu B, Liu M, Liu L. et al. SHMT2 desuccinylation by SIRT5 drives cancer cell proliferation. Cancer Res. 2018;78:372-86
27. Ye X, Niu X, Gu L, Xu Y, Li Z, Yu Y. et al. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget. 2017;8:6984-93
28. Tang Z, Li L, Tang Y, Xie D, Wu K, Wei W. et al. CDK2 positively regulates aerobic glycolysis by suppressing SIRT5 in gastric cancer. Cancer Sci. 2018;109:2590-8
29. Wang C, Zhang C, Li X, Shen J, Xu Y, Shi H. et al. CPT1A-mediated succinylation of S100A10 increases human gastric cancer invasion. J Cell Mol Med. 2019;23:293-305
30. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
31. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
32. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
33. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-76
34. Carr BI. Hepatocellular carcinoma: current management and future trends. Gastroenterology. 2004;127:S218-24
35. El-Serag HB. Hepatocellular Carcinoma. N Engl J Med. 2011;365:1118-27
36. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. The Lancet. 2003;362:1907-17
37. Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271-89
38. Bringman-Rodenbarger LR, Guo AH, Lyssiotis CA, Lombard DB. Emerging roles for SIRT5 in metabolism and cancer. Antioxid Redox Signal. 2018;28:677-90
39. Chang L, Xi L, Liu Y, Liu R, Wu Z, Jian Z. SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol Med Rep. 2018;17:342-9
40. Zamzami N, Marchetti P, Castedo M, Decaudin D, Macho A, Hirsch T. et al. Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J Exp Med. 1995;182:367-77
41. Zamzami N, Marchetti P, Castedo M, Zanin C, Vayssiere JL, Petit PX. et al. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J Exp Med. 1995;181:1661-72
42. Lim ML, Lum MG, Hansen TM, Roucou X, Nagley P. On the release of cytochrome c from mitochondria during cell death signaling. J Biomed Sci. 2002;9:488-506
43. Guan D, Lim JH, Peng L, Liu Y, Lam M, Seto E. et al. Deacetylation of the tumor suppressor protein PML regulates hydrogen peroxide-induced cell death. Cell Death Dis. 2014;5:e1340
44. Juarez-Hernandez E, Motola-Kuba D, Chavez-Tapia NC, Uribe M, Barbero Becerra V. Biomarkers in hepatocellular carcinoma: an overview. Expert Rev Gastroenterol Hepatol. 2017;11:549-58
45. Pan JH, Zhou H, Zhao XX, Ding H, Li W, Qin L. et al. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: Potential in diagnosis and antitumour treatments (Review). Int J Mol Med. 2018;41:1809-16
46. Osborne B, Bentley NL, Montgomery MK, Turner N. The role of mitochondrial sirtuins in health and disease. Free Radic Biol Med. 2016;100:164-74
47. Mei Z, Zhang X, Yi J, Huang J, He J, Tao Y. Sirtuins in metabolism, DNA repair and cancer. J Exp Clin Cancer Res. 2016;35:182
48. Kleszcz R, Paluszczak J, Baer-Dubowska W. Targeting aberrant cancer metabolism - The role of sirtuins. Pharmacol Rep. 2015;67:1068-80
49. Sebastian C, Mostoslavsky R. The role of mammalian sirtuins in cancer metabolism. Semin Cell Dev Biol. 2015;43:33-42
50. Liang F, Wang X, Ow SH, Chen W, Ong WC. Sirtuin 5 is anti-apoptotic and anti-oxidative in cultured SH-EP neuroblastoma cells. Neurotox Res. 2017;31:63-76
51. Song CL, Tang H, Ran LK, Ko BC, Zhang ZZ, Chen X. et al. Sirtuin 3 inhibits hepatocellular carcinoma growth through the glycogen synthase kinase-3beta/BCL2-associated X protein-dependent apoptotic pathway. Oncogene. 2016;35:631-41
52. Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174-9
53. Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790-801
54. Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR. et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920-33
Author contact
![]() Corresponding author: Liming Wang, Email: wangbcc259com; Tel #: +86-17709870007
Corresponding author: Liming Wang, Email: wangbcc259com; Tel #: +86-17709870007

 Global reach, higher impact
Global reach, higher impact