Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(17):3893-3898. doi:10.7150/jca.31362 This issue Cite
Research Paper
CircABCC2 Regulates Hepatocellular Cancer Progression by Decoying MiR-665
1. Department of General Surgery, Xiangya Hospital, Central South University, Changsha, China
2. Department of XIMC Outpatient, Xiangya Hospital, Central South University, Changsha, China
Received 2018-11-9; Accepted 2019-4-26; Published 2019-6-10
Abstract
Background: Numerous studies have shown that circular RNAs (circRNAs) play vital roles in tumor progression. However, how circRNAs function in hepatocellular cancer (HCC) remains mostly unclear.
Methods: We analyzed HCC circRNA expression via a microarray, and the expression of an upregulated circRNA, circABCC2, was detected. We next explored the function of circABCC2 in HCC via a series of experiments. We performed RNA immunoprecipitation (RIP) and luciferase assays to explore the competing endogenous RNA (ceRNA) function of circABCC2 in HCC.
Results: qRT-PCR verified that circABCC2 was overexpressed in HCC. Inhibition of circABCC2 suppressed HCC cell proliferation and invasion, but promoted apoptosis. Luciferase assays and RIP showed that circABCC2 and ABCC2 could directly bind to miR-665 and that circABCC2 could regulate ABCC2 expression by sponging miR-665.
Conclusions: In summary, circABCC2 regulates ABCC2 expression and HCC progression by sponging miR-665. circABCC2 could be used as a biomarker and therapeutic target in HCC.
Keywords: hepatocellular cancer, circular RNAs, miR-665, ABCC2, competitive endogenous RNAs
Introduction
Liver cancer is a common cancer with poor outcomes [1]. Hepatocellular carcinoma (HCC) is the most frequent primary liver cancer [2]. Despite the progress that has been made in HCC treatment, prognosis remains poor. Exploring the molecular mechanisms that regulate HCC progression would help develop novel therapeutic targets and prognostic predictors for HCC.
Circular RNAs (circRNAs) have been implicated to be closely associated with cancer development [3]. In HCC, the circular RNA cSMARCA5 was reported to inhibit cancer growth and metastasis [4]. Furthermore, hsa_circ_0064428 has been identified as a potential immune-associated prognosis biomarker [5]. However, the functions of most circRNAs remain unknown in HCC.
circRNAs are reported to regulate gene expression as competitive endogenous RNAs (ceRNAs) or miRNA sponges [6]. In glioblastoma, circNT5E promotes tumorigenesis by acting as a sponge of miRNA-422a [7]. In lung cancer, the circRNA circPRKCI promotes tumor growth by sponging miR-545 and miR-589 [8]. In pancreatic cancer, circ-PDE8A promotes invasive growth by acting as a ceRNA of miR-338 [9]. These findings indicate that circRNAs could regulate cancer progression via a ceRNA mechanism.
Here, we reanalyzed the HCC circRNA expression microarray from our previous study [10]. We found that a circRNA, circABCC2, was upregulated in HCC cell lines and tissues. Inhibition of circABCC2 suppressed cell proliferation and invasion, but induced apoptosis. Moreover, circABCC2 could bind to miR-665 and regulate the expression of ABCC2. Therefore, circABCC2 could be used as a therapeutic target and biomarker for HCC.
Material and Methods
Cell culture
Cell lines were bought from ATCC (USA) and passaged within six months. Cells were cultured according to the manufacturer's instructions and re-authenticated by STR (short tandem repeat DNA profiling) before use.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cytoplasmic and nuclear RNA was isolated with a PARIS™ Kit (Invitrogen). Total RNA was isolated with TRIzol (Invitrogen, USA). cDNA was synthesized with a PrimeScript RT reagent kit (Takara, China), and SYBR Premix Ex Taq (Takara) and a CFX96 Real-time PCR system (Bio-Rad, USA) were used for the RT-PCR assay. The relative fold-change was calculated by the 2-ΔΔCt method. The primers were synthesized by Invitrogen (see Supplementary Table 1).
Cell counting kit-8 (CCK8) assay
Cells (1 × 103) were seeded 24 hours before transfection. CCK-8 solution (10 μL) was added 48 hours after transfection. Two hours later, a Spectra Max 250 spectrophotometer (Molecular Devices, USA) was used to measure the absorbance at 490 nM.
Apoptosis assay
The apoptotic rate of the modulated cells was detected. An Annexin V-fluorescein isothiocynate Apoptosis Detection Kit (KeyGen, Nanjing, China) was used to perform the Annexin V/propidium iodide staining and flow cytometry.
Transwell assay
To the lower chamber, 20% fetal bovine serum was added, and the modulated cells were suspended in FBS-free culture medium and seeded onto the upper chamber covered with EC matrix (Millipore, Germany). After 48 hours, the Matrigel in the upper chamber was removed while the invading cells adhering to the bottom were fixed, stained with crystal violet and imaged under a microscope.
Mouse xenograft assay
We subcutaneously injected 2 × 106 7402 cells into 4-week old BALB/c nude mice (to provide a power of 90% for a significance level of 0.05 with a two-tailed t test, a sample size of three mice per group was needed). The mice were then treated with 40 μL si-NC or si-circABCC2 by intratumoral injection.
For metastasis assays, 7402 cells were inoculated through the tail vein into nude mice (to provide a power of 90% for a significance level of 0.05 with a two-tailed t test, a sample size of three mice per group was needed). Four weeks later, mice were sacrificed and lung metastatic nodules were counted.
Luciferase assay
Luciferase reporter vectors with the 3′-UTR of ABCC2 or circABCC2 were constructed. Cells were co-transfected with miR-665 mimics and luciferase reporter vectors using Lipofectamine 2000 (Invitrogen). Then, 48 hours after incubation, a dual-luciferase reporter assay (Promega, USA) was used to measure the luciferase activities according to the manufacturer's guidance.
RNA immunoprecipitation (RIP) assay
Cells were co-transfected with control MS2bs-Rluc, MS2bs-circABCC2mt or MS2bs-circABCC2 by Lipofectamine 2000. RIP was performed with a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) 48 hours later. For the RIP assay on Ago2, RIP was performed with an anti-Ago2 antibody (Millipore) 48 hours after transfection.
Western blot analysis
Proteins were separated by 10% SDS-PAGE, transferred to a PVDF membrane (Millipore) and incubated with 5% skim milk before incubation with antibody against ABCC2 (1:500, Abcam, USA). Anti-β-actin antibody (1:1000, Affinity, USA) was used as an internal control. The membrane was incubated with secondary antibody (1:3000, CST, USA) and detected by chemiluminescence.
Statistical analysis
SPSS 22.0 software was used to perform the statistical analysis. Differences between groups were compared by t tests and χ2 tests. P < 0.05 was considered statistically significant. Unless otherwise indicated, data are presented as the mean ± s.e.m. of triplicate independent experiments.
Results
circABCC2 is upregulated and promotes HCC progression in vitro
We reanalyzed the HCC circRNA expression microarray [10] and found that a circRNA, hsa_circ_0019456, was upregulated in HCC. Based on the human reference genome (GRCh37/hg19), hsa_circ_0019456 is located at chr10:101565138-101594292 and is assumed to derive from the gene ABCC2 (ATP binding cassette subfamily C member 2), located on chromosome 10q24.2. Therefore, we termed hsa_circ_0019456 as “circABCC2”. To explore the expression of circABCC2 in HCC, qRT-PCR was performed and circABCC2 was overexpressed in HCC cell lines (Figure 1A) and tissues (Figure 1B).
Since circABCC2 was upregulated, we knocked down circABCC2 to explore its function in HCC. Figure 1C showed that the inhibition of circABCC2 was successful. The CCK-8 assay showed that inhibition of circABCC2 inhibited cell proliferation (Figure 1D). The apoptosis assay revealed that inhibition of circABCC2 led to increased apoptosis (Figure 1E). The Transwell assay showed that inhibition of circABCC2 suppressed cell invasion (Figure 1F). All of these findings indicate that inhibition of circABCC2 suppresses cell proliferation and invasion, but induces cell apoptosis in HCC in vitro.
circABCC2 promotes HCC progression in vivo
To further explore the function of circABCC2 in vivo, we performed mouse xenograft assays. Knockdown of circABCC2 resulted in decreased tumor growth (Figure 2A). Moreover, lung metastatic nodules decreased after inhibition of circABCC2 (Figure 2B). All of these findings indicate that inhibition of circABCC2 suppresses cell proliferation and invasion in HCC in vivo.
circABCC2 acts as a sponge for miR-665
We explored the intracellular location of circABCC2 and found that circABCC2 was mostly distributed in the cytoplasm (Figure 3A), suggesting that circABCC2 might act as an miRNA sponge to decoy miRNAs. Therefore, we investigated the potential circRNA/miRNA interaction using the Circular RNA Interactome (https://circinteractome.nia.nih.gov/index.html) and found a complementary site for miR-665 within the circABCC2 sequence (Figure 3B). Moreover, the expression of miR-665 was downregulated in HCC cell lines and (Figure 3C) and tissues (Figure 3D). Next, luciferase assays were performed to explore whether miR-665 could bind to circABCC2. The luciferase intensity decreased when co-transfected with luciferase reporters and miR-665 mimics (Figure 3E). To confirm the direct binding of circABCC2 and miR-665, a MS2bp-MS2bs based RIP assay was performed. We found that miR-665 was mostly enriched in the MS2bs-circABCC2 group, indicating the specific interaction of circABCC2 and miR-665 (Figure 3F). Together, our data confirmed that circABCC2 could interact with miR-665 and act as a decoy for miR-665.
circABCC2 regulates ABCC2 via miR-665
To investigate whether circABCC2 decoyed miR-665 to regulate the expression of target genes, we predicted ABCC2 to be a target by using TargetScan to assess the target genes of miR-665 (Figure 4A). We detected the ABCC2 expression level and found that ABCC2 was overexpressed in HCC cell lines (Figure 4B) and tumor tissues (Figure 4C). Next, we performed a luciferase reporter assay and found that the luciferase activity significantly decreased when co-transfected with miR-665 mimics and luciferase reporter (Figure 4D). The expression of ABCC2 was inhibited by miR-665 (Figure 4E).
circABCC2 is upregulated and promotes HCC progression in vitro A. The expression of circABCC2 in HCC cell lines. B. The expression of circABCC2 in HCC tissues. C. Cells were transfected with si-NC or si-circABCC2, and qRT-PCR analysis was used to assess the efficacy. D. Cell proliferation was assessed via CCK8 assay. E. Cell apoptosis was stained and detected by flow cytometry 48 hours after transfection. F. Transwell assay was performed and imaged. Original magnification, ×200. **P < 0.01.
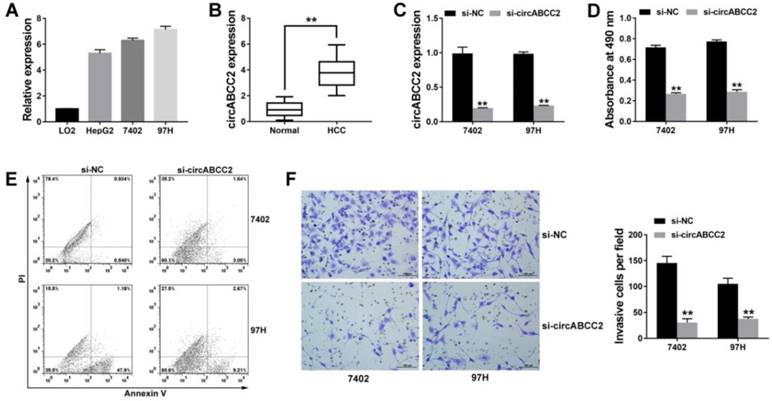
circABCC2 promotes HCC progression in vivo A. Representative images of tumor xenografts in nude mice are shown in the left panel (three mice per group). The weights of xenograft tumors are summarized in the right panel. B. Microscopic images of lung metastatic nodules (HE stained) are shown in the left panel. The number of metastatic nodules was summarized in the right panel (three mice per group). *P < 0.05, **P < 0.01.
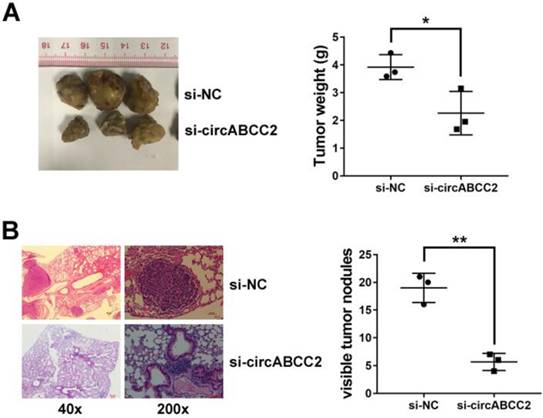
circABCC2 acts as a sponge for miR-665 A. The expression levels of GAPDH, U6 and circABCC2 in nuclear and cytoplasmic fractions were measured by qRT-PCR. B. The possible binding sites of miR-665 within circABCC2. C. The expression of miR-665 in HCC cell lines. D. The expression of miR-665 in HCC tissues. E. Luciferase reporter assay of cells co-transfected with miR-665 mimics and luciferase reporters containing circABCC2 (wt) or mutant construct (mut). F. MS2bp-MS2bs based RIP assay in cancer cells transfected with MS2bs-circABCC2, MS2bs-circABCC2mt or MS2bs-Rluc. **P < 0.01.
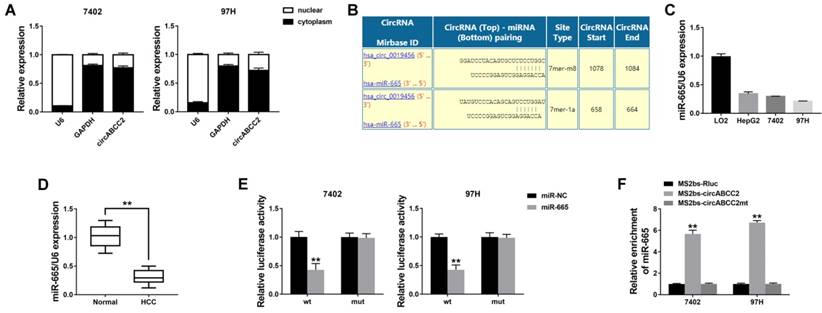
Next, a RIP assay on Ago2 showed that circABCC2, ABCC2 and miR-665 were mainly enriched to Ago2 (Figure 4F), indicating that circABCC2 and ABCC2 are recruited to an Ago2-related RISC where they interact with miR-665. In addition, inhibition of circABCC2 decreased the enrichment of Ago2 to circABCC2, but increased the enrichment of Ago2 to ABCC2 (Figure 4G), indicating that circABCC2 could function as a ceRNA and compete with ABCC2 for miRNA binding. Therefore, we explored the expression of ABCC2 after circABCC2 knockdown and found it to be decreased, but miR-665 LNA could reverse this repression (Figure 4H). These results indicate that circABCC2 can regulate ABCC2 expression as a ceRNA by decoying miR-665.
Discussion
Numerous recent studies have indicated that circRNAs play vital roles in cancer progression [11]. In lung cancer, circRNAs have been reported to modulate cell migration, invasion, apoptosis, cell cycle progression and cellular proliferation [12]. In colorectal cancer, the circRNA circITGA7 inhibits cancer growth and metastasis [13]. However, only a few reports have explored circRNAs in HCC. Therefore, we studied the circRNA expression microarray of HCC and found circABCC2 to be upregulated in HCC. Subsequent experiments revealed that inhibition of circABCC2 suppressed cell proliferation and invasion, but induced cell apoptosis, indicating that circABCC2 could regulate HCC progression. Therefore, circABCC2 could be a therapeutic target and potential biomarker.
Previous studies have reported that circRNAs regulate gene expression by sponging miRNA [14]. In colorectal cancer, circHIPK3 promoted metastasis and cancer growth by sponging miR-7 [15]. Dai X et al reported that circRNA_100284 affected cell proliferation and the cell cycle, and led to malignant transformation by sponging miRNA-217 [16]. Here, we found that circABCC2 could bind to miR-665 and acted as an miRNA sponge. In gastric cancer, miR-665 was downregulated and acted as a tumor suppressor [17]. In breast cancer, miR-665 has been reported to inhibit B7-H3 expression [18]. In HCC, miRNAs are also involved in cancer progression, such as cell proliferation, migration and invasion [19-22]. Here, we also found that miR-665 was downregulated in HCC.
In addition, we found that ABCC2 was a target gene of miR-665. ABCC2, also known as MRP2 (multidrug resistance-associated protein 2), belongs to the MRP family. It has been reported that ABCC2 is overexpressed in multiple cancers and confers resistance to several anticancer agents, such as MTX and cisplatin [23]. The expression of ABCC2 was positively correlated with shorter survival in lung cancer [24, 25], breast cancer [26], pancreatic cancer [27] and ovarian carcinoma [28]. We found that ABCC2 was also overexpressed in HCC, consistent with previous studies [29]. Subsequent experiments suggest that circABCC2 acted as a decoy for miR-665 to regulate ABCC2 expression. Inhibition of circABCC2 reduced ABCC2 expression, which could be reversed by inhibition of miR-665.
Conclusions
In summary, we found that circABCC2 was overexpressed in HCC. Inhibition of circABCC2 suppressed cell proliferation and invasion, but induced cell apoptosis. CircABCC2 regulated ABCC2 expression by sponging miR-665. circABCC2 could be a potential biomarker and therapeutic target in HCC.
Abbreviations
ceRNA: competing endogenous RNA; HCC: hepatocellular cancer; qRT-PCR: quantitative real-time polymerase chain reaction; CCK8: cell counting kit-8; circRNAs: circular RNAs; RIP: RNA immunoprecipitation; MS2bp: MS2-binding protein; MS2bs: MS2-binding sequences.
circABCC2 regulates ABCC2 via miR-665 A. The possible binding sites of miR-665 within ABCC2. B. The expression of ABCC2 in HCC cell lines was determined by Western blot (right) and qRT-PCR (left). C. The expression of ABCC2 in HCC tissues. D. Luciferase reporter assay of cells co-transfected with miR-665 mimics and luciferase reporter containing mutant construct (mut) or ABCC2 3′-UTR (wt). E. qRT-PCR (left) and Western blot (right) were used to evaluate the expression of ABCC2 after transfection, as previously described. F. RIP assay showing the enrichment of circABCC2, ABCC2 and miR-665 on Ago2 relative to IgG. G. RIP assay on Ago2. H. The expression of ABCC2 was determined by qRT-PCR after cell transfection. **P < 0.01.
Supplementary Material
Supplementary tables.
Acknowledgements
This study was approved by the Ethics Committees of Xiangya Hospital, Central South University and conducted according to the Declaration of Helsinki. Informed consent was obtained from all patients. The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of Xiangya Hospital, Central South University and conducted according to the IACUC protocol.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-14
3. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31-44
4. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ. et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214-27
5. Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W. et al. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2018
6. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
7. Wang R, Zhang S, Chen X, Li N, Li J, Jia R. et al. CircNT5E Acts as a Sponge of miR-422a to Promote Glioblastoma Tumorigenesis. Cancer research. 2018;78:4812-25
8. Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z. et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer research. 2018;78:2839-51
9. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237-50
10. Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao Y. et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37:172
11. Yang Z, Xie L, Han L, Qu X, Yang Y, Zhang Y. et al. Circular RNAs: Regulators of Cancer-Related Signaling Pathways and Potential Diagnostic Biomarkers for Human Cancers. Theranostics. 2017;7:3106-17
12. Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang F. et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18-27
13. Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W. et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018
14. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Molecular cell. 2018;71:428-42
15. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T. et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell death & disease. 2018;9:417
16. Dai X, Chen C, Yang Q, Xue J, Chen X, Sun B. et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell death & disease. 2018;9:454
17. Chen J, Sun D, Chu H, Gong Z, Zhang C, Gong B. et al. Screening of differential microRNA expression in gastric signet ring cell carcinoma and gastric adenocarcinoma and target gene prediction. Oncol Rep. 2015;33:2963-71
18. Nygren MK, Tekle C, Ingebrigtsen VA, Makela R, Krohn M, Aure MR. et al. Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br J Cancer. 2014;110:2072-80
19. Changjun L, Feizhou H, Dezhen P, Zhao L, Xianhai M. MiR-545-3p/MT1M axis regulates cell proliferation, invasion and migration in hepatocellular carcinoma. Biomed Pharmacother. 2018;108:347-54
20. Fu Y, Sun LQ, Huang Y, Quan J, Hu X, Tang D. et al. miR-142-3p Inhibits the Metastasis of Hepatocellular Carcinoma Cells by Regulating HMGB1 Gene Expression. Curr Mol Med. 2018
21. Liang H, Sun H, Yang J, Yi C. miR1455p reduces proliferation and migration of hepatocellular carcinoma by targeting KLF5. Mol Med Rep. 2018;17:8332-8
22. Liu Z, Ma M, Yan L, Chen S, Li S, Yang D. et al. miR-370 regulates ISG15 expression and influences IFN-alpha sensitivity in hepatocellular carcinoma cells. Cancer Biomark. 2018;22:453-66
23. Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92:1295-302
24. Filipits M, Haddad V, Schmid K, Huynh A, Dunant A, Andre F. et al. Multidrug resistance proteins do not predict benefit of adjuvant chemotherapy in patients with completely resected non-small cell lung cancer: International Adjuvant Lung Cancer Trial Biologic Program. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3892-8
25. Sun Z, Chen J, Aakre J, Marks RS, Garces YY, Jiang R. et al. Genetic variation in glutathione metabolism and DNA repair genes predicts survival of small-cell lung cancer patients. Ann Oncol. 2010;21:2011-6
26. Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M. et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287-93
27. Tanaka M, Okazaki T, Suzuki H, Abbruzzese JL, Li D. Association of multi-drug resistance gene polymorphisms with pancreatic cancer outcome. Cancer. 2011;117:744-51
28. Surowiak P, Materna V, Kaplenko I, Spaczynski M, Dolinska-Krajewska B, Gebarowska E. et al. ABCC2 (MRP2, cMOAT) can be localized in the nuclear membrane of ovarian carcinomas and correlates with resistance to cisplatin and clinical outcome. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:7149-58
29. Nies AT, Konig J, Pfannschmidt M, Klar E, Hofmann WJ, Keppler D. Expression of the multidrug resistance proteins MRP2 and MRP3 in human hepatocellular carcinoma. Int J Cancer. 2001;94:492-9
Author contact
![]() Corresponding authors: Xinying Li and Xiaogang Li, Department of General Surgery, Xiangya Hospital, Central South University; 87 Xiangya Road, Changsha, Hunan, 410008, P.R. China. E-mail: lixinyingcnedu.cn and lixiaogangcom.cn; Tel: +8613975804018
Corresponding authors: Xinying Li and Xiaogang Li, Department of General Surgery, Xiangya Hospital, Central South University; 87 Xiangya Road, Changsha, Hunan, 410008, P.R. China. E-mail: lixinyingcnedu.cn and lixiaogangcom.cn; Tel: +8613975804018

 Global reach, higher impact
Global reach, higher impact