3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(17):3926-3932. doi:10.7150/jca.31345 This issue Cite
Research Paper
GPC6 Promotes Cell Proliferation, Migration, and Invasion in Nasopharyngeal Carcinoma
1. NHC Key Laboratory of Carcinogenesis (Central South University) and Hunan Key Laboratory of Translational Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, China
2. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute and Basic Medical Science, Central South University, Changsha, Hunan, China
3. Hunan Key Laboratory of Nonresolving Inflammation and Cancer, Disease Genome Research Center, The Third Xiangya Hospital, Central South University, Changsha, Hunan, China.
4. Department of Chemistry, University of North Dakota, Grand Forks, North Dakota, USA.
Received 2018-11-8; Accepted 2019-5-7; Published 2019-6-24
Abstract
Nasopharyngeal carcinoma (NPC) is a highly metastatic tumor that occurs frequently in Southeast Asia, particularly including southern China. Epstein-Barr virus infection is well established as a primary cause of NPC; nevertheless, the mechanisms underlying NPC pathogenesis remain largely unknown. In our previous study, we conducted whole-genome sequencing to screen for genomic variations that were associated with NPC. Of the resultantly identified variations, glypican-6 (GPC6), was shown, for the first time, to be frequently mutated in NPC. In the present study, we verified this finding and conducted a series of functional experiments, which demonstrated that GPC6 promotes the migration, invasion, and proliferation of NPC cells in vitro. Thus, the present study identified novel biological functions for GPC6 in NPC, and thus, showed that GPC6 may be a promising potential therapeutic target for this disease.
Keywords: NPC, GPC6, genomic variations, migration, invasion, proliferation
Introduction
Nasopharyngeal carcinoma (NPC) is a polygenic hereditary disease that arises in nasopharyngeal epithelial cells, and is common in southern China [1-3]. While Epstein-Barr virus infection, environmental factors, and genetic susceptibility have been shown to promote NPC progression [4-10], the molecular basis underlying NPC pathogenesis has not yet been fully elucidated.
Genetic susceptibility is strongly implicated in NPC pathogenesis [11-13]. In fact, many previous studies have revealed frequent NPC-causative chromosome rearrangements (of chromosomes 1 and 12) and deletions (of chromosomes 3, 6, 11, and 14) [14-19]. Moreover, novel NPC-susceptibility loci have recently been discovered, such as TNFRSF19 at 13q12, MECOM at 3q26m, and CDKN2A/2B at 9p21 [11]. Similarly, various single-nucleotide variants (SNVs) have been shown to be associated with a range of immune diseases and cancers, in which they often represent somatic mutations relative to normal tissue. With the rapid development of second-generation sequencing technologies, it is now possible to perform a comprehensive, multi-level, whole-genome analysis of the genetic factors underlying NPC pathogenesis [20-22]. For example, in our previous study, we performed a genome-wide, high-throughput resequencing of genomic DNA in paired NPC tissue and peripheral blood (control) samples to explore the molecular basis of NPC. We resultantly identified a large number of NPC tumor-specific mutations, including SNVs, insertions, deletions, and genomic-structural and copy-number variants [23]. In the present study, we selected a gene called glypican-6 (GPC6) that has not been previously shown to be associated with NPC, but was found to be frequently mutated in NPC tumor tissues (i.e. 8.3% and 58.3% of the analyzed NPC tumor samples were shown to harbor SNVs and deletions in GPC6, respectively) for further analysis.
Materials and Methods
PCR and Sanger-sequencing analyses
PCR primers targeted to the identified GPC6 SNVs were designed using primer 3 software, synthesized (Tsingke Company), amplified via PCR, and subjected to Sanger sequencing (Tsingke Company). The utilized Sanger-sequencing primers comprised (forward) 5′-TCTCATTGCAGGGGAACACT-3′ and (reverse) 5′-TCTCACAGGCAAAGCACAAA -3′.
Clinical tissue samples
A total of 37 NPC and 19 adjacent non-tumor nasopharyngeal epithelium tissue samples were collected from patients who were newly diagnosed with NPC at the Affiliated Tumor Hospital of Central South University and who had not undergone either radio- or chemotherapy, or surgery. Relevant clinical data were also collected. All patients provided written informed consent for their participation in the study, which was approved by the Ethics Committee of the Health Authority of Central South University, China.
Cell lines, plasmids, and transfection
The NPC cell lines used in this study, HNE2 and 5-8F, were maintained (37°C, 5% CO2) in RPMI 1640 medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Shanghai, China). They were then cultured to 50-60% confluency in 6-well plates, and transfected with an pIRESneo3 empty (Clontech, Mountain View, CA, USA) or pIRESneo3-GPC6 GPC6-overexpression (containing the full-length GPC6 open reading frame) vector using Lipofectamine 3000 (Life Technologies, Grand Island, NY) according to the manufacturer's instructions[24-27].
RNA isolation and quantitative qRT-PCR
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions and reverse transcribed into complementary DNA (cDNA) using 5× All-in-One RT MasterMix (Applied Biological Materials, Richmond, BC, Canada). quantitative real-time (qRT)-PCR was performed using the MiniOpticon system (Bio-Rad, Hercules, CA, USA) with EvaGreen 2× qPCR MasterMix (Applied Biological Materials) and primers for GPC6 (forward, 5′-ACCGAGTTTCCAAGGTCAGC-3′; reverse, 5′-CAGCCTGATTTGCCAAGCAG-3′) and the selected normalizing control, GAPDH (forward, 5′-TCTCTGCTCCTCCTGTTC-3′; reverse, 5′-GTTGACTCCGACCTTCAC-3′) (Life Technologies, Carlsbad, CA, USA).
Wound healing assay
Cells were transfected with an empty vector or pIRESneo3-GPC6 when they reached 50-60% confluency. A 10 μL pipette tip was used to scratch the monolayers at a consistent width (and each original scratch was normalized prior to the statistical analysis). After scratching, images were captured at 0, 12, 24, and 36 h with an inverted-phase contrast microscope. The width of the healed scratch in different groups was measured and analyzed using Image Pro Plus 6.0 software[28-30].
MTT assay
A total of 800 cells in 200 µl medium were added to 96-well plates after transfection. Proliferation was determined by incubating the cells (4 h) in 20 µl of 1 mg/ml tetrazolium salt MTT reagent (Sigma Chemicals, St Louis, MO, USA) and then dissolving them in dimethyl sulfoxide (DMSO). The absorbance of the resultant suspension was measured at 490 nm simultaneously each day on days 0, 1, 2, 3, 4, and 5.
Matrigel invasion assay
The invasive capacity of the cells was determined using transwell chambers (8-µm pores; Corning, NY, USA) in 24-well plates. The chambers were evenly coated with Matrigel (BD Biosciences, San Jose, CA, USA) diluted 1:8 in serum-free medium. A total of 20,000 cells suspended in 200 µl of serum-free medium were added to the upper chamber, and the lower chamber was filled with 20% FBS to induce cell invasion. Following 24-36 h of incubation, cells that had invaded through the filter pores were fixed with methanol and stained with 5% crystal violet. The number of invasive tumor cells from five randomly selected 20× fields was counted using Image Pro Plus 6.0 software.
Colony formation assay
A total of 800-1,000 transfected cells were seeded into 6-well culture plates. After 7 days, the cultures were harvested, fixed (15 min) with 4% paraformaldehyde, and stained (15 min) with hematoxylin. The number of colonies formed was counted using Image Pro Plus 6.0 software.
Statistical analyses
A Student's t-test was used to assess differences between two groups. All data were obtained from three independent replicates of each experiment and expressed as the mean ± standard deviation. A P value <0.05 was considered to indicate statistical significance. All statistical analyses were performed using SPSS 13.0 and GraphPad 5 software.
Results
Validation of GPC6 somatic mutations in NPC
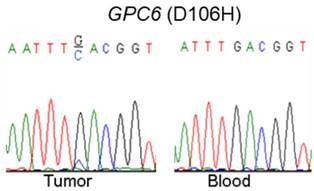
Via our previously conducted study, we showed that GPC6 SNV and deletion mutations occurred in a high proportion (8.3% and 58.3%, respectively) of the analyzed NPC cases. To verify this finding, we used a combination of PCR and Sanger-sequencing techniques to detect and analyze somatic GPC6 mutations in the present patient cohort. We resultantly showed that GPC6 G-C mutations occurred more frequently in the NPC tumor than in blood (control) samples, following the change of the 106th amino acid of GPC6 protein from Aspartic acid to Histidine (Fig. 1).
GPC6 is highly expressed in NPC tissues
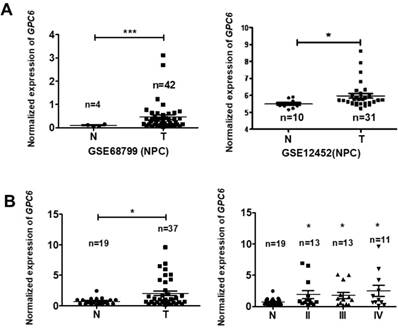
To explore the potential biological function of GPC6 in NPC, we analyzed GPC6 expression levels using the online Gene Expression Omnibus (GEO) database. By retrieving and analyzing GPC6- associated datasets (GSE68799 and GSE12452), we showed that GPC6 is more highly expressed in NPC than in adjacent control tissues (Fig. 2A). Since these datasets did not include patient clinical data, and to verify this finding, we collected and evaluated GPC6 expression in 37 NPC and 19 normal control tissues from newly enrolled patients via qRT-PCR. The results of this analysis showed that GPC6 expression was significantly elevated in NPC tumor tissues, and furthermore, that the increased GPC6 expression was associated with higher TNM tumor stages (Fig. 2B).
GPC6 promotes NPC cell proliferation
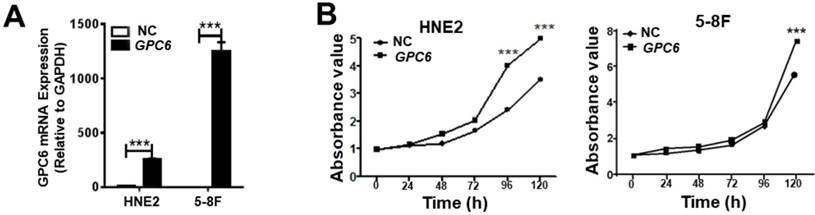
After cloning the full length GPC6 open reading frame sequence into the pIRESneo3 expression vector, HNE2 and 5-8F cells were transfected with pIRESneo3-GPC6 and confirmed to overexpress GPC6 mRNA (Fig. 3A). The subsequently conducted MTT assays demonstrated that pIRESneo3-GPC6-transfected 5-8F and HNE2 cells exhibited a higher absorbance value than the corresponding negative control (NC; empty vector) cells. This finding indicates that the induced GPC6 overexpression promoted NPC cell proliferation (Fig. 3B).
GPC6 increases NPC cell colony-formation and invasion capacities
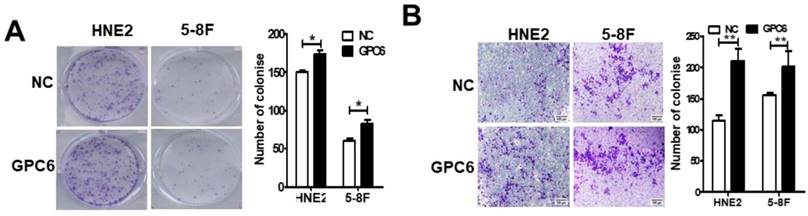
We next conducted colony formation and Matrigel invasion assays to examine whether changes to GPC6 expression affected the ability of NPC cells to form colonies and/or become invasive. The results of these assays showed that overexpressing GPC6 increased colony formation in both 5-8F and HNE2 cells compared to that in the controls (Fig. 4A), and similarly, that NPC cells transfected with pIRESneo3-GPC6 exhibited an increased capacity for invasion (Fig. 4B).
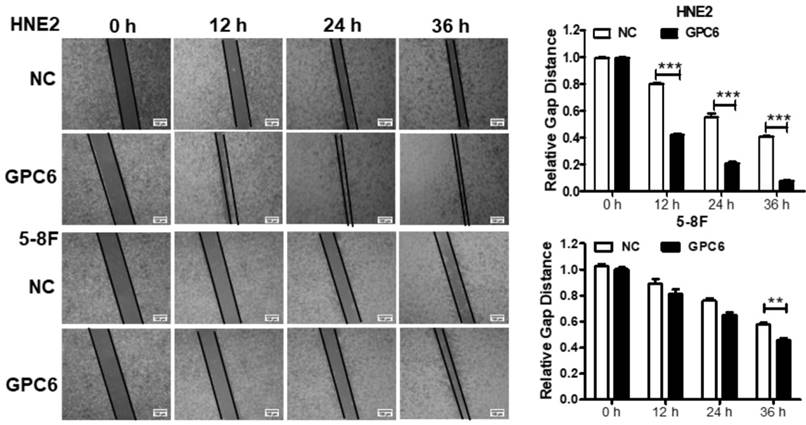
GPC6 promotes NPC cell migration
We also examined whether GPC6 overexpression impacted NPC cell migration. Cells were seeded in 6-well plates and transfected with pIRESneo3-GPC6 or an empty vector, before their migration capacity was evaluated via a wound-healing assay. The results of this analysis showed that the migratory potential of the GPC6-overexpressing cells was dramatically increased compared to that of the NC NPC cells (Fig. 5).
Validation of somatic mutations in nasopharyngeal carcinoma (NPC). A combination of polymerase chain reaction (PCR) and Sanger-sequencing techniques was used to validate previously identified somatic mutations in tumor and blood (negative control) samples collected from patients with NPC. The depicted GPC6 G-C mutation was found to occur more frequently in the analyzed tumor than in blood samples.
GPC6 is highly expressed in nasopharyngeal carcinoma (NPC). (A) GPC6 expression was shown to be significantly increased in NPC biopsies via the analysis of publicly available datasets GSE68799 (Normal, n = 4; Tumor, n = 42; p = 0.0006) and GSE12452 (Normal, n = 10; Tumor, n = 31; p = 0.041) that were sourced from the Gene Expression Omnibus database. (B) GPC6 expression was measured by RT-qPCR and was shown to be significantly increased in 37 NPC compared to 19 non-tumor tissues collected from newly enrolled patients (left), and this increase in GPC6 expression was shown to be positively associated with higher TNM stages (right). N, normal tissue sample; T, tumor tissue sample.
GPC6 overexpression promotes nasopharyngeal carcinoma (NPC) cell proliferation. (A) Transfection of an GPC6 overexpression construct, pIRESneo3-GPC6, dramatically increased GPC6 expression in 5-8F and HNE2 cells, as shown via qRT-PCR. (B) Cells were seeded in 6-well plates, transfected with pIRESneo3-GPC6 or an empty vector (negative control, NC), and allowed to proliferate for a further 24 h. They were then digested, counted, suspended (800 cells/200 µl), added to a 96-well plate, and subjected to an MTT assay. The results of this analysis showed that GPC6 overexpression significantly promoted the proliferation of NPC cells.
GPC6 increased the capacity of nasopharyngeal carcinoma (NPC) cells to form colonies and become invasive in vitro. 5-8F and HNE2 cells were transfected with pIRESneo3-GPC6 or an empty vector (negative control, NC), and allowed to proliferate for 24 h before being subjected to (A) a colony formation and (B) a Matrigel invasion assay. The results of these analyses showed that GPC6 overexpression increased both the number of colonies that were formed by the cells, and the invasive capacity of the cells, respectively.
Overexpression of GPC6 promoted nasopharyngeal carcinoma (NPC) cell migration. 5-8F and HNE2 cells were transfected with pIRESneo3-GPC6 or an empty vector (negative control, NC), and cultured to 100% confluency before being subjected to a wound healing assay. After scratching, images were captured at 0, 12, 24, and 36 h with an inverted phase contrast microscope. Data are presented as the width ratio of migratory promotion.
Discussion
NPC is a highly metastatic tumor that occurs more or less frequently with remarkable racial and regional specificity and that is most common in South China, Southeast Asia, and North Africa [31-33]. Because of its concealed location in the body, lymph- node and/or distant metastases are often already present at the time of NPC diagnosis. The current clinical treatment for NPC mainly comprises a combination of radiotherapy and chemotherapy [34, 35]; however, these therapies incur severe side-effects, and treatment is frequently hindered by tumor heterogeneity [36-38]. Therefore, recent studies have focused on screening for novel NPC pathogenic mutations with the aim of identifying potential drug targets and/or molecular prognostic markers that can be used to improve current detection and treatment methods, and thus improve the outcome of patients with this disease [8, 39-46].
In recent years, whole-genome sequencing has been used to identify pathogenic mutations in various tumor types [47-49]; however, to date, NPC-mutation screening has only been conducted at the exon level. For example, Lin et al.[50] performed exon sequencing for NPC cases from Singapore and identified a large number of novel "driver" genes, many of which were found to be mutated and dysfunctional in NPC cells, and shown to thus mediate NPC pathogenesis. Following this, our previous study employed whole- genome sequencing to screen for recurring pathogenic NPC mutations. We resultantly showed that GPC6 is frequently mutated in NPC, and thus, it was selected for further research in the present study. The conducted in vitro functional experiments herein showed, for the first time, that GPC6 likely performs a regulatory role in NPC.
The conducted analysis of publicly reported NPC gene expression profile data (in the GEO datasets GSE12452 and GSE68799) and the q-PCR examination of the present patient cohort indicated that GPC6 expression was upregulated in NPC compared to adjacent non-tumor samples, suggesting that it may promote oncogenesis. GPC6 is a member of the GPC family of thiohepatic glycoproteins and is a cell surface phosphatidylinositol glycan [51]. To date, six members (comprising GPC1-6) of this family have been identified in mammals and Drosophila. Glypicans are primarily expressed during development and play a pivotal role in morphogenesis [52]. While previous studies have shown that GPC6 mutations and copy-number variations frequently occur in various tumor types, the function of GPC6 in human cancers remains controversial. For example, GPC6 mutations have been identified in terminal cases of prostate cancer (via exon sequencing) [53], and GPC6 copy-number variations have been suggested to promote the progression of non-small cell lung cancer (via a whole-genome microarray analysis) [54]. Moreover, GPC6 was found to be frequently mutated and aberrantly methylated in colorectal cancer (in 12 paired colorectal tumors and adjacent healthy mucosal tissues using the Illumina Infinium Human Methylation 450 BeadChip) [55]. Similarly, a previous study demonstrated that increased GPC6 expression was significantly correlated with a reduced progression-free survival time in patients with melanoma, and furthermore, that this effect occurred in an HIF1α-dependent/hypoxia- responsive manner (as shown via a ROC curve) [56]. Likewise, high GPC6 expression has been shown to be closely associated with gastric cancer progression [57]. In contrast, GPC6 has been found to function as a tumor suppressor, and high GPC6 expression has been shown to be positively correlated with T-lymphocyte infiltration and increased overall survival in patients with early-stage ovarian cancer [58]. Likewise, reduced GPC6 mRNA expression in retinoblastoma (RB) is associated with non-random allelic loss at 13q31, which is thought to promote RB progression [59]. Nevertheless, to date, the function of GPC6 is unknown in NPC. The in vitro functional assays conducted by the present study suggest that GPC6 likely promotes NPC cell proliferation, migration, and invasion, supporting the previously reported and present genome sequencing results. Taken together, our findings implicate GPC6 as a likely oncogenic factor in NPC, and thus, as a promising prognostic biomarker. Further studies are needed to elucidate the mechanisms by which GPC6 mediates NPC pathogenesis. Notably, GPC6 has been shown to promote breast cancer progression by inhibiting β-catenin and Wnt signaling pathways. This effectively upregulates Wnt5A signaling, which, in turn, inhibits JNK and p38 MAPK signaling, thereby promoting cancer-cell metastasis and invasion [60]. Moreover, mounting evidence suggests that GPCs may regulate several Hedgehog signaling pathways [61]. Thus, further research is required to examine the functional roles of GPC6 in NPC.
Aspartic acid is an important amino acid. In the synthesis of amino acids, aspartic acid can function as a precursor of various amino acids; in the degradation of amino acids, it participates in the urea cycle and in the malic acid shuttle system. It also facilitates the transfer of NADHs from the mitochondrial membrane to the mitochondrial matrix. Our results indicated the presence of a G/C mutation in NPC samples that led to the change of the 106th amino acid of GPC6 protein from aspartic acid to histidine. Histamine, a well-known mediator of inflammation, is released from mast cells and basophils. The histamine- producing enzyme, histidine decarboxylase (HDC), is commonly induced at inflammatory sites, enabling the regulation of various functions through the production of cytokines/chemokines/growth factors [62]. Baly et al. introduced a His19 to Ala mutant of melanoma growth-stimulating activity (MGSA) to antagonize the CXCR2 receptor, which offers the opportunity to design antagonists of MGSA that could have clinical utility [63]. Interestingly, accumulating evidence suggests that histamine synthesis induced in several types of tumor tissues modulates tumor immunity via inducing IFN production [64, 65]. His15 residue is necessary for tumor necrosis factor-alpha (TNF) to exert its cytotoxic activity [66]. Moreover, histidine can promote vasodilation. Formation of histamine by histidine decarboxylase contributes to histamine-mediated vasodilation following aerobic exercise [67]. Histidine is involved in structural rearrangement in the heme pocket and signal transduction [68]. Other studies have shown that histidine catabolism is a major determinant of methotrexate sensitivity [64]. The proliferation of different cancer cells requires the consumption of a large number of specific amino acids, and we speculated whether this mutation (aspartic acid to histidine) promotes the malignancy of tumor cells. The search for an analog of this amino acid, a metabolic antagonist, will provide effective means to treat tumors.
In summary, we herein employed a series of functional assays to validate the role of GPC6 in NPC, and resultantly showed that increased GPC6 expression promotes the migration, invasion, and proliferation of NPC cells in vitro. These findings provide an invaluable foundation for further research into the association of genomic variations with NPC development, and into the molecular mechanisms and signaling pathways underlying NPC pathogenesis.
Abbreviations
NPC: nasopharyngeal carcinoma; EBV: Epstein- Barr virus; GPC6: glypican-6; NC: negative control; MTT: 3-(4,5-dimethylthiazole-2-yl)-2.5-diphenyl-tetrazolium bromide; DMSO: dimethyl sulfoxide; GEO: Gene Expression Omnibus; SNV: single-nucleotide variant.
Acknowledgements
This work has been supported by the National Natural Science Foundation of China (81672683, 81672993, 81772928, 81702907, 31741049, 21806186 and 81872278), the Natural Science Foundation of Hunan Province (2016JC2035, 2017SK2100, 2018SK21210, 2018SK21211, 2018JJ3815 and 2018JJ3704), the 111 Project (111-2-12), the Open Sharing Fund for the Large-scale Instruments and Equipment of Central South University (CSUZC201947), the Fundamental Research Funds for the Central Universities of Central South University (2019zzts089), the financial assistance from the special scholarship of Central South University for students studying abroad and the outstanding youth exchange program of China Association for Science and Technology (2018CASTQNJL56).
Ethics approval
All patients provided written informed consent for their participation in the present study, the design of which (including the use of human tissues) was approved by the Ethics Committee of the Health Authority of Central South University, Changsha, Hunan, China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wu C, Li M, Meng H. et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62:640-7
2. He Y, Jing Y, Wei F. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235-47
3. Wei F, Wu Y, Tang L. et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60:1271-5
4. He B, Li W, Wu Y. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353-65
5. Song Y, Li X, Zeng Z. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783-99
6. Zeng Z, Huang H, Huang L. et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014;57:315-26
7. Wang M, Zhao J, Zhang L. et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8:761-73
8. Zhong Y, Du Y, Yang X. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79-90
9. Fan C, Tang Y, Wang J. et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9:2852-64
10. Deng X, Xiong F, Li X. et al. Application of atomic force microscopy in cancer research. J Nanobiotechnology. 2018;16:102-107
11. Fan CM, Wang JP, Tang YY. et al. circMAN1A2 may serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019 doi: 10.1111/cas.14034. [Epub ahead of print]
12. Xiong F, Deng S, Huang HB. et al. Effects and mechanisms of innate immune molecules on inhibiting nasopharyngeal carcinoma. Chin Med J (Engl). 2019;132:749-52
13. Bo H, Fan L, Li J. et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer. 2018;9:4677-83
14. Wei F, Jing YZ, He Y. et al. Cloning and characterization of the putative AFAP1-AS1 promoter region. J Cancer. 2019;10:1145-53
15. Cheung AK, Lung HL, Ko JM. et al. Chromosome 14 transfer and functional studies identify a candidate tumor suppressor gene, mirror image polydactyly 1, in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2009;106:14478-83
16. Wu Y, Wei F, Tang L. et al. Herpesvirus acts with the cytoskeleton and promotes cancer progression. J Cancer. 2019;10:2185-93
17. Wang H, Zhou Y, Oyang L. et al. LPLUNC1 stabilises PHB1 by counteracting TRIM21-mediated ubiquitination to inhibit NF-κB activity in nasopharyngeal carcinoma. Oncogene. 2019;38:5062-75
18. Or YY, Chung GT, To KF. et al. Identification of a novel 12p13.3 amplicon in nasopharyngeal carcinoma. J Pathol. 2010;220:97-107
19. Tse KP, Su WH, Yang ML. et al. A gender-specific association of CNV at 6p21.3 with NPC susceptibility. Hum Mol Genet. 2011;20:2889-96
20. Xiong W, Zeng ZY, Xia JH. et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972-4
21. Mo Y, Wang Y, Xiong F. et al. Proteomic analysis of the molecular mechanism of lovastatin inhibiting the growth of nasopharyngeal carcinoma cells. J Cancer. 2019;10:2342-9
22. Tu C, Zeng Z, Qi P. et al. Genome-Wide Analysis of 18 Epstein-Barr Viruses Isolated from Primary Nasopharyngeal Carcinoma Biopsy Specimens. J Virol. 2017:91 pii: e00301-17
23. Tu C, Zeng Z, Qi P. et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole genome sequencing. Carcinogenesis. 2018;39:1517-1528
24. Yu J, Liu Y, Gong Z. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621-32
25. Yu J, Liu Y, Guo C. et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523-30
26. Li Q, Chen P, Zeng Z. et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37:12503-12
27. Liang F, Li Q, Li X. et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49:1046-56
28. He R, Liu P, Xie X. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145-57
29. Lian Y, Xiong F, Yang L. et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37:253-70
30. Fan C, Wang J, Tang Y. et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2019;40:380-391
31. Wei F, Tang L, He Y. et al. BPIFB1 (LPLUNC1) inhibits radioresistance in nasopharyngeal carcinoma by inhibiting VTN expression. Cell Death Dis. 2018;9:432-44
32. Wei F, Wu Y, Tang L. et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br J Cancer. 2018;118:233-47
33. Tang Y, He Y, Zhang P. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77-87
34. Tang L, Wei F, Wu Y. et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87-102
35. Wang JP, Tang YY, Fan CM. et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2018;9:12487-502
36. Yan Q, Zeng Z, Gong Z. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766-82
37. Yang L, Tang Y, He Y. et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97-103
38. Tang Y, He Y, Shi L. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001-11
39. Fan C, Tang Y, Wang J. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130-141
40. Wang Y, Mo Y, Gong Z. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25-33
41. Zhang Y, Xia M, Jin K. et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45-59
42. Tang Y, Wang J, Lian Y. et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol Cancer. 2017;16:42-50
43. Zhou R, Wu Y, Wang W. et al. Circular RNAs (circRNAs) in cancer. Cancer lett. 2018;425:134-42
44. Wang Y, Xue D, Li Y. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
45. Yang L, Tang Y, Xiong F. et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2018;9:1426-43
46. Wang YA, Li XL, Mo YZ. et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17:168-83
47. Duan S, Guo W, Xu Z, Natural killer group 2D receptor, its ligands in cancer immune escape. Mol Cancer. 2019;18:29.
48. Kan Z, Zheng H, Liu X. et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422-33
49. Xiao K, Yu Z, Li X. et al. Genome-wide Analysis of Epstein-Barr Virus (EBV) Integration and Strain in C666-1 and Raji Cells. J Cancer. 2016;7:214-24
50. Lin DC, Meng X, Hazawa M. et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866-71
51. Xiao L, Wei F, Liang F. et al. TSC22D2 identified as a candidate susceptibility gene of multi-cancer pedigree using genome-wide linkage analysis and whole exome sequencing. Carcinogenesis. 2019 pii: bgz095. doi: 10.1093/carcin/bgz095. [Epub ahead of print]
52. Filmus J. Glypicans in growth control and cancer. Glycobiology. 2001;11:19R-23R
53. Kumar A, White TA, MacKenzie AP. et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087-92
54. Mo Y, Wang Y, Zhang L. et al. The role of Wnt signaling pathway in tumor metabolic reprogramming. J Cancer. 2019;10:3789-97
55. Farkas SA, Vymetalkova V, Vodickova L. et al. DNA methylation changes in genes frequently mutated in sporadic colorectal cancer and in the DNA repair and Wnt/beta-catenin signaling pathway genes. Epigenomics. 2014;6:179-91
56. Loftus SK, Baxter LL, Cronin JC. et al. Hypoxia-induced HIF1alpha targets in melanocytes reveal a molecular profile associated with poor melanoma prognosis. Pigment Cell Melanoma Res. 2017;30:339-52
57. Dinccelik-Aslan M, Gumus-Akay G, Elhan AH. et al. Diagnostic and prognostic significance of glypican 5 and glypican 6 gene expression levels in gastric adenocarcinoma. Mol Clin Oncol. 2015;3:584-90
58. Karapetsas A, Giannakakis A, Dangaj D. et al. Overexpression of GPC6 and TMEM132D in Early Stage Ovarian Cancer Correlates with CD8+ T-Lymphocyte Infiltration and Increased Patient Survival. Biomed Res Int. 2015;2015:712438
59. Lau CS, Yu CB, Wong HK. et al. Allelic imbalance at 13q31 is associated with reduced GPC6 in Chinese with sporadic retinoblastoma. Br J Ophthalmol. 2010;94:357-62
60. Yiu GK, Kaunisto A, Chin YR. et al. NFAT promotes carcinoma invasive migration through glypican-6. Biochem J. 2011;440:157-66
61. Filmus J, Capurro M. The role of glypicans in Hedgehog signaling. Matrix Biol. 2014;35:248-52
62. Wang W, Zhou R, Wu Y. et al. PVT1 promotes cancer progression via microRNAs. Front Oncol. 2019;9:609
63. Baly DL, Horuk R, Yansura DG. et al. A His19 to Ala mutant of melanoma growth-stimulating activity is a partial antagonist of the CXCR2 receptor. J Immunol. 1998;161:4944-9
64. Kanarek N, Keys HR, Cantor JR. et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559:632-6
65. Jiang X, Wang J, Deng X. et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10-27
66. Bo H, Fan L, Gong Z. et al. Upregulation and hypomethylation of lncRNA AFAP1?AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. 2019;41:2431-9
67. Xia M, Zhang Y, Jin K. et al. Communication between mitochondria and other organelles: a brand-new perspective on mitochondria in cancer. Cell Biosci. 2019;9:27
68. Herzik MA Jr, Jonnalagadda R, Kuriyan J. et al. Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins. Proc Natl Acad Sci U S A. 2014;111:E4156-64
Author contact
![]() Corresponding author: zengzhaoyangedu.cn
Corresponding author: zengzhaoyangedu.cn

 Global reach, higher impact
Global reach, higher impact