Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(17):4000-4008. doi:10.7150/jca.29029 This issue Cite
Research Paper
Knockdown of LncRNA RHPN1-AS1 Inhibits Cell Migration, Invasion and Proliferation in Head and Neck Squamous Cell Carcinoma
1. Department of Stomatology, Second Xiangya Hospital, Central South University, Changsha 410011, Hunan, China;
2. Department of Stomatology, Haikou people's Hospital, Haikou 570208, Hainan, China;
3. Xiangya Stomatological Hospital & School of Stomatology, Central South University, Changsha 410008, Hunan, China;
4. Department of Laboratory Medicine, Second Xiangya Hospital, Central South University, Changsha 410011, Hunan, China.
Received 2018-8-6; Accepted 2019-5-26; Published 2019-7-5
Abstract
Previous studies have revealed that long non-coding RNAs (lncRNAs) are involved in head and neck squamous cell carcinoma (HNSCC) progression. However, the detailed roles of lncRNA RHPN1-AS1 remain to be elucidated. In this study, by analyzing online RNA-Seq data, we found that RHPN1-AS1 was upregulated in HNSCC tissues and that its expression level was associated with neoplasm histologic grade. High expression of RHPN1-AS1 was also confirmed in HNSCC tissues. Knockdown of RHPN1-AS1 inhibited tumor cell migration, invasion and proliferation in HNSCC. Furthermore, inhibition of RHPN1-AS1 suppressed the expression of epithelial-mesenchymal transition (EMT)-related genes (β-Catenin, Claudin-1 and Vimentin) in HNSCC cells. Collectively, our results suggest that RHPN1-AS1, acting as an oncogene, may be a potential diagnostic and therapeutic target in HNSCC.
Keywords: Long non-coding RNA, RHPN1-AS1, HNSCC, EMT
Introduction
Squamous cell carcinoma is one of the most aggressive cancers worldwide and has a poor prognosis despite recent advances in therapeutics [1]. Head and neck squamous cell carcinoma (HNSCC) accounts for up to 90% of all head and neck cancers derived from the oral cavity, oropharynx and hypopharynx and is the sixth leading cancer worldwide, with an estimated 600,000 new cases annually [2, 3]. Patients usually have no obvious signs in the early stage of the disease, resulting in delayed diagnosis. Although surgery, radiation therapy and adjuvant chemotherapy are used to treat HNSCC according to National Comprehensive Cancer Network (NCCN) guidelines, tumor control and patient survival are poor [4]. Therefore, identification of early biomarkers and elucidation of the tumor process of HNSCC will be helpful for improving survival while maintaining patient quality of life.
Long non-coding RNAs (lncRNAs) are newly discovered non-coding RNAs with more than 200 nucleotides and no protein coding ability [5]. In recent years, accumulating evidence has revealed that lncRNAs function as tumor suppressors or promoters in carcinogenesis and cancer progression among different cancer types [6-10]. The lncRNA AFAP1-AS1 is upregulated and acts as an oncogene in a variety of cancers [11]. The lncRNAs CCAT1 and CCAT2, located in the 8q24.21 locus, were highly overexpressed in colorectal cancer and were significantly associated with recurrence-free survival (RFS) and overall survival (OS), which serve as important prognostic biomarkers in colorectal cancer [12]. The lncRNA DANCR increased cancer stem cell function by upregulating AXL/PI3K-Akt via competitive binding to miR-33a-5p in osteosarcoma [13]. We analyzed the TANRIC online HNSCC data [14] and found that RHPN1-AS1 was significantly upregulated in HNSCC tissues, suggesting that RHPN1-AS1 may function as a tumor promoter in this cancer. However, the relationship between RHPN1-AS1 and HNSCC has not been previously reported.
In this article, we identified the RHPN1-AS1 expression level and detailed functions in HNSCC. Our study might provide a promising new strategy for RNA-based diagnosis and therapies in HNSCC.
Materials and methods
Data mining and analysis
The online data of RHPN1-AS1 expression level and clinical characteristics were obtained from the TANRIC platform (http://ibl.mdanderson.org/tanric/_design/basic/index.html). The search strategy was described in a previous report [14]. The correlation analysis between RHPN1-AS1 and the β-Catenin, Claudin-1 and Vimentin expression levels in HNSCC was analyzed by using the Gene Expression Profilling Interactive Analysis (GEPIA) database [15] (http://gepia.cancer-pku.cn/index.html).
Tissue specimens and cell lines
HNSCC tissues and adjacent normal tissues (located > 1 cm away from the tumor) were obtained from 26 patients who underwent HNSCC resection at Second Xiangya Hospital, Central South University. These tissues were immediately frozen in liquid nitrogen and stored at -80°C until analysis. Informed consent was obtained from each patient, and the study was approved by the Ethics Committee of Second Xiangya Hospital, Central South University. The human HNSCC cell lines Cal-27 and Tca8113 were maintained in DMEM and RPMI-1640 medium supplemented with 10% fetal bovine serum and antibiotics (100 units/ml penicillin and 100 µg/ml streptomycin). Cells were incubated at 37°C in a humidified incubator containing 5% CO2.
Short interfering RNA (siRNA) and cell transfection
Knockdown of RHPN1-AS1 was performed using siRNAs provided by RiboBio (Guangzhou, China). The siRNA sequences were as follows: RHPN1-AS1-siRNA1-sense: CCGAAUCUCUUUACUUCCAdTdT, RHPN1-AS1-siRNA1-antisense: UGGAAGUAAAGAGAUUCGGdTdT; RHPN1-AS1-siRNA2-sense: CUCAAACUUUGAGGGUCAUdTdT, RHPN1-AS1-siRNA2-antisense: AUGACCCUCAAAGUUUGAGdTdT. The cells were cultured overnight and transfected with siRNAs using Lipofectamine RNAiMAX Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. For validation of the transfection efficiency, lncRNA expression was examined by quantitative real-time PCR (qPCR).
QPCR
Total RNA from cells and tissues was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Reverse-transcribed RHPN1-AS1 cDNA was obtained using a Revert Aid™ First Strand cDNA synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.). QPCR analyses were performed according to a previous study [11]. Samples were compared using the relative CT method, where the relative expression of RHPN1-AS1 was normalized to that of β-Actin. The primer sequences were as follows: RHPN1-AS1 forward, 5'- GCTCCTGGTCATCAAGTTCCTCT-3' and reverse, 5'- GCACAGGCACCAGAATGATCC-3'; β-Actin forward, 5'-TCACCAACTGGGACGACATG-3' and reverse, 5'-GTCACCGGAGTCCATCACGAT-3'.
Cell proliferation assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to measure cell proliferation. After transfection, 5 × 103 cells/well cells were seeded in 96-well plates and cultured for 1, 2, 3 and 4 days, and then, the supernatant was removed, and MTT (5 mg/ml, 20 μl) was added to each well at 37˚C. After 4 h, 100 μl dimethyl sulfoxide (DMSO) was added to each well. The optical density (OD) value was determined at 490 nm by a microplate reader and used to construct a growth curve to assess cell proliferation.
Τranswell assays
Transwell migration and invasion assays were performed to measure the migration and invasive capacity of the transfected cells. In brief, cells were plated on 8-µm pore size membranes with or without Matrigel (2 × 104 cells/well in serum-free medium), which were in turn placed in the top chamber of 24-well transwell plates. The bottom chamber contained 800 µl chemotactic factor. After 24 h, cells on the upper surface were removed, while cells attached to the membranes were fixed in 4% paraformaldehyde for 20 min and stained with hematoxylin. The results of the transwell assay were imaged, and the number of invasive cells was evaluated by manual counting.
Flow cytometry analysis
For analysis of the cell cycle, cells were collected after transfection for 24 h and fixed in precooled 75% ethanol overnight at 4°C. After the cells were washed with 1 × phosphate buffer solution (PBS) (1,000 rpm, 5 min), they were resuspended in a solution of 800 µl of 1 × PBS containing 1% bovine serum albumin (BSA). After addition of 100 µl of PI dye (3.8 × 10-2 M sodium citrate, pH 7.0) and 100 µl of RNase A (10 mg/ml), cells were incubated at 37°C for 30 min in the dark. Cell cycles were analyzed by flow cytometry. For detection of apoptosis, cells were digested with 0.25% EDTA-free trypsin and collected by centrifugation at 1,000 rpm for 5 min. After cells were washed with 1 × PBS, 2 × 105 cells were suspended in 500 µl of binding buffer. Then, 5 µl of annexin V-FITC was added, followed by the addition of 5 µl of propidium iodide, and the samples were incubated at room temperature for 10 min in the dark room. Apoptotic cells were sorted by flow cytometry.
Western blot assay
Cells were collected 48 h after transfection with RHPN1-AS1 siRNA or control siRNA. Then, the cells were washed twice with precooled PBS. After RIPA cleaved the cells, we used a BCA kit to measure the protein concentration. Denatured proteins (30 μg) were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). After completion of electrophoresis, the protein was transferred onto polyvinylidene fluoride (PVDF) membranes. After the membranes were blocked with 5% milk for 1 h, they were incubated with primary anti-β-catenin, claudin-1, and vimentin antibodies at 4°C overnight. The next day, membranes were washed with PBS and incubated with secondary antibodies at 37°C for 1 h. Intensity of protein expression was detected by ECL chemiluminescence.
Statistical analysis
All experiments were repeated at least three times. Statistical analyses were carried out using SPSS 19.0 software (SPSS, Chicago, IL, USA). The results were assessed by Student's t-test. All statistical tests were two-sided, and a P-value of < 0.05 was considered statistically significant.
Results
LncRNA RHPN1-AS1 was upregulated in HNSCC tissues
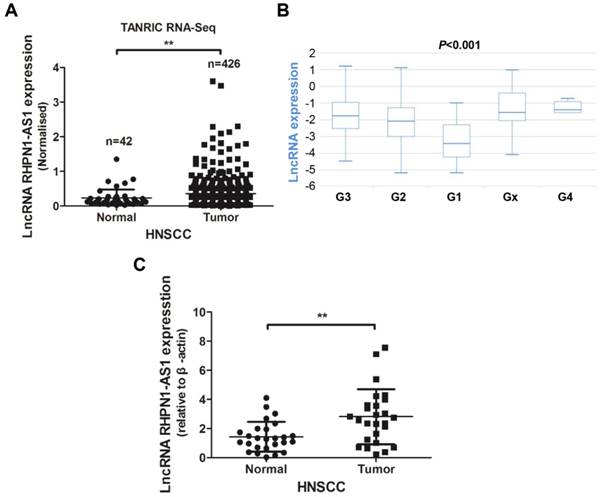
LncRNA RHPN1-AS1 was recently identified as an oncoRNA in uveal melanoma (UM) [16]. We analyzed the RHPN1-AS1 expression levels in HNSCC tissues from the TANRIC platform. The analysis results revealed that RHPN1-AS1 was upregulated in HNSCC tissues (426 tumor tissues vs. 42 normal tissues; Fig. 1A, P < 0.01), and the RHPN1-AS1 level was correlated with neoplasm histologic grade (Fig. 1B, P < 0.001). In addition, patients with high RHPN1-AS1 expression had poor survival compared to patients with low RHPN1-AS1 expression; however, the P value was greater than 0.05 (P = 0.086, data not shown). To further verify the expression of RHPN1-AS1, we obtained 26 pairs of HNSCC and adjacent normal tissues (Table 1) to measure the lncRNA expression levels. QPCR results revealed that RHPN1-AS1 was remarkably upregulated in HNSCC tissues (Fig. 1C, P < 0.01), which was in accord with our above results.
Knockdown of RHPN1-AS1 inhibited the migration and invasion of HNSCC cells
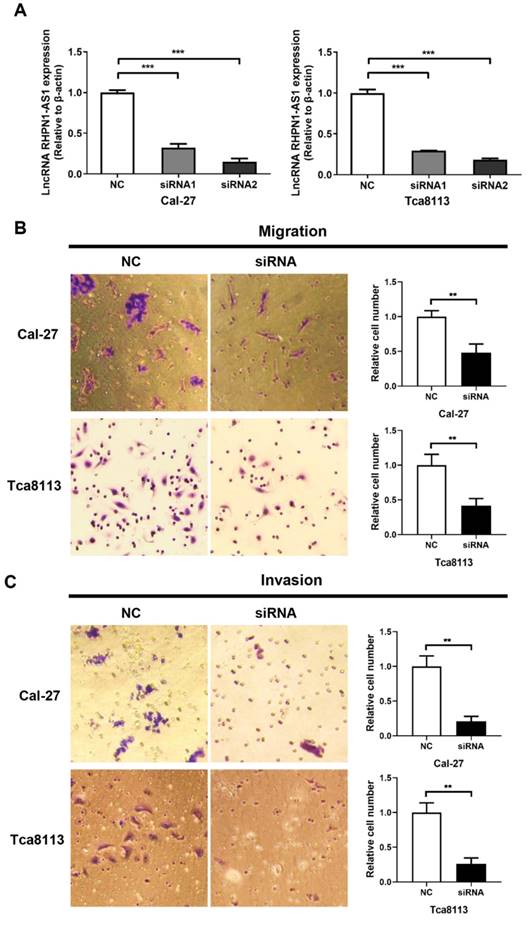
To investigate whether RHPN1-AS1 affected the malignant behavior of HNSCC cells, we transfected siRNAs targeting RHPN1-AS1 (si-RHPN1-AS1) and negative control siRNA (NC) into Cal-27 and Tca8113 cells. Transfection efficiency was evaluated by qPCR (Fig. 2A, P < 0.001), and the second siRNA had the strongest effect. Therefore, we chose this siRNA in our subsequent experiments. Transwell assays were used to investigate cell migration and invasion. As shown in Fig. 2B, RHPN1-AS1 knockdown significantly inhibited migration by approximately 50% in Cal-27 cells and by nearly 60% in Tca8113 cells (P < 0.01). As expected, a significant inhibition of invasion was observed after transfecting RHPN1-AS1 siRNA into Cal-27 and Tca8113 cells compared with that of the NC group (Fig. 2C, P < 0.01).
The clinicopathological information of 26 HNSCC patients
| No. | Gender | Age (years) | Diagnosis | Clinical stage | Histological grade |
|---|---|---|---|---|---|
| 1 | M | 53 | TSCC | IV (T2N2M0) | Moderate |
| 2 | M | 63 | TSCC | II (T2N0M0) | Moderate |
| 3 | M | 55 | BSCC | II (T2N0M0) | Moderate |
| 4 | M | 65 | TSCC | III (T3N1M0) | Well/Moderate |
| 5 | M | 61 | TSCC | III (T2N1M0) | Well/Moderate |
| 6 | M | 66 | BSCC | IV (T3N2M0) | Moderate |
| 7 | F | 53 | TSCC | III (T2N1M0) | Moderate /Poor |
| 8 | M | 52 | TSCC | II (T2N0M0) | Moderate |
| 9 | M | 47 | TSCC | II (T2N0M0) | Well/Moderate |
| 10 | M | 68 | TSCC | II (T2N0M0) | Well/Moderate |
| 11 | M | 46 | TSCC | I (T1N0M0) | Well |
| 12 | M | 44 | TSCC | I (T1N0M0) | Well |
| 13 | F | 38 | BSCC | II (T2N0M0) | Well/Moderate |
| 14 | M | 52 | BSCC | II (T2N0M0) | Well |
| 15 | M | 37 | TSCC | IV (T2N2M0) | Poor |
| 16 | M | 50 | TSCC | II (T2N0M0) | Well |
| 17 | M | 55 | TSCC | I (T1N0M0) | Moderate/Poor |
| 18 | M | 62 | TSCC | IV (T2N2M0) | Well/Moderate |
| 19 | M | 54 | TSCC | III (T3N0M0) | Well/Moderate |
| 20 | M | 57 | TSCC | II (T2N0M0) | Well |
| 21 | M | 53 | TSCC | IV (T3N2M0) | Well/Moderate |
| 22 | M | 36 | TSCC | II (T2N0M0) | Moderate |
| 23 | M | 45 | TSCC | IV (T2N2M0) | Well/Moderate |
| 24 | M | 51 | BSCC | III (T3N0M0) | Well |
| 25 | M | 46 | TSCC | I (T1N0M0) | Well |
| 26 | M | 44 | TSCC | I (T1N0M0) | Well |
Abbreviations: M, male; F, female; TSCC, tongue squamous cell carcinoma; BSCC, buccal squamous cell carcinoma.
LncRNA RHPN1-AS1 was upregulated in HNSCC (A) - (B) Data on RHPN1-AS1 expression in HNSCC and normal tissues obtained from the TANRIC platform were analyzed, and RHPN1-AS1 was upregulated in HNSCC tissues compared to normal tissues (426 tumor cases vs. 42 normal cases; P < 0.01), and RHPN1-AS1 expression level was associated with neoplasm histologic grade (P < 0.001). (C) Relative expression of RHPN1-AS1 in HNSCC tissues compared with that of adjacent normal tissues (n = 26, P < 0.01). RHPN1-AS1 expression was determined using qPCR and was normalized to β-actin expression. The data represent the mean ± SD of 3 replicates. ** P < 0.01.
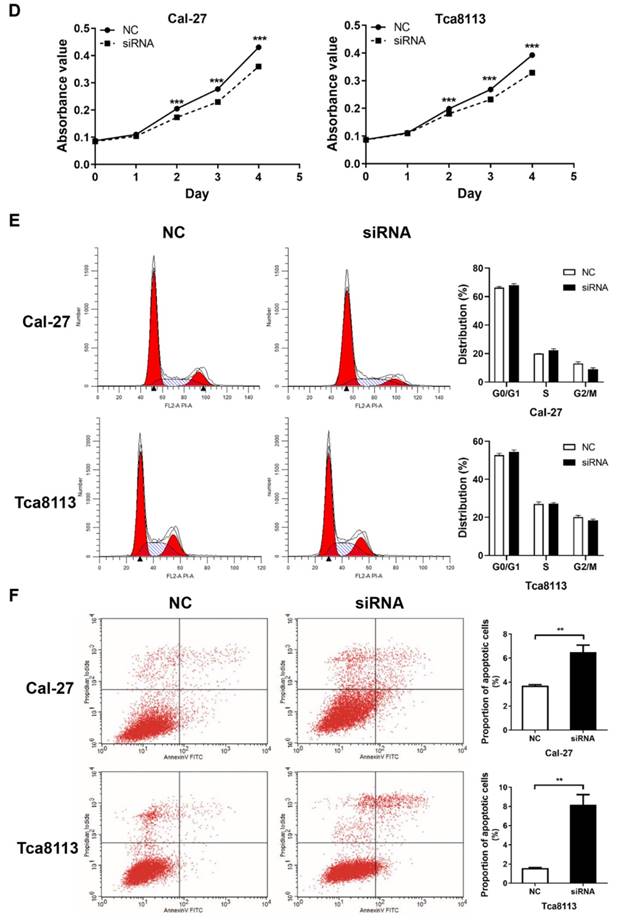
Knockdown of RHPN1-AS1 blocked cell proliferation and promoted cell apoptosis in HNSCC
An MTT assay was performed to evaluate cell viability. The si-RHPN1-AS1 group of both Cal-27 and Tca8113 cells showed slow proliferation compared with the control group (Fig. 2D, P < 0.001). Flow cytometric analysis was performed to detect the cell cycle and apoptosis. Transfection of RHPN1-AS1-siRNA did not affect the cell cycle distribution of Cal-27 and Tca8113 cells (Fig. 2E). However, the percentage of apoptotic cells was significantly increased after silencing RHPN1-AS1 in both Cal-27 and Tca8113 cells (Fig. 2F, P < 0.01).
RHPN1-AS1 regulated the expression of epithelial-mesenchymal transition (EMT)-related genes
EMT is known to facilitate tumor migration and invasion [17]. In a previous study, a gene expression profile analysis revealed that RHPN1-AS1 may participate in the EMT process through regulation of TGF-β signaling in UM [16]. To illuminate the functional mechanisms of RHPN1-AS1, we detected the expression levels of several EMT-related genes by western blot analyses. The results showed that RHPN1-AS1 knockdown significantly downregulated the expression of β-catenin, claudin-1 and vimentin in Cal-27 and Tca8113 cells (Fig. 3A). In addition, the correlation analysis between RHPN1-AS1 and the β-Catenin, Claudin-1 and Vimentin expression levels in HNSCC was performed by using the GEPIA database. The results indicated that RHPN1-AS1 expression is positively correlated with β-Catenin, Claudin-1 and Vimentin expression (Fig. 3B-D).
Discussion
LncRNAs originate from different regions of the genome, including the promoters or untranslated regions of protein-coding genes, the antisense strand or the introns of protein-coding genes, or even independent transcripts within and outside of protein-coding genes [18, 19]. These RNAs were previously considered as genetic byproducts and did not attract much attention in the past few decades because of the absence of biological functions [20, 21]. However, recent reports have demonstrated that the majority of the human genome is transcribed into lncRNAs, and increasing evidence suggests that lncRNAs play a critical role in the regulation of diverse pathological and physiological processes [22]. Currently, an increasing number of studies have identified “oncogene” or “tumor suppressor” lncRNAs in cancer [23, 24]. To date, the dysregulated expression and involvement of lncRNAs have been reported in diverse cancers, including HNSCC [11, 24-26]. For example, high expression of the lncRNA KCNQ1OT1 was closely correlated with poor prognosis in tongue squamous cell carcinoma (TSCC) and promoted TSCC cell proliferation [27]. Upregulation of HOTAIR promoted HNSCC cell invasion and metastasis and was associated with poor prognosis of patients [28, 29]. UCA1 was not only correlated with the lymph node metastasis of TSCC but could also enhance the migration of TSCC cells [30]. In addition, Cao et al. [31] reported that KTN1-AS1, LINC00460 and RP5-894A10.6 act as novel biomarkers for the accurate prognostic prediction of patients with HNSCC by analyzing RNA-seq data derived from the TANRIC database. Therefore, lncRNAs have promising applications in new diagnostic and therapeutic strategies in HNSCC.
RHPN1-AS1 knockdown inhibited HNSCC cell migration, invasion and proliferation (A) Cal-27 and Tca8113 cells were transfected with RHPN1-AS1 siRNA1, RHPN1-AS1 siRNA2, and a negative control (NC). QPCR was used to detect the efficacy of the RHPN1-AS1 knockdown. (B) - (C) Cell migration and invasion were examined in Cal-27 and Tca8113 cells transfected with RHPN1-AS1 siRNA or NC by Transwell assays without and with Matrigel coating in the upper compartment. (D) Measurement of growth curves of Cal-27 and Tca8113 cells transfected with RHPN1-AS1 siRNA or NC by MTT assays. (E) - (F) Cell cycle distribution and apoptosis were determined by flow cytometry. The data represent the mean ± SD of 3 replicates. ** P < 0.01; ***P < 0.001.
RHPN1-AS1, a 2030-bp transcript originating from human chromosome 8q24.3, is an antisense lncRNA derived from the promoter region of RHPN1. A previous study reported that RHPN1-AS1 was upregulated in the UM and that knockdown of RHPN1-AS1 significantly inhibited UM cell proliferation, migration and invasion [16]. In another study, the authors developed a lncRNA network-based prioritization approach, named “LncNetP”, and predicted that RHPN1-AS1 may serve as a novel disease risk factor for hepatocellular carcinoma diagnosis and prognosis [32]. We first found that this lncRNA has abnormal expression levels in HNSCC based on large-scale samples by analyzing RNA-Seq data, strongly implying that RHPN1-AS1 may function as a tumor promoter in HNSCC. Thus, fresh HNSCC tissues were collected, and cell lines were used for further investigation. As expected, RHPN1-AS1 was significantly overexpressed in HNSCC tissues compared with adjacent normal tissues. Inhibition of RHPN1-AS1 in Cal-27 and Tca8113 cells suppressed migration, invasion and cell viability, which is consistent with the findings of a previous study of UM [16]. In addition, knockdown of RHPN1-AS1 promoted HNSCC cell apoptosis, but it had no effect on the cell cycle. Collectively, our results suggest that RHPN1-AS1, acting as an oncogene, may be a potential diagnostic and therapeutic target in HNSCC.
Cell migration and invasion are significant aspects of cancer progression, and EMT plays an important role in tumor cell migration and invasion [33]. EMT represents a transformation program underpinning the invasive and metastatic phenotype of cancers, which could be interpreted as a state of de-differentiation [34]. This process can be induced by multiple signaling pathways, including the Wnt/β-catenin signaling pathway [35-37]. Accumulating evidence indicates that many important oncogenes or tumor suppressors regulate the EMT program and thus are implicated in tumor cell migration and invasion [38]. In addition, lncRNAs also participate in this regulatory process [34]. The lncRNA NKILA inhibited migration and invasion of TSCC cells by suppressing EMT [39]. MALAT1 promoted TSCC cell growth and metastasis by regulating the activity of the Wnt/β-catenin signaling pathway [40, 41]. In the present study, we tested the effect of RHPN1-AS1 knockdown on EMT-related genes. Compared with the control group, the RHPN1-AS1 knockdown group showed decreased protein expression of β-catenin, claudin-1 and vimentin in Cal-27 and Tca8113 cells, which are critical for EMT. In addition, we performed the correlation analysis between RHPN1-AS1 and the β-Catenin, Claudin-1 and Vimentin expression levels in HNSCC by using the GEPIA database. The results indicated that RHPN1-AS1 expression is positively correlated with the expression of β-Catenin, Claudin-1 and Vimentin in HNSCC. The above data indicated that RHPN1-AS1 could regulate the expression of EMT-related genes in HNSCC cells.
RHPN1-AS1 knockdown suppressed the expression of EMT-related genes (A) The protein levels of β-catenin, vimentin and claudin-1 were detected in Cal-27 and Tca8113 cells transfected with RHPN1-AS1 siRNA or NC by western blotting. The expression of β-actin was detected as a protein loading control. (B)-(D) The correlation analysis between RHPN1-AS1 and β-Catenin, Claudin-1 and Vimentin expression levels in HNSCC was performed by using the GEPIA database.
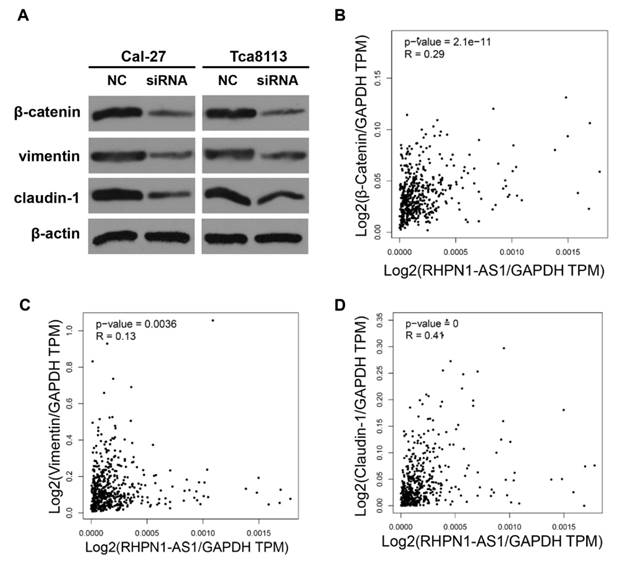
Studies have revealed that lncRNAs function by direct regulation or indirect interaction with other molecules, such as protein, DNA, and RNA [22, 42-44]. For example, lncRNAs may exert their functions through interactions with regulatory proteins, such as chromatin remodeling proteins [45]. The lncRNA HOTAIR has been shown to interact with Polycomb Repressive Complex 2 components such as Ezh2 to regulate gene expression [46]. The lncRNA GClnc1 could be used as a molecular scaffold to recruit WDR5 and KAT2A complexes for specific modifications of histones [47]. Additionally, lncRNAs can interact with RNA molecules and thus regulate gene expression as competing endogenous RNAs (ceRNAs), as described previously [22, 48]. For instance, the lncRNA SNHG15 acts as a ceRNA to regulate the YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma [49]. H19 functions as a ceRNA to sponge the miRNA let-7 family, leading to an increase in the expression of let-7 targets in several cancers [48]. In this study, our preliminary exploration suggests that RHPN1-AS1 promotes tumor cell migration and invasion by regulating the expression of EMT-related genes in HNSCC cells. However, the mechanisms underlying how RHPN1-AS1 regulates the expression of EMT-related genes remain unclear and should be further studied.
Every study, including the present study, has its limitations. First and foremost, there were no reciprocal experiments to examine the effects of RHPN1-AS1 overexpression on the malignant phenotype of HNSCC cells. Additionally, we confirmed that RHPN1-AS1 promotes tumor cell migration and invasion by regulating the expression of EMT-related genes in HNSCC cells. However, the mechanisms underlying RHPN1-AS1-mediated regulation of EMT-related gene expression remain unclear.
Conclusion
In conclusion, this study demonstrates that lncRNA RHPN1-AS1 was upregulated in HNSCC and that its expression level was correlated with neoplasm histologic grade. Knockdown of RHPN1-AS1 inhibited the migration, invasion and proliferation of HNSCC cells. RHPN1-AS1 promoted tumor development by regulating the expression of EMT-related genes. Collectively, our results suggest that RHPN1-AS1, acting as an oncogene, may be a potential diagnostic and therapeutic target in HNSCC.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81772901 and 81301757) and the Natural Science Foundation of Hunan Province (Grant No. 2015JJ2191).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bhardwaj R, Bhardwaj G, Gautam A, Karagozian R. Upper Gastrointestinal Bleed as a Manifestation of Poorly Differentiated Metastatic Squamous Cell Carcinoma of the Lung. J Clin Diagn Res. 2017;11:OD13-OD4
2. Pi L, Zhu G, She L, Wei M, Liu G, Chen C. et al. Elevated expression of Derlin-1 associates with unfavorable survival time of squamous cell carcinoma of the head and neck and promotes its malignance. J Cancer. 2017;8:2336-45
3. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
4. Argiris A. EGFR inhibition for recurrent or metastatic HNSCC. Lancet Oncol. 2015;16:488-9
5. Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15:62
6. Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen Z. et al. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch Biochem Biophys. 2017;615:1-9
7. Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118:4836-43
8. Wang R, Yan B, Li Z, Jiang Y, Mao C, Wang X. et al. Long non-coding RNA HOX transcript antisense RNA promotes expression of 14-3-3sigma in non-small cell lung cancer. Exp Ther Med. 2017;14:4503-8
9. Wang Y, Fu L, Sun A, Tang D, Xu Y, Li Z. et al. C/EBPbeta contributes to transcriptional activation of long non-coding RNA NEAT1 during APL cell differentiation. Biochem Biophys Res Commun. 2018;499:99-104
10. Liu CW, Liu D, Peng D. Long non-coding RNA ZFAS1 regulates NOB1 expression through interacting with miR-646 and promotes tumorigenesis in osteosarcoma. Eur Rev Med Pharmacol Sci. 2019;23:3206-16
11. Bo H, Fan L, Li J, Liu Z, Zhang S, Shi L. et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer. 2018;9:4677-83
12. Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H. et al. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 'gene desert', serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28:1882-8
13. Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J. et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46-55
14. Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y. et al. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75:3728-37
15. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
16. Lu L, Yu X, Zhang L, Ding X, Pan H, Wen X. et al. The Long Non-Coding RNA RHPN1-AS1 Promotes Uveal Melanoma Progression. Int J Mol Sci. 2017:18
17. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666-81
18. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-61
19. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-63
20. Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103-5
21. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556-65
22. Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J. et al. LncRNA Structural Characteristics in Epigenetic Regulation. Int J Mol Sci. 2017:18
23. Gong Z, Zhang S, Zhang W, Huang H, Li Q, Deng H. et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120-4
24. Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683-95
25. Gong Z, Zhang S, Zeng Z, Wu H, Yang Q, Xiong F. et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9:e110674
26. Nohata N, Abba MC, Gutkind JS. Unraveling the oral cancer lncRNAome: Identification of novel lncRNAs associated with malignant progression and HPV infection. Oral Oncol. 2016;59:58-66
27. Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q. et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742
28. Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J. et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64-70
29. Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X. et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46:2586-94
30. Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89-95
31. Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T. et al. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94-101
32. Xu C, Ping Y, Zhao H, Ning S, Xia P, Wang W. et al. LncNetP, a systematical lncRNA prioritization approach based on ceRNA and disease phenotype association assumptions. Oncotarget. 2017;8:114603-12
33. Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ. et al. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547:E1-E3
34. Luo X, Qiu Y, Jiang Y, Chen F, Jiang L, Zhou Y. et al. Long non-coding RNA implicated in the invasion and metastasis of head and neck cancer: possible function and mechanisms. Mol Cancer. 2018;17:14
35. Taciak B, Pruszynska I, Kiraga L, Bialasek M, Krol M. Wnt signaling pathway in development and cancer. J Physiol Pharmacol. 2018:69
36. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442-54
37. Foroutan M, Cursons J, Hediyeh-Zadeh S, Thompson EW, Davis MJ. A Transcriptional Program for Detecting TGFbeta-Induced EMT in Cancer. Mol Cancer Res. 2017;15:619-31
38. Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med. 2018;12:361-73
39. Huang W, Cui X, Chen J, Feng Y, Song E, Li J. et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7:62520-32
40. Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y. et al. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci Rep. 2015;5:15972
41. Liang J, Liang L, Ouyang K, Li Z, Yi X. MALAT1 induces tongue cancer cells' EMT and inhibits apoptosis through Wnt/beta-catenin signaling pathway. J Oral Pathol Med. 2017;46:98-105
42. Fenner A. Kidney cancer: AR promotes RCC via lncRNA interaction. Nat Rev Urol. 2016;13:242
43. Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y. et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704-16
44. Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z. et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976-87
45. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667-78
46. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-93
47. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC. et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784-801
48. Wang P, Li X, Gao Y, Guo Q, Wang Y, Fang Y. et al. LncACTdb 2.0: an updated database of experimentally supported ceRNA interactions curated from low- and high-throughput experiments. Nucleic Acids Res. 2018
49. Wu DM, Wang S, Wen X, Han XR, Wang YJ, Shen M. et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9:947
Author contact
![]() Corresponding author: Zhaojian Gong, Department of Stomatology, The Second Xiangya Hospital of Central South University, No. 139 Middle Renmin Road, Furong District, Changsha City, Hunan Province 410011, China. E-mail address: gongzhaojian4458edu.cn.
Corresponding author: Zhaojian Gong, Department of Stomatology, The Second Xiangya Hospital of Central South University, No. 139 Middle Renmin Road, Furong District, Changsha City, Hunan Province 410011, China. E-mail address: gongzhaojian4458edu.cn.

 Global reach, higher impact
Global reach, higher impact