3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(17):4009-4016. doi:10.7150/jca.27529 This issue Cite
Research Paper
The Prognostic Value of the MiR-200 Family in Colorectal Cancer: A Meta-analysis with 1882 Patients
1. Department of General Surgery, The First Affiliated Hospital of Nanchang University, Nanchang, China.
2. Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Nanchang University, Nanchang, China
*These authors contributed equally to this paper
Received 2018-5-28; Accepted 2019-5-13; Published 2019-7-8
Abstract
Background: MicroRNAs are small non-coding RNAs containing 18-22 nucleotides which play a role in RNA silencing and post-transcriptional regulation of their target genes. The MiR-200 family comprises miR-141, miR-200a, miR-200b, miR-200c and miR-429. Increasing evidence indicates that miR-200 microRNAs play a role in cancer metastasis. For example, miR-200 microRNAs were reported to influence the prognosis in colorectal cancer patients by regulating the expression of genes related to the epithelial-mesenchymal transition6. Previous studies have shown that the high expression of miR-200 microRNAs has an impact on the overall survival and Relapse-free Survival of CRC patients. However, the study results were inconsistent.
Results: Data from a total of 1882 patients from 9 studies was included in the meta-analysis. Poorer Relapse-free Survival (RFS) was observed in patients with high expression levels of miR-200 microRNAs (HR=1.13, 95% CI 1.04-1.23). Additionally, subgroup analysis of sample types revealed a significant association between higher expression of the miR-200 family in the plasma and poorer OS (HR=1.23, 95% CI 1.08-1.41) and RFS (HR=2.39, 95% CI 1.20-4.77), which indicates that the miR-200 family can be used as an easily detectable biomarker for evaluation of the prognosis of patients with colorectal cancer.
Conclusions: High expression levels of miR-200 microRNAs were associated with poor clinical outcomes in colorectal cancer patients. The miR-200 family can therefore potentially serve as a prognostic biomarker. Further studies should be performed to verify the clinical utility of the miR-200 family in colorectal cancer.
Keywords: MiR-200 family, Colorectal cancer, Prognostic Value, Meta-analysis
Introduction
Colorectal cancer (CRC) is one of the most common cancers, responsible for 715,000 worldwide deaths in 2010 [1]. It is the second most common cause of cancer death in the USA [2]. In China, colorectal cancer is the fourth most common cause of death and the fifth most common cancer type [3]. In spite of the development of novel treatment options and early screening, the prognosis of CRC patients has not improved markedly. Furthermore, the incidence of CRC in China has been rising rapidly in recent years [4]. Hence, novel biomarkers with clinical value are urgently needed to improve the compliance rate.
MicroRNAs are small non-coding RNAs containing 18-22 nucleotides which play a role in RNA silencing and post-transcriptional regulation of targeted genes. The miR-200 family contains miR-141, miR-200a, miR-200b, miR-200c and miR-429. Increasing evidence suggests miR-200 microRNAs played a role in cancer metastasis [5-14]. In CRC, miR-200 microRNAs were reported to influence the prognosis of cancer patients by regulating genes related to the epithelial to mesenchymal transition [10, 15-19]. Previous studies have shown that the high expression of miR-200 microRNAs has an impact on the overall- and Relapse-free Survival of CRC patients. However, the results were inconsistent. This paper aims to investigate the prognostic value of the MiR-200 family in patients with colorectal cancer via a meta-analysis of available literature.
Materials and Methods
Literature search
A comprehensive literature search was conducted in PMC (PubMed Central), PubMed, Web of Science, Embase, and Cochrane library, with a cut-off date of May 11, 2018, to obtain potentially eligible studies. The keywords for the search were (“miR-141” OR “miR-200a” OR “miR-200b” OR “miR-200c” OR “miR-429”) AND (“colorectal cancer” OR “Colorectal Tumor” OR “Colorectal Neoplasm” OR “Colorectal Carcinoma”) AND (“prognosis” OR “survival”). Additionally, other relevant articles were also obtained by manually screening the reference lists.
Inclusion and exclusion criteria
Inclusion criteria for the articles were as follows: (1) the association between miR-200 family expression and overall survival (OS)/Relapse-free Survival (RFS) was investigated in patients with colorectal cancer; (2) the studies provided hazard ratios (HRs) and their 95% CIs of OS or RFS; (3) the patients were divided into low- and high expression groups according to the expression level of miR-200 microRNAs. Exclusion criteria for the articles were as follows: (1) duplicate publications; (2) studies with unusable data; and (3) reviews, letters, case reports and expert opinions.
Data extraction and quality assessment
Two investigators (H-W and R-S) collected the data and information from all included studies. The following information was extracted from each study: first author name, publication year, total number of patients, outcome measures, the criteria for high miR-200 microRNA expression, determination method, hazard ratios (HRs) and corresponding 95% CIs. For all included studies, the results of multivariate analysis were only selected because of its increased precision for interpreting confounding factors. If a study provided the results of both multivariate and univariate outcomes, we chose the former. If a study only provided Kaplan-Meier survival curves, we extracted the corresponding survival data by using Engauge Digitizer V4.1 [20]. In the event of a disagreement, a third investigator (W-W) was consulted to reach a consensus. The quality of all included studies was assessed by using the Newcastle-Ottawa Scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The NOS scores ranged from 0 to 9 and studies with NOS scores ≥ 6 were considered to be of high quality. The quality of all included studies varied from 7 to 9, with a mean value of 7.77 (Table 2).
Statistical methods
The meta-analysis was conducted by using Stata SE12.0 (Stata Corp, College Station, TX) and R software 3.5.3 (Microsoft Revolution Analytics). The heterogeneity across studies was assessed by using the Chi square-based Q test and I2 statistics [21]. A P-value of less than 0.05 for the Q test or an I2 value of more than 50 % was considered to indicate a significant difference. If the heterogeneity across studies was small (Ph > 0.05, I2 < 50%), the fixed effects model was adopted. Otherwise, the random effects model was applied (Ph ≤ 0.05, I2 ≥ 50%). Potential publication bias was calculated using Begg's test and Egger's test [22]. A sensitivity analysis was also conducted to evaluate the stability of the results. Differences with P-values of less than 0.05 were considered statistically significant.
Results
Study characteristics
The detailed process of literature retrieval is shown in Figure 1. A total of 1882 cancer patients in 9 studies [23-31] were ultimately included in the meta-analysis. The mean sample size was 209 patients (from 78 to 543). The included studies were from America, Asia and Europe (Table 1).
Characteristics of studies included in the meta-analysis.
| Study | Year | Sample size | Country | RNA | HR (OS) | LCI (OS) | UCI (OS) | HR (RFS) | LCI (RFS) | UCI (RFS) | Sample type |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cheng | 2010 | 102 | USA | miR-141 | 1.36 | 0.45 | 4.14 | plasma | |||
| Cheng | 2010 | 156 | China | miR-141 | 3.41 | 1.36 | 8.56 | plasma | |||
| DIAZ | 2014 | 127 | Spain | miR-429 | 0.28 | 0.05 | 0.42 | tumor tissues | |||
| Dong | 2016 | 78 | China | miR-429 | 2.296 | 1.10 | 4.52 | tumor tissues | |||
| Li | 2013 | 110 | China | miR-429 | 2.165 | 1.015 | 4.618 | tumor tissues | |||
| Maierthaler | 2016 | 543 | Germany | miR-141 | 1.105 | 0.904 | 1.35 | 1.085 | 0.89 | 1.321 | plasma |
| Maierthaler | 2016 | 543 | Germany | miR-200a | 1.227 | 1.008 | 1.495 | 1.2 | 0.989 | 1.456 | plasma |
| Maierthaler | 2016 | 543 | Germany | miR-200b | 1.208 | 0.975 | 1.497 | 1.143 | 0.919 | 1.422 | plasma |
| Maierthaler | 2016 | 543 | Germany | miR-200c | 1.152 | 0.975 | 1.362 | 1.1 | 0.93 | 1.302 | plasma |
| Maierthaler | 2016 | 543 | Germany | miR-429 | 1.006 | 0.845 | 1.198 | 1.078 | 0.91 | 1.277 | plasma |
| Roh | 2018 | 109 | Korea | miR-200c | 2.22 | 0.79 | 6.26 | 1.96 | 0.77 | 5.02 | tumor tissues |
| Toiyama | 2014 | 446 | Japan | miR-200c | 2.67 | 1.28 | 5.67 | 3.01 | 1.08 | 8.39 | plasma |
| Yu | 2014 | 103 | China | miR-200c | 1.665 | 1.13 | 1.135 | plasma | |||
| Shelton | 2018 | 108 | USA | miR-200b | 0.459 | 0.284 | 0.74 | tumor tissues | |||
| Shelton | 2018 | 106 | USA | miR-200a | 0.444 | 0.216 | 0.793 | tumor tissues |
Flowchart presenting the steps of literature search and selection.
The expression of miR-200 microRNAs and overall survival
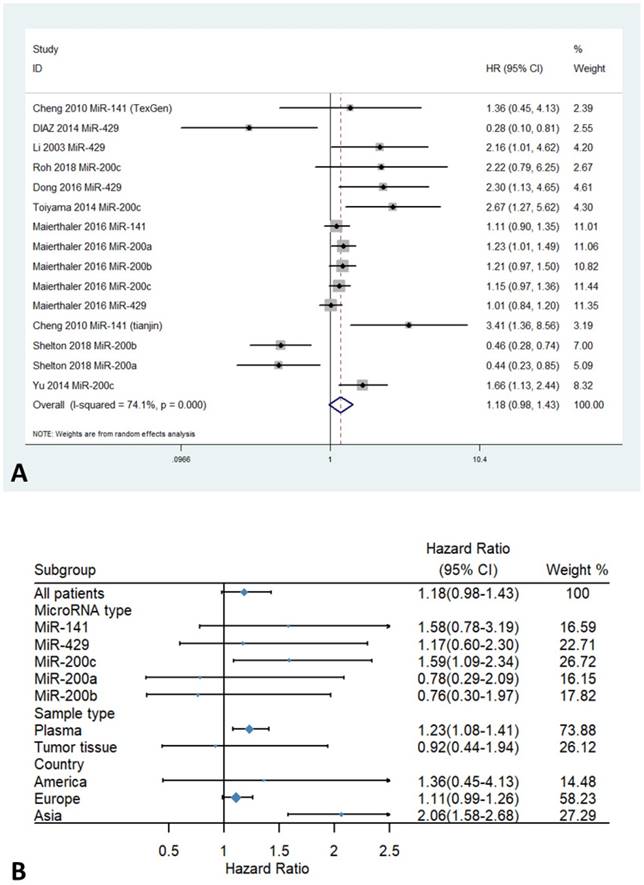
The forest plots for the association between miR-200 expression and overall survival of patients with colorectal cancer are shown in Figure 2. Overall, The HRs of the high miR-200 microRNAs expression group versus the low miR-200 microRNAs expression group was 1.18 (95% CI 0.98-1.43, Figure 2A). Stratified analysis by sample types showed a significant association between enhanced expression of miR-200c in plasma and poor OS (HR=1.23, 95% CI 1.08-1.41, Figure 2B). Moreover, a significant association was found between higher expression of the miR-200 family and poorer OS in the Asian population (HR=2.06, 95% CI 1.58-2.68), compared with the European population (HR=1.11, 95% CI 0.99-1.26, Figure 2B) (Table 3).
The expression of miR-200 microRNAs and Relapse-free Survival
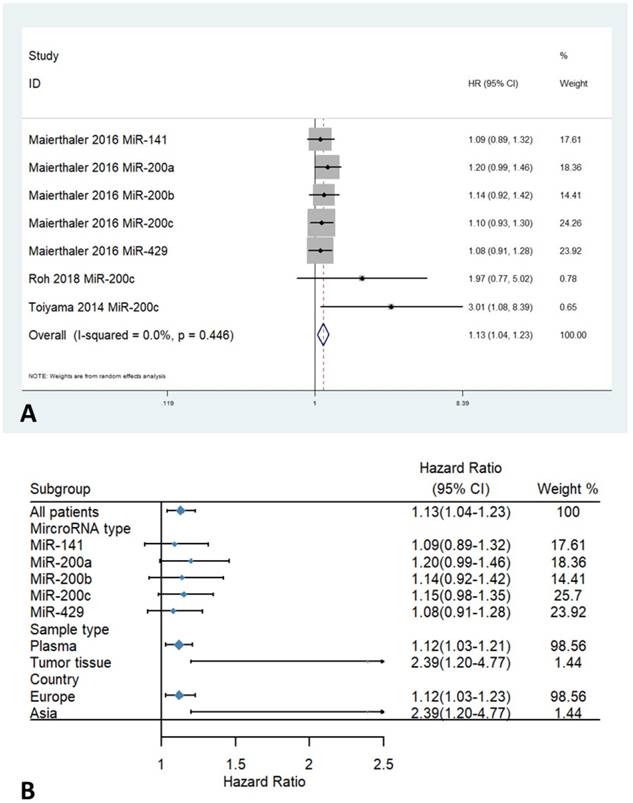
Only three studies with a total of 1098 patients contained data on Relapse-free Survival (RFS). No obvious statistical heterogeneity was observed among the included studies (I2 = 0 %; Ph = 0.446). The fixed-effects model was adopted to assess the pooled hazard ratios (HRs) and corresponding 95% confidence intervals (CIs). The overall result demonstrated a strong association between high expression levels of miR-200 microRNAs and poor RFS (HR=1.13, 95% CI 1.04-1.23, Figure 3A). Stratified analysis by country revealed a poorer RFS in patients with high expression levels of miR-200 microRNAs from Asia. Furthermore, stratified analysis by sample types found a significant association between higher expression of the miR-200 family and poor RFS in plasma (HR= 1.12, 95% CI 1.03-1.21, Figure 3B) (Table 3).
Methodological quality of studies included in the meta-analysis.
| Study | Cheng | DIAZ | Dong | Li | Maierthaler | Roh | Toiyama | Yu | Shelton |
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of the exposed cohort | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Selection of the non-exposed cohort | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Ascertainment of exposure | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Demonstration that outcome of interest was not present at start of study | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Comparability of cohorts on the basis of the design or analysis | ☆☆ | ☆ | ☆☆ | ☆☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ |
| Assessment of outcome | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Was follow-up long enough for outcomes to occur | - | ☆ | - | ☆ | ☆ | ☆ | - | - | ☆ |
| Adequacy of follow-up of cohorts | - | ☆ | - | ☆ | ☆ | ☆ | - | ☆ | - |
| Stars | 7 | 8 | 7 | 9 | 8 | 8 | 7 | 8 | 8 |
Results of quantitative analysis
| Subgroup | HR | 95% CI | I2 (%) | PH |
|---|---|---|---|---|
| OS | ||||
| Overall | 1.18 | 0.98-1.43 | 74.1 | <0.001 |
| Member type | ||||
| miR-141 | 1.58 | 0.78-3.19 | 64.1 | 0.061 |
| miR-200a | 0.78 | 0.29-2.09 | 88.4 | 0.003 |
| miR-200b | 0.76 | 0.30-1.79 | 92.3 | <0.001 |
| miR-200c | 1.59 | 1.09-2.34 | 63.3 | 0.043 |
| miR-429 | 1.17 | 0.60-2.30 | 79.1 | 0.002 |
| Location | ||||
| Asia | 2.06 | 1.58-2.68 | 0 | 0.71 |
| Europe | 1.11 | 0.99-1.26 | 46.8 | 0.094 |
| America | 0.56 | 0.33-0.91 | 40.5 | 0.186 |
| Sample type | ||||
| Plasma | 1.23 | 1.08-1.41 | 52.3 | 0.033 |
| Tumor tissues | 0.92 | 0.44-1.94 | 83.9 | <0.001 |
| RFS | ||||
| Overall | 1.13 | 1.04-1.23 | 0 | 0.446 |
| Member type | ||||
| miR-200c | 1.15 | 0.98-1.35 | 59.4 | 0.085 |
| Location | ||||
| Asia | 2.39 | 1.20-4.77 | 0 | 0.549 |
| Europe | 1.12 | 1.03-1.21 | 0 | 0.931 |
| Sample type | ||||
| Plasma | 1.12 | 1.03-1.21 | 0 | 0.549 |
| Tumor tissues | 2.39 | 1.20-4.77 | 0 | 0.931 |
Sensitivity analysis
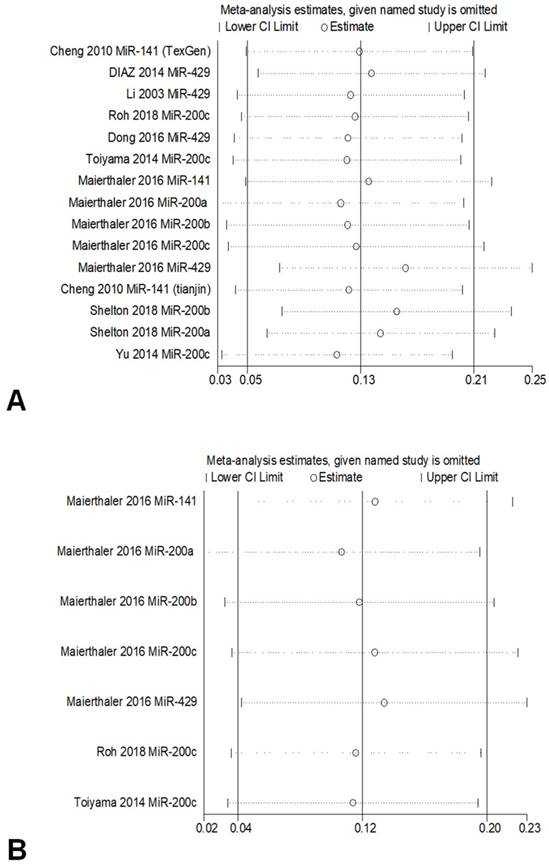
To investigate the correlation between the expression of miR-200 microRNAs and OS, sensitivity analysis was conducted by sequentially omitting each study from the pooled analysis, one at a time. The result was not obviously influenced by the exclusion of any study, which underscored the robustness of the results (Figure 4).
Publication bias
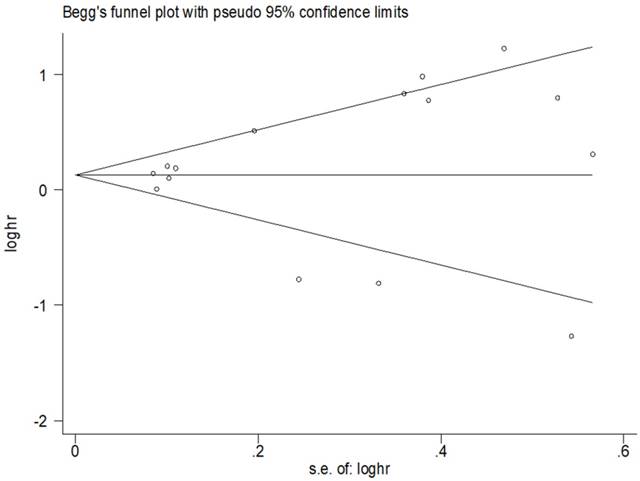
For the relationship between miR-200 microRNA expression levels and OS, Begg's test and Egger's test were applied to test for publication bias. No publication bias was observed in the included studies (Figure 5).
Forest plots of HR for the relationship between high miR-200 microRNA expression and OS: (A) overall, (B) stratified by country, (C) stratified by miRNA type, (D) stratified by sample type.
Forest plots of HR for the relationship between high miR-200 microRNA expression and RFS: (A) overall, (B) stratified by country, (C) stratified by miRNA type, (D) stratified by sample type.
Discussion
MiR-200 microRNAs were found to act together with ZEB1 and ZEB2 proteins to regulate the epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) in the development of CRC and influence its prognosis[15, 32]. Some studies showed that high expression of MiR-200 microRNAs was associated with improved OS in CRC patients [15, 29]. However, other studies demonstrated that the high expression of MiR-200 microRNAs was related to poor OS in comparable patients [24-28]. Although the inconsistent results of these studies have not yet been explained in detail, the discrepancy might partly attributed to the different results of microarray analysis, since these miRNA profiling studies were conducted with different microarray platforms. Furthermore, small differences of experimental design across studies could also affect the observed results. Notably, the insufficient number of tumor samples in the inclusive studies could have led to limited statistical power, diminishing the observed prognostic value of the miR-200 family.
In the current paper, we assessed the relationship between the survival of patients with colorectal cancer and miR-200 expression. The pool analysis showed that higher expression of miR-200 family RNAs was observed to be related to poor RFS in patients with colorectal cancer. Furthermore, the subgroup analysis of sample types found a significant association between higher expression of the miR-200 family in the ser poorer OS and RFS, which implied that the miR-200 family might become an easily detectable biomarker for the evaluation of the prognosis of patients with colorectal cancer.
Sensitivity analyses of studies regarding overall survival (a) and Relapse-free Survival (b).
However, there are some limitations to our meta-analysis. Firstly, the total sample size was relatively small and we failed to detect a relationship between the expression of miR-200 microRNAs and clinicopathological parameters. Secondly, there was significant heterogeneity across studies regarding the association of the miR-200 family with OS. Subgroup analysis demonstrated that the country of origin of the inclusive studies might be the main source of heterogeneity. Thirdly, the number of inclusive studies is small, whose statistical power is still limited. Although the result of publication bias demonstrated that there did not exist publication bias, there may exist publication bias because of limited inclusive studies. Fourthly, there still exists small differences in the role of each member of miR-200 family in colorectal cancer, which may cause also affect the observed results. Fifth, estimating HRs from Kaplan Meier curves can become inaccurate when there are very small differences in overall OS and RFS, which may also have impacted the results of this meta-analysis. Furthermore, the different methods/platforms used for mRNA detection, inconsistent definition of OS/RFS, various sample sources and different types of disease might cause a deviation of the results of this meta-analysis. Finally, this paper did not assess the prognostic value of a combination of miR-200 and other miRNA markers. Therefore, larger-size, multi-center and higher-quality studies with a unified criterion for determining the expression of miR-200 family microRNAs are necessary to validate the results in this study.
Results of publication bias for OS.
Acknowledgements
We wish to thank the participants in this study. This work was supported by grants from the National Natural Science Foundation of China (Nos. 81160248 and 81560464 to DYL).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-128
2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11-30
3. Zhang YL, Zhang ZS, Wu BP, Zhou DY. Early diagnosis for colorectal cancer in China. World journal of gastroenterology. 2002;8:21-5
4. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69-90
5. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283:14910-4
6. Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472-98
7. Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C. et al. Tumour angiogenesis regulation by the miR-200 family. Nature communications. 2013;4:2427
8. Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer letters. 2014;344:166-73
9. Chen Y, Zhang L. Members of the microRNA-200 family are promising therapeutic targets in cancer. Experimental and therapeutic medicine. 2017;14:10-7
10. Le MT, Hamar P, Guo C, Basar E, Perdigao-Henriques R, Balaj L. et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. The Journal of clinical investigation. 2014;124:5109-28
11. Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J. et al. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173-86
12. Bracken CP, Li X, Wright JA, Lawrence DM, Pillman KA, Salmanidis M. et al. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. The EMBO journal. 2014;33:2040-56
13. Zhang G, Zhang W, Li B, Stringer-Reasor E, Chu C, Sun L. et al. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast cancer research: BCR. 2017;19:73
14. Kim JS, Kurie JM, Ahn YH. BMP4 depletion by miR-200 inhibits tumorigenesis and metastasis of lung adenocarcinoma cells. Molecular cancer. 2015;14:173
15. Pichler M, Ress AL, Winter E, Stiegelbauer V, Karbiener M, Schwarzenbacher D. et al. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. British journal of cancer. 2014;110:1614-21
16. Tseng JH, Bisogna M, Hoang LN, Olvera N, Rodriguez-Aguayo C, Lopez-Berestein G. et al. miR-200c-driven Mesenchymal-To-Epithelial Transition is a Therapeutic Target in Uterine Carcinosarcomas. Scientific reports. 2017;7:3614
17. Tang X, Hou Y, Yang G, Wang X, Tang S, Du YE. et al. Stromal miR-200s contribute to breast cancer cell invasion through CAF activation and ECM remodeling. Cell death and differentiation. 2016;23:132-45
18. Howe EN, Cochrane DR, Richer JK. The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. Journal of mammary gland biology and neoplasia. 2012;17:65-77
19. Muralidhar GG, Barbolina MV. The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer. International journal of molecular sciences. 2015;16:16833-47
20. Wells G. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analysis. http: //www, ohri ca/programs/clinical_epidemiology oxford htm. 2004
21. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557-60
22. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
23. Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR. et al. Plasma miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Annals of surgery. 2014;259:735-43
24. Roh MS, Lee HW, Jung SB, Kim K, Lee EH, Park MI. et al. Expression of miR-200c and its clinicopathological significance in patients with colorectal cancer. Pathology, research and practice. 2018;214:350-5
25. Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P. et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. International journal of cancer. 2017;140:176-87
26. Li J, Du L, Yang Y, Wang C, Liu H, Wang L. et al. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer letters. 2013;329:84-90
27. Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A. et al. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PloS one. 2012;7:e46684
28. Diaz T, Tejero R, Moreno I, Ferrer G, Cordeiro A, Artells R. et al. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. Journal of surgical oncology. 2014;109:676-83
29. Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M. et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PloS one. 2011;6:e17745
30. Yu H, Duan B, Jiang L, Lin M, Sheng H, Huang J. et al. Plasma miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. American journal of translational research. 2013;6:71-7
31. Shelton PM, Duran A, Nakanishi Y, Reina-Campos M, Kasashima H, Llado V. et al. The Secretion of miR-200s by a PKCzeta/ADAR2 Signaling Axis Promotes Liver Metastasis in Colorectal Cancer. Cell reports. 2018;23:1178-91
32. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593-601
Author contact
![]() Corresponding author: Renhua Wan; Email:zww726696com
Corresponding author: Renhua Wan; Email:zww726696com

 Global reach, higher impact
Global reach, higher impact