Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(18):4341-4349. doi:10.7150/jca.31326 This issue Cite
Research Paper
Detection of Plasma EGFR Mutations in NSCLC Patients with a Validated ddPCR Lung cfDNA Assay
1. Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
2. Shanghai Lung Cancer Center, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
3. Bio-Chain Biological Technology Co., Ltd, Shanghai, China
#Qiao-mei Guo and Lin Wang are the first co-authors and they contributed equally to this work.
Received 2018-11-8; Accepted 2019-5-9; Published 2019-7-10
Abstract
Purpose: The clinical utility of cell-free DNA (cfDNA) to assess EGFR mutations is increasing. However, there are limited studies determining their clinical validity and utility. The value of cfDNA assays in cancer management remains controversial.
Methods: In this study, we first evaluated the analytical performance of the ddPCR Lung cfDNA Assay. We next analyzed the concordance of the results with tissue amplification refractory mutation system PCR (ARMS-PCR) and plasma next-generation sequencing (NGS) genotyping. Finally, we assessed its clinical utility by exploring the association of cfDNA EGFR mutations with metastatic sites and the efficacy of EGFR-TKIs treatment.
Results: The ddPCR Lung cfDNA Assay demonstrated a limit of blank of 1 droplet per reaction, an analytical specificity of 100%, and detection limit of 0.05%, 0.05%, and 0.1% for E746_A750del, L858R, and T790M, respectively. With tissue ARMS-PCR as a standard for comparison, the clinical sensitivity and specificity of ddPCR were 62.5% (15/24) and 100% (82/82) for E746_A750del, and 75.0% (15/20) and 94.2% (81/86) for L858R, respectively. The ddPCR showed high concordance with NGS in determining cfDNA EGFR mutations. Patients with bone and/or brain metastasis showed a higher detection rate and mutant abundance of cfDNA EGFR mutations compared to those with other sites of metastasis. Moreover, EGFR-TKIs treatment was effective in patients with sensitive EGFR mutations in either plasma cfDNA or tumor tissue-derived DNA.
Conclusions: We validated in this study that the ddPCR Lung cfDNA Assay is reliable for detection of EGFR mutations in lung cancers, in terms of analytical performance, clinical validity and utility.
Keywords: cell-free DNA, droplet digital PCR, EGFR, NGS, non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. Non-small cell lung cancer (NSCLC) accounts for approximately 80% of lung cancers. The majority of the NSCLC patients are unresectable at their initial diagnosis and suffer from desperate prognosis. In the past decade, the advancement of molecular targeted therapies has greatly improved the survival of patients with advanced NSCLC, in which EGFR-tyrosine kinase inhibitors (EGFR-TKIs) was a tremendous success. Epidermal growth factor receptor (EGFR) is a transmembrane protein that plays a complicated role in signal transduction and cellular processes. EGFR mutations have been recently reported to occur in 10% of NSCLC patients in Western countries and 30-50% of cases in East Asia [1]. The most common EGFR mutations are located in the tyrosine kinase domain of the EGFR gene, including deletions in exon 19 (45%) and a missense mutation (L858R) in exon 21 (40-45%) [2, 3]. These alterations result in constitutive activation of the downstream signaling and serve as the driver of neoplastic transformation and progression. The presence of these mutations are associated with EGFR-TKI sensitivity and hence may serve as a predictive biomarker of response to the related targeted therapies [4, 5]. Despite showing a response rate of over 70% in patients harboring EGFR driving mutations, the diseases eventually progress at a median time ranging from approximately 10-13 months [4, 6, 7]. The most common mode of acquired resistant in EGFR mutation-positive patients is the development of a secondary point mutation in the EGFR active domain, substituting a bulky methionine amino acid for threonine (T790M) and inhibiting the binding of EGFR-TKIs. Currently, the T790M mutation was estimated to represent 50-60% of resistance to first- and second-generation TKIs [8-10]. Hence, it is necessary to detect and monitor EGFR mutations throughout the treatment and surveillance of patients with advanced NSCLC.
In clinical practice, tumor tissues are insufficient for EGFR genotyping in at least 20% of advanced NSCLC patients for various reasons including insufficient availability of neoplastic tissue, lack of appropriate tumor tissue for biopsy, or that a biopsy is not technically feasible [11, 12]. Beyond that, a single biopsy snapshot could not represent the global feature of a heterogeneous tumor and repeat biopsy for treatment monitoring is challenging. Recently, circulating tumor DNA (ctDNA) as a non-invasive “real-time” biomarker is being extensively studied, aiming to substitute tissue genotyping and provide an insight into the tumor heterogeneity. These may allow the molecular analysis in patients with tissue sample unavailable and offer a chance to monitor the mutational evolution of the tumors during treatment, possibly predicting disease progression. In fact, ctDNA has been studied in several specific areas to be an alternative surrogate for molecular analysis in cancer patients [13-15].
Nevertheless, the mutation analysis and mutant abundance quantification of the trace amounts of ctDNA still pose a challenge to the plasma DNA-based detection techniques. The commonly used methods for plasma ctDNA EGFR mutation analysis include amplification refractory mutation system PCR (ARMS-PCR), droplet digital PCR (ddPCR), and next-generation sequencing (NGS) [16-19]. ARMS-PCR was considered to be better suited for detection of tissue samples where mutant allele fraction (MAF) was usually higher than 1%. Testing of EGFR mutations as part of targeted NGS panels allows various driver mutations to be analyzed simultaneously, however, the bioinformatic pipeline to call mutations from large amounts of raw sequencing data is complicated. Digital PCR now represents a precise and sensitive single-molecule counting strategy to detect extremely low levels of genetic materials. The performance of digital PCR surpasses many quantitative methods, enabling the detection of rare mutations originating from tumor cells mixed in a large background of homologous sequences [20]. Researches regarding ctDNA assays by ddPCR are rapidly growing, but there is a lack of study assessing its analytical validity, clinical validity, and utility in the real-world setting.
Here, we established a lung cancer ctDNA detection platform named ddPCR Lung cfDNA Assay. We assessed the analytical and clinical performance of the platform by evaluating DNA derived from mutant cell lines at specified MAF and clinical specimens. The clinical utility of the platform was also explored by analyzing the association between EGFR mutation status and metastatic sites in 108 metastatic NSCLC patients. Furthermore, the efficacy of EGFR-TKIs treatment in 122 newly diagnosed patients with sensitive EGFR mutations was also evaluated.
Material and Methods
Patients
The study was approved by the Ethics Committee of Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China. From June 2016 to March 2018, NSCLC patients who were subjected to peripheral blood cfDNA EGFR analyses at the Shanghai Chest Hospital were enrolled in this study. Healthy individuals with similar age and gender distributions who performed regular physical check-up in our hospital and were not diagnosed with any malignant diseases were recruited and served as a control group. Signed written informed consent was obtained from each participant.
Cell lines
Three human lung cancer cell lines including PC-9, H1975, and A549 were purchased from the American Type Culture Collection. The cells were cultured at 37°C in a 5% CO2 humidified atmosphere with RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 50 U/mL penicillin, 50 µg/mL streptomycin, and 2 mmol/L L-glutamine (Gibco). Among them, PC-9 contains E746_A750del mutation, H1975 contains L858R and T790M mutations, and A549 contains wild-type EGFR. The genomic DNA from these cell lines were extracted for mutation analysis in our study.
Plasma collection and DNA extraction
Blood samples were collected into two 5-mL EDTA vacutainer tubes from each participant after informed consent had been obtained. Blood samples were immediately spun into plasma with two-step centrifuges within 2 hours. Firstly, the whole blood was centrifuged at 1600g for 10 mins at 4°C, then the supernatant was centrifuged at 16000g for 10 mins at 4°C. Cell-free DNA (cfDNA) was extracted from 4 mL of plasma using QIAamp Circulating Nucleic Acid Kit (Qiagen). Genomic DNA was extracted from lung cancer cell lines using TIANamp Genomic DNA Kit (Tiangen) according to the manufacturer's instructions.
ddPCR
ddPCR was performed using the QX200 Droplet Digital PCR system (Bio-Rad Laboratories). Samples were prepared by mixing 10 µL ddPCR Supermix for probes (No dUTP, Bio-Rad Laboratories), 2 µL ddPCR™ probe assay Kit (Bio-Rad Laboratories) which consist of forward and reverse PCR primers and FAM or HEX-labeled fluorescent probe (specific for each mutation assay), and 8 µL of template DNA in a final reaction volume of 20 μL. Droplets were generated by a QX200 droplet generator. Endpoint PCR was performed on a T100 Thermal Cycler (Bio-Rad Laboratories). Thermal cycling profile for EGFR mutation assay was starting with a hot start denaturation step of 10 mins at 95°C, followed by 40 cycles of: 94°C for 30 s, 70°C for 1 min. These cycles were followed by 98°C for 10 mins and then 4°C hold. Then, PCR products were loaded into the QX200 droplet reader and analyzed by QuantaSoft version 1.7.4.0917 (Bio-Rad Laboratories). For each assay: water without templates (NTC) served as a control for detecting environmental contamination; a negative control (genomic DNA from EGFR wild-type A549 cell line) was used to estimate the false-positive rates; and a positive control containing genomic DNA from mutant (MT) cell lines (H1975 or PC-9) was used to verify the assay performance and determine the threshold value of fluorescent signals. Poisson distribution was used to determine the concentration of mutant DNA and calculate the MAF. The MAF is equal to the number of droplets positive for mutant FAM probe / total number of positive for mutant FAM probe plus wild-type HEX probe. The measured MAF was expressed in percentage.
NGS
Plasma ctDNA libraries were prepared with a Firefly NGS DNA Library Prep Reagent Set (AccuraGen) as described in a previous study [21]. The Firefly NGS is an amplicon-based lung cancer panel targeting about 68 hotspots in EGFR, KRAS, and BRAF genes. The platform had a detection limit of 0.1%. All samples were sequenced using Illumina Miseq (Illumina) in a 150 bp paired-end pattern. Mutation calls data were analyzed by CometScope software (AccuraGen).
ARMS-PCR
The paired tumor tissue DNA samples were tested by using the EGFR 29 Mutation Detection Kit (Amoy Diagnostics) according to the manufacturer's instructions. The kit detects 29 clinically-relevant EGFR mutations in lung cancer. Mutation status of deletions in exon 19, L858R and T790M mutations were used for the analysis in this study.
Statistical analysis
Coincidence rates between different detection platforms were calculated by using Cohen's kappa. The average MAF between patients with bone and/or brain metastasis (B/BM) and patients with other sites of metastases (OSM) were compared by using independent sample t-test. Progression-free survival (PFS) was defined as the period from the date of initiation of treatment to the date of disease progression or death due to any cause. Survival curves for PFS were generated by using the Kaplan-Meier method. Log-rank tests were used to compare the survival curves among different subgroups. P values < 0.05 were considered to be statistically significant. All statistical analyses were performed by using SPSS software (version 24.0, SPSS Inc.) or GraphPad Prism for Windows (version 6.07, GraphPad Software).
Results
Patients' characteristics
A total of 201 NSCLC patients were subjected to peripheral blood cfDNA EGFR analyses. 82.6% of patients had adenocarcinoma, 11.4% had NSCLC not otherwise specified, and 6.0% had squamous cell carcinoma. The majority of the patients were stage IV (85.6%) and only 14.4% were stage I to III. Of these, 68 patients were resistant to EGFR-TKIs, 133 patients were EGFR-TKI treatment-naïve, and 106 patients were identified retrospectively to have matched tumor tissue EGFR genotyping results. The schema for patient screening is shown in Figure 1, and the detailed clinical characteristics of the enrolled patients are summarised in Table 1.
Analytical validity
Firstly, we intended to evaluate the analytical performance of the ddPCR Lung cfDNA Assay by determining the Limit of Blank (LoB) for single variants, and defining the detection limit, analytical specificity, and linearity of the assay. By using plasma cfDNA from 20 healthy donors, the LoB was decided by fitting a Poisson model to the false-positive frequency distribution for each target and evaluating the 95% one-tailed upper limit of the model distribution as previously described [22]. The number of false-positive droplet event is 1 for all of the E746_A750del, L858R, and T790M (Figure 2A). Therefore, samples were considered “positive” if assays showed equal to or more than 2 droplets at the expected amplitude for all the three types of mutations (Figure 2B).
Then, the detection limit was assessed by using cell line-derived genomic DNA with known variants at specified MAF of 1%, 0.5%, 0.1%, and 0.05%. A total of 50 ng of input DNA with varying proportions of mutant DNA mixed into wild-type DNA was subjected to ddPCR. To evaluate the robustness of the assay, all reactions were repeated 20 times at each MAF. The minimum MAF that our system can reliably detect (CV<20%) was considered as the detection limit. Results indicated that the detection limit was 0.05% for E746_A750del and L858R, and 0.1% for T790M, respectively (Figure 2C).
The analytical specificity was evaluated by testing different input amounts of wild-type genomic DNA mixed into potentially interfering substances such as bacterial genomic DNA. No false-positive events above the defined threshold of 2 droplets per reaction were observed for each type of mutation. In addition, the three positive DNA templates were not cross-reacting or interfering with each other (Table 2). In summary, the analytical specificity of the ddPCR assay was 100%.
Clinical characteristics of the 201 enrolled advanced NSCLC patients.
| Characteristic | Parameter value | |
|---|---|---|
| Gender; No. (%) | ||
| Male | 96 (47.8) | |
| Female | 105 (52.2) | |
| Age, years; means (range) | 61.5 (23-87) | |
| Histology; No. (%) | ||
| Adenocarcinoma (AC) | 166 (82.6) | |
| Squamous cell carcinoma (SCC) | 12 (6.0) | |
| NSCLC not otherwise specified | 23 (11.4) | |
| Stage; No. (%) | ||
| Ⅰ-Ⅲ Ⅳ | 29 (14.4) 172 (85.6) | |
| Tissue mutation status; No. | ||
| 19dels | 24 | |
| L858R | 20 | |
| Wild type | 62 | |
| Unavailable | 95 | |
| Prior treatment status; No. | ||
| Acquired resistant to EGFR-TKIs | 68 | |
| EGFR-TKIs naïve patients | 133 | |
| Metastasis in TKI-naïve patients; No. | ||
| Bone and/or brain (B/BM) | 67 | |
| Other sites of metastasis (OSM) Without metastasis | 41 25 | |
The schema for patient screening.
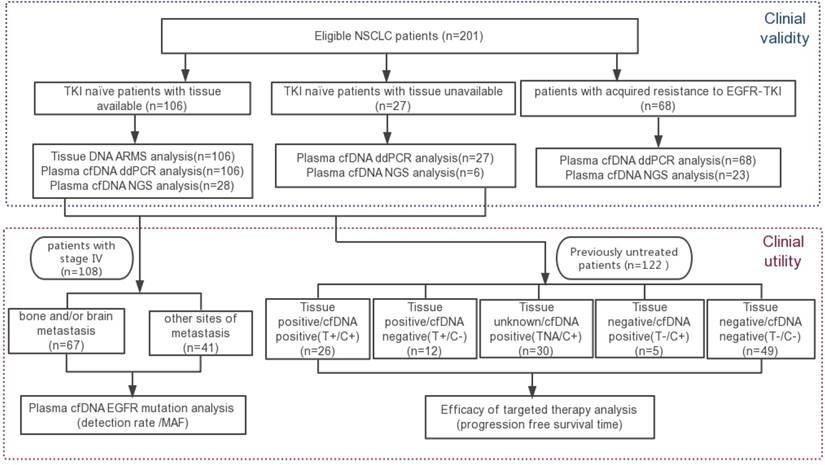
The analytical validity of the ddPCR Lung cfDNA Assay. (A) Determination of the limit of blank (LOB) of E746_A750del, L858R, and T790M. The LOBs were determined as 1 event per reaction for all the three types of mutation from the 95% confidence interval of the Poisson model fit. (B) The representative positive result detected by ddPCR. Droplets containing mutative targets are double positive for FAM and HEX (shown in the upper right quadrant). (C-D) The analytical sensitivity and linearity of E746_A750del, L858R, and T790M. The serially diluted positive cell line DNA with different MAF was duplicated 20 times for sensitivity and coefficient of variation (CV%) was calculated and exhibited. For linearity assay, the serially diluted positive cell line DNA were tested in triplicates at each concentration.
The analytical specificity of the ddPCR assay.
| Template | Mutant allele fraction (mutant/wild-type) | ||
|---|---|---|---|
| Ex19del | T790M | L858R | |
| Low concentration of wild-type genomic DNA | 0/1780 | 0/1840 | 0/1900 |
| Medium concentration of wild-type genomic DNA | 0/7400 | 3.6/7720 (0 event) | 0/7960 |
| High concentration of wild-type genomic DNA | 0/13780 | 1.8/13800 (0 event) | 0/1442 |
| L858Rand T790M positive genomic DNA | 0/11720 | - | - |
| E746_A750del positive genome DNA | - | 5.6/20120 (1event) | 0/2038 |
| DNA from Staphylococcus aureus | 0/0 | 0/0 | 0/0 |
| DNA from Escherichia coli | 0/0 | 0/1.8 | 0/2 |
To determine the linearity, cell line-derived genomic DNA containing the desired EGFR mutations was serially diluted with wild-type DNA to obtain samples with MAF at 50%, 10%, 5%, 1%, 0.5%, 0.1%, 0.05%, and 0.01%. All reactions for each MAF were performed in triplicates. On average, all three types of mutations showed good linearity (R2>0.99, Figure 2D) and wide dynamic range (from 1:20000 to 2000:20000 copies).
Clinical validity
The clinical validity of ddPCR Lung cfDNA Assay was first evaluated in terms of the concordance of EGFR mutations detected between paired tumor tissue specimens and plasma samples obtained from 106 EGFR-TKIs treatment-naïve patients. The median amount of input cfDNA per reaction was 5.66 ng (range: 2.21-76 ng). With tissue genotyping results as a standard for comparison, the sensitivity and specificity of ddPCR were 62.5% (15/24) and 100% (82/82) for E746_A750del, and 75.0% (15/20) and 94.2% (81/86) for L858R, respectively (Table 3).
Performance of the ddPCR assay for detecting EGFR mutations in comparison with Tissue ARMS-PCR genotyping.
| cfDNA ddPCR | Tissue ARMS-PCR | Performance of ddPCR | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Sensitivity (%) | Specifity (%) | Concordance (%) | ||
| E746_A750del | Positive | 15 | 0 | 62.5 | 100 | 91.5 |
| Negative | 9 | 82 | ||||
| L858R | Positive | 15 | 5 | 75.0 | 94.2 | 90.6 |
| Negative | 5 | 81 | ||||
Performance of the ddPCR assay for detecting cfDNA EGFR mutations in comparison with NGS.
| cfDNA ddPCR | NGS | Concordance | Kappa | P | ||
|---|---|---|---|---|---|---|
| Positive | Negative | (%) | ||||
| E746_A750del | Positive | 16 | 3 | 89.5 | ||
| Negative | 3 | 35 | 0.763 | <0.001 | ||
| L858R | Positive | 14 | 1 | 98.3 | ||
| Negative | 0 | 42 | 0.954 | <0.001 | ||
| T790M | Positive | 6 | 2 | |||
| Negative | 2 | 47 | 93.0 | 0.709 | <0.001 | |
In view of the biological discrepancies between plasma and tissue samples, we next compared our ddPCR Lung cfDNA Assay with a commercial NGS platform for the detection of EGFR mutations in cfDNA from 57 patients. The concordance rates between the two platforms were 89.5% (51/57) for E746_A750del, 98.3% (56/57) for L858R, and 93.0% (53/57) for T790M, respectively (Table 4). Furthermore, ddPCR Lung cfDNA Assay showed a 42.7% (29/68) positive rate for T790M in cases with acquired resistance to TKI, which is consistent with previous findings of ASTRIS and AURA 17 studies [17, 23, 24].
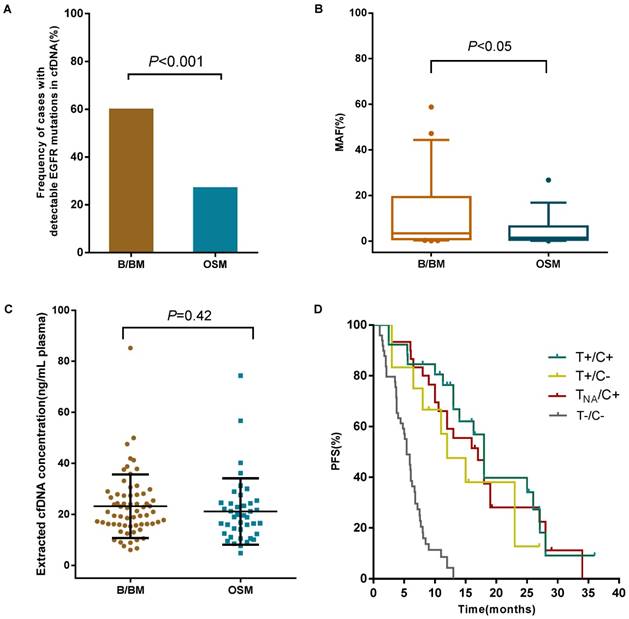
Clinical utility of cfDNA EGFR mutation detection by the ddPCR assay. (A) The frequency of cases with detectable EGFR mutations in cfDNA between B/BM group and OSM group was presented. (B) The mutant allele fraction (MAF) in patients with B/BM and OSM. The average percentage of MAF in plasma EGFR is shown by a midline (outliers excluded). (C) The extracted cfDNA concentration between B/BM group and OSM group. The mean concentration of cfDNA was analyzed in 67 B/BM patients and 41 OSM patients. (D) Survival curves of progression-free survival (PFS) in 122 newly diagnosed NSCLC patients.
Clinical Utility
Correlation of plasma EGFR mutations with metastases
B/BM are common metastatic sites in patients with lung cancer. The prognosis of these patients is poor with a median survival of less than 1 year [25]. We retrospectively investigated the relationship between metastases and EGFR mutation status in cfDNA from 108 TKI-naïve patients with stage IV NSCLC. For this cohort of patients, 59.7% (40 out of 67 patients who developed B/BM) had detectable sensitive EGFR mutations in cfDNA, in contrast, only 26.8% (11/41) with OSM had detectable sensitive EGFR mutations in cfDNA (P<0.001, Figure 3A). Then, we analyzed whether EGFR mutant abundance in plasma was correlated with the occurrence of B/BM. In 51 patients with sensitive EGFR mutations in cfDNA, the average MAF in patients with B/BM was 9.9%, which was obviously higher than that in patients with OSM (3.5%, P<0.05, Figure 3B), even though the extracted cfDNA concentration was similar between the two groups (23.3 ng/mL vs 21.2 ng/mL, P=0.42, Figure 3C). Moreover, patients who developed brain metastases harbored higher EGFR mutant abundance with an average MAF of 18.2%, whereas patients who developed only bone metastases carried slightly lower EGFR mutant abundance with an average MAF of 6.6% (P<0.05). Interestingly, 8 out of 11 patients (72.7%) with brain metastases accompanied by bone metastases. The above results indicated that B/BM in lung cancer patients was associated with positive EGFR mutations in cfDNA and that the higher abundance of sensitive EGFR mutations in cfDNA might be a risk factor for developing brain metastases.
Prognostic significance of plasma EGFR mutations detected by ddPCR
To emphasize the clinical significance of different EGFR mutation status, we analyzed the progression-free survival (PFS) in 122 newly diagnosed patients treated with EGFR-TKIs or standard chemotherapy. Patients were stratified into subgroups based on the genotyping result: Group A, Tissue-positive/cfDNA-positive EGFR mutation (T+/C+); Group B, Tissue-positive/ctDNA-negative EGFR mutation (T+/C-); Group C, Tissue-unknown/ctDNA-positive EGFR mutation (TNA/C+); Group D, Tissue-negative/ctDNA-negative EGFR mutation (T-/C-); and Group E, Tissue-negative/ctDNA-positive EGFR mutation (T-/C+) . The PFS of each group was investigated (Figure 3D). Patients with EGFR mutations in either tissue or cfDNA (Group A-C) had a significantly improved PFS (13 months, n=68) compared to patients harbored wild-type EGFR in both tissue and cfDNA (Group D: 5.4 months, n=49, P<0.001). The median PFS of Group A was 15 months, which was slightly longer than that of Group B and Group C, (11.5 and 13 months, respectively), however, no statistical difference was reached (P=0.202). Furthermore, we observed the two of five patients with Tissue-negative /cfDNA-positive EGFR mutation (T-/C+, group E) showed a PFS of 8 and 14 months, respectively. These results suggest that EGFR mutation status in either tissue and cfDNA was associated with clinical response to TKIs. EGFR analysis in cfDNA is a potential alternative method for those patients who cannot obtain sufficient tumor tissue sample. Plasma cfDNA-based EGFR mutations analysis by ddPCR is useful in guiding clinical decisions in patients with insufficient or unavailable tumor specimens.
Discussion
Plasma cfDNA genotyping has already been utilized in guiding clinical decision-making in advanced NSCLC patients. The joint assessment from experts of American Society of Clinical Oncology and the College of American Pathologists has declared that there is insufficient evidence of analytical validity, clinical validity, and utility for the majority of ctDNA assays in cancer [26]. We, therefore, established the ddPCR Lung cfDNA Assay and assessed its performance according to the suggestions. Our results showed that the detection limit of the ddPCR Lung cfDNA Assay was 0.05% for E746_A750del and L858R, and 0.1% for T790M, respectively. Using the defined 2 events per reaction as the threshold, the analytical specificity was 100% for all three types of EGFR mutations.
In a real-world setting, the practical sensitivity of the ddPCR assay is strongly dependent on the amount of available DNA sample [27, 28]. To yield sufficient input DNA for ddPCR assay, we optimized the pre-analytical specimen processing. A sequential pair of centrifugations of 4 mL peripheral anticoagulant venous blood was performed within 2 hours after collection [26]. The median amount of cfDNA is 5.22 ng per reaction and 95% of the samples are over 3 ng (equivalent to about 1000 copies of the genome), which will generate a theoretical sensitivity of 0.1%. Zhang, et al. demonstrated that the sensitivity of ddPCR was more than 60% for the cases with cfDNA inputs of 2-5 ng per reaction when tumor-tissue EGFR mutation served as a standard for comparison [27]. Our results were in agreement with their findings.
The ddPCR Lung cfDNA Assay exhibited a high concordance with tissue ARMS-PCR result for the detection of E746_A750del and L858R, which was consistent with the previously reported concordance rate ranging from 70% to 90% [13, 21, 29]. However, the clinical sensitivity of the ddPCR Lung cfDNA Assay for detecting E746_A750del (62.5%, 15/24) was modest. The false-negative results may be arise from the different coverage of the platforms, the heterogeneity of tumor, the complexity of cfDNA shedding, and other clinical factors. Particularly, the tissue ARMS-PCR assay used in the present study covered multiple types of exon 19 deletion mutations, not just E746_A750del. This could be the critical reason for the decrease in sensitivity of detecting plasma E746_A750del by the assay. In addition, we found that 29.0% (11/38) of patients who have discordant EGFR mutations performed their tumor tissue ARMS-PCR detection at least one months earlier than plasma ddPCR detection, suggesting that the discrepancies may be caused by molecular evolution of tumors.
For cfDNA T790M mutation detection, the ddPCR Lung cfDNA assay shows a high concordance with a well-developed commercial NGS platform, proving the reliability of the cfDNA Lung ddPCR Assay in T790M mutation detection. Moreover, in the present study, the positive rate of plasma T790M in patients with acquired resistance to EGFR-TKIs was 42.7%, which was consistent with the previously reported detection rate. Several famous studies based on the NSCLC population in the Asia-Pacific, namely ASTRIS and AURA 17, had also reported similar positive rates of plasma T790M with our current study, ranging from 36.1% to 56%. The difference in the T790M detection rates may be due to clinical factors such as chemotherapy, smoking, different types of primary mutation, targeted therapy, the duration of systemic treatment, and etc. [9, 30-32].
For clinical utility, we found that patients with B/BM demonstrated a higher detection rate of sensitive EGFR mutations in plasma cfDNA than those with OSM. Patients with B/BM carried a higher abundance of sensitive EGFR mutations in cfDNA. Subgroup analysis revealed that MAF in patients with brain metastasis was obviously higher than that in patients with only bone metastasis and that in patients with OSM. In addition, most of the brain metastasis patients were accompanied by bone metastasis. Survival analysis revealed that patients with either Tissue-positive or cfDNA-positive EGFR mutation had a longer PFS after EGFR-TKIs treatment compared to those with wild-type EGFR mutations receiving conventional standard treatments, suggesting that the combined use of tissue and cfDNA assays may be a better strategy to select patients eligible for EGFR-TKIs treatment in a real-world clinical setting.
Our study has several limitations. First, self-made materials were used to establish the analytical sensitivity of the ddPCR platform instead of commercial cfDNA reference which might be more reliable. Second, the sample size is too small in the subgroup of patients with Tissue-negative /cfDNA-positive EGFR mutation (T-/C+). Several studies also reported the occurrence of T-/C+ patients [33-36], however, they accounted for only 4.0% of the study population (155 in 3834 cases). The efficacy of EGFR-TKIs to those patients was also observed as reported by Mok, et al. [37]. Besides, the sensitivity of ddPCR is highly depended on the input amount of DNA template. Enrichment or pre-amplification is required before ddPCR assay for low concentration samples. Despite these limitations, our study is valuable in terms of the well-validated analytical performance and clinical utility in real-world setting.
In conclusion, we have developed the ddPCR Lung cfDNA Assay for detecting EGFR mutations with both high sensitivity and specificity, and with the robustness needed in clinical practice. The assay could be valuable in the detection of actionable EGFR mutations in patients who are unable to undergo repeat biopsies and possibly detecting “missed” mutations by standard tissue genotyping due to tumor heterogeneity. Furthermore, this approach would be particularly useful in predicting the B/BM in NSCLC patients.
Abbreviations
EGFR: epidermal growth factor receptor; ddPCR: droplet digital polymerase chain reaction; NSCLC: non-small cell lung cancer; EGFR-TKIs: epidermal growth factor receptor tyrosine kinase inhibitors; ctDNA: circulating tumor DNA; cfDNA: cell-free DNA; LOB: limit of blank; ARMS: amplification refractory mutation system; NGS: next-generation sequencing; MAF: mutant allele fraction; PFS: progression-free survival.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (81672833), the Ministry of Science and Technology (2014DFA33010), Shanghai Municipal Health Commission Collaborative Innovation Cluster Project (2019CXJQ03), and Shanghai Collaborative Innovation Center for Translational Medicine (TM201610).
Author contributions
Conception and design: Jia-tao Lou, Lin Wang, Qiao-mei Guo, Yun-hua Xu. Provision of study patients: Qiao-mei Guo, Wen-jun Yu, Li-hua Qiao, Ming-na Zhao. Performing experiments: Qiao-mei Guo, Lin Wang, Yun-hua Xu, Sheng Ni. Collection and assembly of data: Qiao-mei Guo, Xiao-meng Hu, Lin Wang, Ya-meng Sun. Manuscript writing: Qiao-mei Guo and Lin Wang. Final approval of manuscript: All authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587-95
2. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367-80
3. Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR. et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2-16
4. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947-57
5. Linardou H, Dahabreh IJ, Bafaloukos D, Kosmidis P, Murray S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 2009;6:352-66
6. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169-81
7. Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T. et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327-34
8. Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26
9. Matsuo N, Azuma K, Sakai K, Hattori S, Kawahara A, Ishii H. et al. Association of EGFR Exon 19 Deletion and EGFR-TKI Treatment Duration with Frequency of T790M Mutation in EGFR-Mutant Lung Cancer Patients. Sci Rep. 2016;6:36458
10. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786-92
11. Hiley CT, Le Quesne J, Santis G, Sharpe R, de Castro DG, Middleton G. et al. Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet. 2016;388:1002-11
12. Bernabe R, Hickson N, Wallace A, Blackhall FH. What do we need to make circulating tumour DNA (ctDNA) a routine diagnostic test in lung cancer? Eur J Cancer. 2017;81:66-73
13. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-86
14. Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108-12
15. Goto K, Ichinose Y, Ohe Y, Yamamoto N, Negoro S, Nishio K. et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7:115-21
16. Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J. et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17:7808-15
17. Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med. 2017;6:154-62
18. Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y. et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn. 2015;17:265-72
19. Pirker R, Herth FJ, Kerr KM, Filipits M, Taron M, Gandara D. et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010;5:1706-13
20. Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. 2015;48:948-56
21. Xu T, Kang X, You X, Dai L, Tian D, Yan W. et al. Cross-Platform Comparison of Four Leading Technologies for Detecting EGFR Mutations in Circulating Tumor DNA from Non-Small Cell Lung Carcinoma Patient Plasma. Theranostics. 2017;7:1437-46
22. Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X. et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722-31
23. Zhou C, Wang M, Cheng Y, Chen Y, Zhao Y, Shi Y-K. et al. P3.02b-096 Osimertinib (AZD9291) in Asia-Pacific Patients with T790M Mutation-Positive Advanced NSCLC: Open-Label Phase II Study Results: Topic: EGFR RES. Journal of Thoracic Oncology. 2017;12:S1250
24. Li C, Jia R, Liu H, Zhang B, Wang C. EGFR T790M detection and osimertinib treatment response evaluation by liquid biopsy in lung adenocarcinoma patients with acquired resistance to first generation EGFR tyrosine kinase inhibitors. Diagn Pathol. 2018;13:49
25. D'Antonio C, Passaro A, Gori B, Del Signore E, Migliorino MR, Ricciardi S. et al. Bone and brain metastasis in lung cancer: recent advances in therapeutic strategies. Ther Adv Med Oncol. 2014;6:101-14
26. Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ. et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018 JCO2017768671
27. Zhang Y, Xu Y, Zhong W, Zhao J, Chen M, Zhang L. et al. Total DNA input is a crucial determinant of the sensitivity of plasma cell-free DNA EGFR mutation detection using droplet digital PCR. Oncotarget. 2017;8:5861-73
28. Milbury CA, Zhong Q, Lin J, Williams M, Olson J, Link DR. et al. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif. 2014;1:8-22
29. Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H. et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90:509-15
30. Luo YH, Chen YM. Influence of chemotherapy on EGFR mutation status. Transl Lung Cancer Res. 2013;2:442-4
31. Bai H, Wang Z, Chen K, Zhao J, Lee JJ, Wang S. et al. Influence of chemotherapy on EGFR mutation status among patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:3077-83
32. Nosaki K, Satouchi M, Kurata T, Yoshida T, Okamoto I, Katakami N. et al. Re-biopsy status among non-small cell lung cancer patients in Japan: A retrospective study. Lung Cancer. 2016;101:1-8
33. Wang S, Han X, Hu X, Wang X, Zhao L, Tang L. et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta. 2014;430:63-70
34. Uchida J, Kato K, Kukita Y, Kumagai T, Nishino K, Daga H. et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin Chem. 2015;61:1191-6
35. Weber B, Meldgaard P, Hager H, Wu L, Wei W, Tsai J. et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer. 2014;14:294
36. Qian X, Liu J, Sun Y, Wang M, Lei H, Luo G. et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget. 2016;7:29154-65
37. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J. et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res. 2015;21:3196-203
Author contact
![]() Corresponding authors: Yun-hua Xu, Shanghai Lung Cancer Center, Shanghai Chest Hospital, Shanghai Jiao Tong University, No.241 Huai Hai Road(W), Shanghai, China, 200030, Fax: 86-21-62808279, E-mail: xuyunhua1015com. Jia-tao Lou, Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, No.241 Huai Hai Road(W), Shanghai, China, 200030, Fax: 86-21-62808279, Tel: 86-21-22200000-1601, E-mail: loujiataocom
Corresponding authors: Yun-hua Xu, Shanghai Lung Cancer Center, Shanghai Chest Hospital, Shanghai Jiao Tong University, No.241 Huai Hai Road(W), Shanghai, China, 200030, Fax: 86-21-62808279, E-mail: xuyunhua1015com. Jia-tao Lou, Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, No.241 Huai Hai Road(W), Shanghai, China, 200030, Fax: 86-21-62808279, Tel: 86-21-22200000-1601, E-mail: loujiataocom

 Global reach, higher impact
Global reach, higher impact