Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(18):4350-4356. doi:10.7150/jca.35205 This issue Cite
Review
MicroRNA networks regulate the differentiation, expansion and suppression function of myeloid-derived suppressor cells in tumor microenvironment
1. Department of Histology and embryology, Shangdong First Medical University & Shangdong Academy of Medical Sciences, Taian, Shangdong, China.
2. National Engineering Lab for Druggable gene and protein screening, Northeast Normal University, Changchun, Jilin, China.
3. Department of pharmacy, Changchun University of Chinese Medicine, Changchun, Jilin, China.
4. National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Received 2019-3-25; Accepted 2019-6-3; Published 2019-7-10
Abstract
Myeloid-derived suppressor cells (MDSCs), one heterogeneous population of immature myeloid cells, have suppressive function on immune response during tumor, inflammation, infection and autoimmune diseases. The molecular mechanism underlying expansion and function of MDSCs is becoming appreciated to manipulate immune response in the diseases. MicroRNA (miRNAs) as one short noncoding RNAs, are involved in regulating cell proliferation, differentiation and maturation. However, it needs to be further studied how miRNAs mediate the development and function of MDSC in association with cancer and other diseases. In the review, we report and discuss recent studies that miRNAs networks regulate the differentiation, expansion and suppression function of MDSCs in tumor microenvironment or other diseases through different signaling pathways. Those studies may provide one novel potential approach for tumor immunotherapy.
Keywords: MicroRNA, Myeloid-derived suppressor cells, Tumor
Introduction
Myeloid-derived suppressor cells (MDSCs) are the progenitors of myeloid cells with suppressive function on immune response in tumor microenvironment (TME). In tumor bearing-mice, MDSCs are generally characterized as GR-1+CD11b+ cells, which are further divided as two subtypes: CD11b+Ly6G-Ly6Chigh monocytic MDSCs and CD11b+Ly6G+ Ly6Clow granulocytic MDSCs [1, 2]. They utilize different suppressive mechanisms to inhibit the antitumor immune response. Monocytic MDSCs regulates immune suppression through the production of NO and arginase[3]. In contrast, the inhibition of granulocytic MDSCs is regulated via ROS and H2O2 [4]. In patients with cancer, there are different types of MDSCs. In general, MDSCs are characterized as CD33+CD15+CD14-HLA-DRlow populations [5]. Many factors, such as cytokines, growth factors and microbial products released in tumor microenvironments have been shown to be involved in the induction and expansion of MDSCs with suppressive activity [6, 7]. Most of these mediators activate signaling pathways in tumor MDSCs that involve NF-κB and Signal transducer and activator of transcription (STATs) [1, 8, 9] . Some novel regulatory mechanisms for the differentiation, expansion and suppression of tumor MDSCs were described recently. There is emerging evidence that microRNAs (miRNAs) cooperate transcriptional factors to become complex regulatory networks which mediate tumor MDSCs [10, 11].
miRNAs are the abundant small, single-stranded, non-coding RNA of about 22 nucleotides. miRNAs base-paired with the complementary sequence within target mRNAs to mediate post-transcriptional gene repression or target mRNA degradation [12]. The stability of the miRNA-mRNA interaction is critical for repressing the potential target [13]. Each mRNA could be targeted by different miRNAs and a single miRNA may target different mRNAs [14, 15]. Gene expression silencing by means of miRNAs and changes in the miRNA expression level regulate various biological processes, including the differentiation, maturation, function of immune cells and maintenance of immune homeostasis [16-19]. MDSCs , an immune-suppressive cell, plays an important role in a wide range of human diseases including cancer, chronic inflammatory and autoimmune diseases. Therefore, both abnormal expression and function of miRNAs in MDSCs were investigated, so that novel miRNAs regulatory mechanisms on MDSCs were displayed.
MicroRNAs regulate the differentiation and activation of tumor MDSCs
1. miRNAs up-regulation on tumor MDSCs
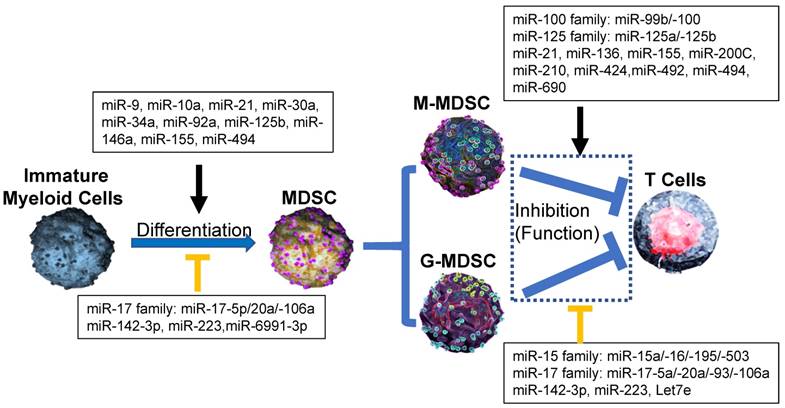
Recent reports demonstrated that miR-494 expression in tumor MDSCs was dramatically induced by tumor-derived factors, such as TGF-β1 to regulate the accumulation and activity of MDSCs by targeting of phosphatase and tensin homolog (PTEN) and activation of the Akt pathway [20]. miR-10a activated AMPK signaling to promote expansion and activation of MDSCs in breast cancer cells with chemotherapy-induced immune resistance [21]. miR-6991-3p could directly target the immune checkpoint gene LGALS9 (Galectin 9) and MiR-6991-3p mimic transfection suppressed expansion and promoted apoptosis of MDSCs through suppressing LGALS9-mediated activation of Janus kinase (JAK) and STAT3 [22]. In B lymphoma -bearing mice, miR-30a expression was increased in both G-MDSCs and M-MDSCs. After the transfection of miR-30a mimics, the differentiation and suppressive abilities of MDSCs were increased via up-regulation of arginase-1. miR-30a also down-regulated suppressor of cytokine signaling 3 (SOCS3) mRNA to activate STAT3 signaling to further promote MDSC differentiation and suppressive activities, indicating that same individual microRNA could regulate differentiation and activity of MDSCs through difference pathways [23, 24] (Figure 1). The inhibition of miR-9 promoted the differentiation of MDSCs with significantly reduced immunosuppressive function via by targeting the runt-related transcription factor 1 (Runx1), an essential transcription factor in regulating MDSC differentiation and function [25] (Figure 2).
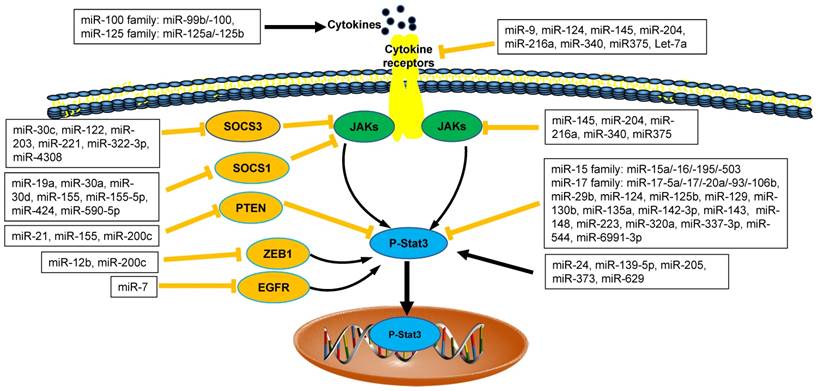
The interaction between microRNAs and Stat3 in tumor microenvironment. MicroRNAs (miRNAs) are emerging as direct or indirect regulators of Janus Kinase (JAK)-Signal Transducer and Activator of Transcription 3 (STAT3) pathways in the pathogenesis of cancer. In each process, microRNAs networks play positive (black line) or negative (orange line) roles. SOCS1: Suppressor Of Cytokine Signaling 1; SOCS3: Suppressor Of Cytokine Signaling 3; PTEN: Phosphatase and Tensin Homologue; ZEB1: Zinc Finger E-Box Binding Homeobox 1; EGFR: Epidermal Growth Factor Receptor.
2. Inhibitory roles of miRNAs on tumor MDSCs
miRNAs also negatively mediated the differentiation and activity of tumor MDSCs. The overexpression of miR-17 family members such as miR-17-5p, miR-20a and miR-106a in human progenitor cells repressed AML1 by binding to its promoter, which resulted in the down-regulation of M-CSFR, thus limiting MDSC differentiation [26] (Figure 2). In LLC and ovarian carcinoma models, miR-223 suppressed differentiation and accumulation of MDSCs by targeting molecule myocyte enhancer factor 2C (MEF2C) [27]. miR-142-3p could prevent MDSC differentiation during tumor-induced myelopoiesis by modulating STAT3 and C/EBPβ signal pathway, indicating that the potential therapeutic application for miR-142-3p oligonucleotide as adjuvant tool for adoptive T cell therapy of cancer [28] (Figure 1).
3. miRNAs from tumor-derived extracellular vesicles
Extracellular vesicles (EVs) were involved in miRNAs regulation on MDSCs. Cancer cells secreted EVs, which were involved in the intercellular transfer of proteins, lipids, and genetic material (such as miRNAs). Those tumor-associated EVs represented an ideal candidate due to their ability to recirculate in body fluids during the process of MDSC generation from bone marrow in tumor microenvironment [29-31]. In melanoma patients, some miRNAs (such as miR-99b, miR-100, miR-125a/-125b, miR-146a/-146b, miR-155, let-7e), which were highly detected in plasma as associated with EVs, mediated the generation and functional features of tumor M-MDSCs [8, 32, 33]. In Acute myeloid leukemia (AML), miR-34a promoted the expansion of MDSCs as the regulatory mechanism by which Mucin 1, cell surface associated (MUC1) drives c-myc expression in Acute AML cells and tumor-derived EVs [30]. In addition, miR-34a also inhibited the apoptosis of MDSCs via targeting N-myc [34] or p2rx7/Tia1 [35] (Figure 2). Those data suggested that miR-34a upregulated the generation and accumulation of tumor MDSC through different pathways as many other miRNAs.
Effect of microRNA networks on MDSCs' differentiation, expansion, activation and function in tumor microenvironment. In tumor microenvironment, MDSCs from Immature myeloid cells are divided as two subtypes: monocytic MDSCs (M-MDSCs) and Granulocytic MDSCs (G-MDSCs), which utilize different suppressive mechanism to inhibit the antitumor ability of T cells. In each process, microRNAs networks play positive (black line) or negative (orange line) roles.
4. miRNAs from tumor-derived exosomes
Exosomes derived from tumor (such as gliomas) were also involved in MDSC differentiation. In glioma-bearing mice, glioma-derived exosomes (GDEs) facilitated the expansion and function of MDSCs. Hypoxia promoted the upregulation of miR-10a and miR-21 expression in GDEs to induce MDSC activation by targeting the IκBα/NF-κB and PTEN/PI3K/AKT pathways. The reduced numbers of MDSCs were observed in the spleens of mice bearing miR-10a or miR-21 knockout glioma cells, compared with those in bearing glioma cells [36]. Those GDEs also regulated the expansion of MDSCs through miRNA-29a/Hbp1 and miRNA-92a/Prkar1a pathways [37], indicating that GDEs could regulate MDSC expansion through difference miRNAs (Table 1).
miRNAs mediate the function of MDSCs in tumor microenvironment
1. miRNAs regulate MDSCs through Stat3 pathway
We ever discussed the findings about the relationship between miRNAs and JAK/STAT3 in cancer [2, 8]. Recently, there were more emerging data about negative and positive regulation of miRNAs networks and JAK/STAT3 signaling pathways via direct and/or indirect regulatory mechanisms in tumor microenvironment (Figure 1). STAT3, as an important transcript factor, is also required for the suppressive function of tumor MDSCs [2, 38-40]. Therefore, we focused on the regulation of miRNAs on tumor MDSC through JAK/STAT3 further.
microRNAs regulation on the differentiation and expansion of tumor MDSCs
| MicroRNAs | Target genes | References |
|---|---|---|
| miR-9 | Runx1 | [25] |
| miR-10a | AMPK | [21] |
| miR-10a/-21 | Rora/NF-κB | [36] |
| miR-17-5p/-20a (miR-17 family) | AML1 | [26] |
| miR-106a (miR-17 family) | AML1 | [26] |
| miR-30a | SOCS3/Stat3 | [24] |
| miR-34a | MUC-1 | [30] |
| miR-34a | N-myc | [34, 35] |
| miR-92a | Prkar1a | [37] |
| miR-125b | TNF | [8] |
| miR-146a | CSF-1R | [33] |
| miR-142-3p | C/EBPβ/Stat3 | [28] |
| miR-155 | C/EBPβ | [46, 47] |
| miR-223 | MEF2C | [27] |
| miR-494 | PTEN | [20] |
| miR-6991-3p | Stat3 | [22] |
Recent data demonstrated that four members of miR-17 family (including miR-17, miR-20a, miR-93, miR-106a) played the inhibitory roles in the function of tumor MDSCs. In tumor microenvironment, tumor-associated factors downregulated the expression of miR-17-5p and miR-20a and promoted the Stat3-associated suppressive function of MDSCs [41]. Thus, miR-17-5p and miR-20a may potentially be used as targets in immunotherapy strategies to inhibit the function of MDSCs via reducing STAT3 expression [42].
The enhanced expression of miR-142-3p reduced the immunosuppressive activity of tumor BM-MDSCs, restoring CD8+ T cell proliferation through inhibiting C/EBPβ/STAT3 pathway [28]. miR223 and Let7e also downregulated the suppressive function of MDSCs through inhibiting the activation of STAT3 in Gliomas [41]. The expression of PD-L1 on tumor MDSCs, which was closely related to the suppressive function of MDSCs was regulated by the miR-93/106b miRNA cluster of miR-17 family through stat3 pathway. Those PD-L1 expression levels on MDSCs could be reduced significantly after treatment of miR-93 mimics [43, 44]. Therefore, those miRNAs above regulated tumor MDSCs plasticity through inhibiting STAT3 pathways (Figure 1).
The regulatory roles of miRNAs on tumor MDSCs are positively involved in STAT3 pathway. miR-200c promoted suppressive potential of tumor MDSCs by targeting PTEN/friend of Gata 2 (FOG2), which led to STAT3 and PI3K/Akt activation [45]. miR-155 and miR-21 demonstrated a synergistic effect on MDSC induction via targeting SHIP-1 and PTEN respectively, leading to Stat3 activation [46]. In a line with this finding above, MDSCs were shown to require miR-155 to facilitate tumor growth [47]. However, recent study revealed the loss of miR-155 in MDSCs enhanced its recruitment and function in solid tumor, which was not consistent with the results above [11, 48].
2. miRNAs regulate MDSCs through PD-L1/PD-1 pathway
The immunotherapy of checkpoints PD-L1/PD-1 on tumor have been broadly applied and those checkpoints were also associated with the tumor MDSCs [1, 8]. But the interaction between miRNAs and checkpoints PD-L1/PD-1 on tumor MDSCs was recently investigated further. Some scientists reported that five members of miR-15 family, which included miR-15a, miR-15b, miR-16, miR-195 and miR-503, activated T cell response by inhibiting the function of MDSCs and/or Tregs in the tumor microenvironment through blocking PD-L1/PD-1 signaling pathway [42, 45, 49, 50], however, miR-424(322), another member of miR-15 family was inversely correlated with PD-L1 pathways. The high level of miR-424 (322) in the tumor was positively correlated with the function of MDSCs and Tregs [51] (Figure 2). The latest results demonstrated that hypoxic tumor-derived exosomes (TEXs) enhanced the suppressive roles of MDSCs on γδ T cells through a miR-21/PTEN/PD-L1 pathway in oral squamous cell carcinoma (OSCC) [52]. Thus, interaction between miRNAs network and PD-L1/PD-1 regulated the expansion and function of tumor MDSCs, providing one novel therapy method for inhibiting MDSC-associated tumor metastasis (Table 2).
3. miRNAs regulate MDSCs through other molecular pathways
miRNAs networks regulate the function of MDSCs through other target genes. Recent studies demonstrated that EL-4 tumor-elicited MDSCs showed increased expression of miR-690 with attenuated C/EBPα expression [50]. Hypoxia-induced miR-210 enhanced the immunosuppressive activity of tumor MDSCs by increasing arginase activity, nitric oxide production and IL-16 [53]. It was reported that the expansion of tumor MDSCs was regulated by miR-494 through PTEN/AKT. The downregulation of PTEN by miR-494 enhanced the activity of AKT to promote the accumulation of MDSCs in tumor tissues [20]. miR-492 also played the similar roles in the suppressive function of MDSCs [54]. In melanoma patients, several miRNAs (including miR-100 family member-miR-99b/-100 and miR-125 family members-miR-125a/-125b) induced the activity and accumulation of MDSCs through IL-6 and CCL2, activating JAK/STAT3 pathway further [32] (Figure 1).
microRNAs mediate the function of MDSCs in tumor microenvironment
| MicroRNAs | Target genes | References |
|---|---|---|
| miR-15a (miR-15 family) | PD-1/PD-L1 | [50] |
| miR-16/195 (miR-15 family) | PD-1/PD-L1 | [49] |
| miR-503 (miR-15 family) | PD-1/Stat3 | [42] |
| miR-424(322) (miR-15 family) | PD-1/PD-L1 | [51] |
| miR-17-5p/-20a (miR-17 family) | Stat3 | [42] |
| miR-93/-106b (miR-17 family) | Stat3 | [43, 44] |
| miR-21 | Stat3 | [46] |
| miR-21 | PTEN/PD-L1 | [52] |
| miR-99b/-100 (miR-100 family) | IL-6/CCL2 | [32] |
| miR-125a/-125b (miR-125 family) | IL-6/CCL2 | [32] |
| miR-136 | NFIA | [61] |
| miR-142-3p | C/EBPβ/Stat3 | [28] |
| miR-155 | Stat3 | [11, 46] |
| miR-155 | MCL-1 | [48] |
| miR-200C | PTEN/FOG2 | [45] |
| miR-210 | NO production | [53] |
| miR-223 | Stat3 | [41] |
| miR-492 | PTEN | [54] |
| miR-494 | PTEN | [20] |
| miR-690 | C/EBPa | [50] |
| Let7e | Stat3 | [41] |
MicroRNAs regulate the expansion and function of MDSCs in inflammation, infection and autoimmune diseases
1. Inflammation and infection
MDSCs also play an important role in other pathological conditions, including inflammation, infection and autoimmune diseases [2, 10, 55]. Thus, we examined if miRNAs had regulatory roles on MDSCs in those diseases. In mouse model with chronic asthma, miR-20b Induced the increased numbers of MDSCs in lung through TGF-β to inhibit airway inflammation[56]. MDSCs enhanced late sepsis development through immunosuppressive function in mice. miR-375 also regulated the function and miR-21 expression of those MDSCs through targeting JAK2 and further impairing STAT3 in the mice with sepsis [57]. miR-21 and miR-181b coupled with NFI-A to promote immunosuppression of MDSCs for improving late-sepsis survival [58]. The overexpression of some miRNAs was induced by the synergistic effect of STAT3 and C/EBPβ, which activated miR-21 and miR-181b promoters after sepsis initiation [59]. The latest results demonstrated that S100A9 stabilized those STAT3/C/EBPβ protein complex to promote MDSC expansion and immunosuppression in late/chronic sepsis by inducing the expression of miR-21 and miR-181b [60]. In inflammation environment, TNFα-mediated miR-136 also targeted NFI-A to induce differentiation and activity of MDSCs [61]. MDSCs and Tregs were developed during chronic hepatitis C virus (HCV) infection. miR-124 downregulated the expression level of STAT3, as well as TGF-β, which were overexpressed in MDSCs to reduce the frequencies of MDSCs and Tregs, thus uncovering a novel mechanism for the expansion of MDSC and Tregs during HCV infection [62].
2. Autoimmune Disease
Recent studies demonstrated that MDSCs were involved in autoimmune diseases. In experimental autoimmune encephalomyelitis (EAE), MDSCs can suppress T cell activities, in which miR-223 downregulate the number and function of MDSCs via STAT3. In miR-223 knockout mice, MO-MDSCs suppressed T cell proliferation in vitro and EAE in vivo more than wild-type MO-MDSCs[63] (Table 3).
microRNAs regulate the expansion and function of MDSCs in inflammation, infection and autoimmune diseases
| MicroRNAs | Diseases/ MDSC plasticity | Target genes | References |
|---|---|---|---|
| miR-20b | Asthma/Expansion | TGFβ | [56] |
| miR-21 | Sepsis/Expansion | NFI-A | [58] |
| miR-124 | HCV/Suppressive function | Stat3 | [62] |
| miR-136 | Inflammation/ differentiation | NFI-A | [61] |
| miR-181b | Sepsis/ Expansion | NFI-A | [58] |
| miR-223 | EAE/Suppressive function | Stat3 | [63] |
| miR-375 | Sepsis/Expansion | Jak2-stat3 | [57] |
Concluding remarks
MDSCs are one of important immune suppressive cells in tumor microenvironment and may be next breakthrough target for tumor immunotherapy [5, 6, 64]. The expansion and function of tumor MDSCs have been widely investigated, however, the regulatory mechanism of MDSCs need be further defined [5, 55, 65]. Recently, the novel research field of miRNA regulation on tumor MDSCs plasticity were opened [32, 66, 67]. The differentiation and function of MDSCs seems to be regulated by multiple miRNAs, some of which were classified by us based on their family members, in order that the scientists may investigate the regulatory roles of other related members of miRNAs family on tumor MDSCs. However, it remains to be clarified how those dysregulated miRNAs were combined in vivo to act on MDSCs on key signaling pathway. In addition, most of research data about miRNA function on tumor MDSCs were gained from murine studies, even though there are a few miRNA data from human patients with cancer. The significant and application of miRNAs for the expansion and function of MDSCs in patients with cancer need be further investigated. Therefore, the interaction of dysregulated miRNAs on MDSCs with transcription factors, cofactors and chromatin modifiers may target specific miRNA-regulated pathways to provide novel ways to treat MDSCs in tumor microenvironment.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81572868, 81803680, 81803796, 81803649); Science Foundation of Shangdong (Grant No. ZR2018LC012). Jilin province science and technology development Project in China (20170307031YY, 20180520050JH); Foundation of Jilin Educational Committee (JJKH20170726KJ); Intramural Research Program at the NCI.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162-74
2. Qu P, Wang LZ, Lin PC. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. 2016;380:253-6
3. Karakasheva TA, Dominguez GA, Hashimoto A, Lin EW, Chiu C, Sasser K. et al. CD38+ M-MDSC expansion characterizes a subset of advanced colorectal cancer patients. JCI Insight. 2018:3
4. Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108-19
5. Tcyganov E, Mastio J, Chen E, Gabrilovich DI. Plasticity of myeloid-derived suppressor cells in cancer. Curr Opin Immunol. 2018;51:76-82
6. Qu P, Du H, Li Y, Yan C. Myeloid-specific expression of Api6/AIM/Sp alpha induces systemic inflammation and adenocarcinoma in the lung. J Immunol. 2009;182:1648-59
7. Yu SJ, Ma C, Heinrich B, Brown ZJ, Sandhu M, Zhang Q. et al. Targeting the crosstalk between cytokine-induced killer cells and myeloid-derived suppressor cells in hepatocellular carcinoma. J Hepatol. 2019;70:449-57
8. Qu P, Boelte KC, Lin PC. Negative regulation of myeloid-derived suppressor cells in cancer. Immunol Invest. 2012;41:562-80
9. Qu P, Yan C, Du H. Matrix metalloproteinase 12 overexpression in myeloid lineage cells plays a key role in modulating myelopoiesis, immune suppression, and lung tumorigenesis. Blood. 2011;117:4476-89
10. Chen S, Zhang Y, Kuzel TM, Zhang B. Regulating Tumor Myeloid-Derived Suppressor Cells by MicroRNAs. Cancer Cell Microenviron. 2015:2
11. Kim S, Song JH, Kim S, Qu P, Martin BK, Sehareen WS. et al. Loss of oncogenic miR-155 in tumor cells promotes tumor growth by enhancing C/EBP-beta-mediated MDSC infiltration. Oncotarget. 2016;7:11094-112
12. O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295-312
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
14. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
15. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460-9
16. Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer. 2018;17:64
17. Raisch J, Darfeuille-Michaud A, Nguyen HT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol. 2013;19:2985-96
18. Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831-42
19. O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111-22
20. Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F. et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. 2012;188:5500-10
21. Rong Y, Yuan CH, Qu Z, Zhou H, Guan Q, Yang N. et al. Doxorubicin resistant cancer cells activate myeloid-derived suppressor cells by releasing PGE2. Sci Rep. 2016;6:23824
22. Sun JP, Ge QX, Ren Z, Sun XF, Xie SP. MiR-6991-3p is identified as a novel suppressor in the expansion and activation of myeloid-derived suppressor cells in hepatoma-bearing mice. Onco Targets Ther. 2019;12:309-17
23. Wang F, Song W, Zhao H, Ma Y, Li Y, Zhai D. et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017;27:416-39
24. Xu Z, Ji J, Xu J, Li D, Shi G, Liu F. et al. MiR-30a increases MDSC differentiation and immunosuppressive function by targeting SOCS3 in mice with B-cell lymphoma. FEBS J. 2017;284:2410-24
25. Tian J, Rui K, Tang X, Ma J, Wang Y, Tian X. et al. MicroRNA-9 Regulates the Differentiation and Function of Myeloid-Derived Suppressor Cells via Targeting Runx1. J Immunol. 2015;195:1301-11
26. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F. et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775-87
27. Liu Q, Zhang M, Jiang X, Zhang Z, Dai L, Min S. et al. miR-223 suppresses differentiation of tumor-induced CD11b(+) Gr1(+) myeloid-derived suppressor cells from bone marrow cells. Int J Cancer. 2011;129:2662-73
28. Sonda N, Simonato F, Peranzoni E, Cali B, Bortoluzzi S, Bisognin A. et al. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity. 2013;38:1236-49
29. Adams KR, Chauhan S, Patel DB, Clements VK, Wang Y, Jay SM. et al. Ubiquitin Conjugation Probed by Inflammation in Myeloid-Derived Suppressor Cell Extracellular Vesicles. J Proteome Res. 2018;17:315-24
30. Pyzer AR, Stroopinsky D, Rajabi H, Washington A, Tagde A, Coll M. et al. MUC1-mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood. 2017;129:1791-801
31. Deng Z, Rong Y, Teng Y, Zhuang X, Samykutty A, Mu J. et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene. 2017;36:639-51
32. Huber V, Vallacchi V, Fleming V, Hu X, Cova A, Dugo M. et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J Clin Invest. 2018;128:5505-16
33. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M. et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189-201
34. Chen S, Huang A, Chen H, Yang Y, Xia F, Jin L. et al. miR-34a inhibits the apoptosis of MDSCs by suppressing the expression of N-myc. Immunol Cell Biol. 2016;94:563-72
35. Huang A, Zhang H, Chen S, Xia F, Yang Y, Dong F. et al. miR-34a expands myeloid-derived suppressor cells via apoptosis inhibition. Exp Cell Res. 2014;326:259-66
36. Guo X, Qiu W, Liu Q, Qian M, Wang S, Zhang Z. et al. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene. 2018
37. Guo X, Qiu W, Wang J, Liu Q, Qian M, Wang S. et al. Glioma exosomes mediate the expansion and function of myeloid-derived suppressor cells through microRNA-29a/Hbp1 and microRNA-92a/Prkar1a pathways. Int J Cancer. 2018
38. Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E. et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693-701
39. Zhang L, Li J, Wang Q, Meng G, Lv X, Zhou H. et al. The relationship between microRNAs and the STAT3-related signaling pathway in cancer. Tumour Biol. 2017;39:1010428317719869
40. Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736-46
41. Goze C, Reynes C, Forestier L, Sabatier R, Duffau H. Pilot Study of Whole Blood MicroRNAs as Potential Tools for Diffuse Low-Grade Gliomas Detection. Cell Mol Neurobiol. 2018;38:715-25
42. Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su X. et al. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J Immunol. 2011;186:4716-24
43. Cioffi M, Trabulo SM, Vallespinos M, Raj D, Kheir TB, Lin ML. et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017;8:21609-25
44. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798-809
45. Mei S, Xin J, Liu Y, Zhang Y, Liang X, Su X. et al. MicroRNA-200c Promotes Suppressive Potential of Myeloid-Derived Suppressor Cells by Modulating PTEN and FOG2 Expression. PLoS One. 2015;10:e0135867
46. Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang CY. et al. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol. 2014;192:1034-43
47. Wang J, Yu F, Jia X, Iwanowycz S, Wang Y, Huang S. et al. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int J Cancer. 2015;136:E602-13
48. Chen W, Han C, Zhang J, Song K, Wang Y, Wu T. Deletion of Mir155 prevents Fas-induced liver injury through up-regulation of Mcl-1. Am J Pathol. 2015;185:1033-44
49. Tao Z, Xu S, Ruan H, Wang T, Song W, Qian L. et al. MiR-195/-16 Family Enhances Radiotherapy via T Cell Activation in the Tumor Microenvironment by Blocking the PD-L1 Immune Checkpoint. Cell Physiol Biochem. 2018;48:801-14
50. Hegde VL, Tomar S, Jackson A, Rao R, Yang X, Singh UP. et al. Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Delta9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer-binding protein alpha by microRNA-690. J Biol Chem. 2013;288:36810-26
51. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T. et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406
52. Li L, Cao B, Liang X, Lu S, Luo H, Wang Z. et al. Microenvironmental oxygen pressure orchestrates an anti- and pro-tumoral gammadelta T cell equilibrium via tumor-derived exosomes. Oncogene. 2019;38:2830-43
53. Noman MZ, Janji B, Hu S, Wu JC, Martelli F, Bronte V. et al. Tumor-Promoting Effects of Myeloid-Derived Suppressor Cells Are Potentiated by Hypoxia-Induced Expression of miR-210. Cancer Res. 2015;75:3771-87
54. Jiang J, Zhang Y, Yu C, Li Z, Pan Y, Sun C. MicroRNA-492 expression promotes the progression of hepatic cancer by targeting PTEN. Cancer Cell Int. 2014;14:95
55. Ben-Meir K, Twaik N, Baniyash M. Plasticity and biological diversity of myeloid derived suppressor cells. Curr Opin Immunol. 2018;51:154-61
56. Ma H, Wang H, Luo Y, Guo S, Song C. Mir-20b-Induced Increase in Myeloid-Derived Suppressor Cells in the Lungs of Mice with Chronic Asthma. Ann Clin Lab Sci. 2017;47:76-82
57. Sheng B, Zhao L, Zang X, Zhen J, Chen W. miR-375 ameliorates sepsis by downregulating miR-21 level via inhibiting JAK2-STAT3 signaling. Biomed Pharmacother. 2017;86:254-61
58. McClure C, Brudecki L, Ferguson DA, Yao ZQ, Moorman JP, McCall CE. et al. MicroRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect Immun. 2014;82:3816-25
59. McClure C, McPeak MB, Youssef D, Yao ZQ, McCall CE, El Gazzar M. Stat3 and C/EBPbeta synergize to induce miR-21 and miR-181b expression during sepsis. Immunol Cell Biol. 2017;95:42-55
60. Alkhateeb T, Kumbhare A, Bah I, Youssef D, Yao ZQ, McCall CE. et al. S100A9 maintains myeloid-derived suppressor cells in chronic sepsis by inducing miR-21 and miR-181b. Mol Immunol. 2019;112:72-81
61. Mei S, Liu Y, Wu X, He Q, Min S, Li L. et al. TNF-alpha-mediated microRNA-136 induces differentiation of myeloid cells by targeting NFIA. J Leukoc Biol. 2016;99:301-10
62. Ren JP, Wang L, Zhao J, Wang L, Ning SB, El Gazzar M. et al. Decline of miR-124 in myeloid cells promotes regulatory T-cell development in hepatitis C virus infection. Immunology. 2017;150:213-20
63. Cantoni C, Cignarella F, Ghezzi L, Mikesell B, Bollman B, Berrien-Elliott MM. et al. Mir-223 regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol. 2017;133:61-77
64. Ostrand-Rosenberg S. Myeloid derived-suppressor cells: their role in cancer and obesity. Curr Opin Immunol. 2018;51:68-75
65. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF. et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150
66. Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH. et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611-21
67. Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN. et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight. 2016:1
Author contact
![]() Corresponding author: Peng Qu, Ph.D., National Cancer Institute, 1050 Boyles St., Bldg 560, Rm 12-34, Frederick, MD 21702, USA. Phone: 301-846-5692; E-mail: peng.qugov or pengquji2000com
Corresponding author: Peng Qu, Ph.D., National Cancer Institute, 1050 Boyles St., Bldg 560, Rm 12-34, Frederick, MD 21702, USA. Phone: 301-846-5692; E-mail: peng.qugov or pengquji2000com

 Global reach, higher impact
Global reach, higher impact