3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(20):4939-4946. doi:10.7150/jca.27616 This issue Cite
Research Paper
PSMC2 is Up-regulated in Pancreatic Cancer and Promotes Cancer Cell Proliferation and Inhibits Apoptosis
1. Department of Medical Oncology, Zhejiang Cancer Hospital, Hangzhou, 310022, P.R. China
2. Department of Oncology, Wuxi Hospital of traditional Chinese Medicine, Wuxi, 214071, P.R. China.
3. Department of Gastroenterology, Wuxi People's Hospital, Affiliated to Nanjing Medical University, Wuxi, 214023, P.R. China.
4. Department of Oncology, Wuxi People's Hospital, Affiliated to Nanjing Medical University, Wuxi, 214023, P.R. China.
Received 2018-6-2; Accepted 2019-6-15; Published 2019-8-27
Abstract
Objective: The lack of effective therapeutic targets poses a leading challenge to prolong survival and improve the quality of life for pancreatic cancer patients. Proteasome 26S subunit ATPase 2 (PSMC2), a recently discovered gene, has been implicated in certain human carcinogenesis. However, limited data is available concerning the functional significance of PSMC2 in cancer cell growth, and whether it plays a role in pancreatic carcinogenesis remains unknown.
Materials and Methods: Quantitative RT-PCR (qRT-PCR) was performed to assess mRNA expression levels of PSMC2 in different pancreatic cancer cell lines. Knockdown of PSMC2 was achieved by using short hairpin RNA (shRNA). The effects of PSMC2 silencing on pancreatic cancer cell proliferation and apoptosis were evaluated by the MTT cell proliferation assay, Celigoassays, Annexin V fluorescence-activated cell sorting (FACS) assay and Caspase-3/7 array.
Results: High expression of PSMC2 was detected in three pancreatic cancer cell lines (SW1990, PANC-1, and AsPC-1). Knockdown of PSMC2 in SW1990 cells inhibited proliferation and enhanced apoptosis.
Conclusions: Our primary study suggests that PSMC2 might be involved in the progression of pancreatic cancer and may serve as a potential therapeutic target.
Keywords: pancreatic cancer, PSMC2, the 26S proteasome, proliferation, apoptosis
1. Introduction
Pancreatic adenocarcinoma remains one of the most rapidly fatal human malignancies [1].Owing to the lack of effective methods of early diagnosis and a high probability of invasion and metastasis, pancreatic adenocarcinoma still has the worst prognosis with a median overall survival(OS) of 6 months and a 5-year survival rate of less than 5% [2]. Therefore, there is an urgency to discover potential novel targets and effective agents to improve the prognosis of pancreatic adenocarcinoma patients.
The 26S proteasome is responsible for protein degradation in the eukaryotic cells. It consists of a 20S core catalytic subunit and a 19S regulatory subunit. The target protein substrate with a polyubiquitin-tag is first recognized by 19S regulatory subunit, and then unfolded and translocated into 20S core subunit for degradation [3]. Either one or two 19S regulatory complexes combine with a single 20S catalytic complex to form, respectively, a singly or doubly capped (26S1 or 26S2) complete 26S proteasome [4]. By specifically eliminating target proteins, the 26S proteasome is involved in almost every biological activity, such as cell division, angiogenesis, immune response, transcription factor activation and post-translational modification of proteins. Given its importance, the 26S proteasome is a multifaceted target for anti-cancer therapies [5]. Indeed, targeting the 26S proteasome has been proved successful in the treatment of aggressive hematopoietic tumors [6].
Proteasome 26S subunit ATPase 2 (PSMC2), a newly identified gene on chromosome 7q22.1-q22.3, encodes an essential member of the 19S proteasome [7, 8]. It is essential for 19S and 26S proteasome assembly [9]. Nijhawan et al. ranked PSMC2 as a top gene in copy number alterations yielding cancer liabilities owing to partial loss (CYCLOPS), which represents a distinct class of cancer-specific liabilities resulting from genome instability and is related to cell proliferation or survival. In ovarian cancer cells, reduction of PSMC2 expression inhibited cell proliferation. In the same study, PSMC2 expression also reportedly correlated with pancreatic cancer cell proliferation. However, no further studies had been carried out [8]. Recently, Song et al. showed high levels of PSMC2 in osteosarcoma samples by tissue microarray analysis and that down-regulation of PSMC2 suppressed osteosarcoma cell proliferation and enhanced apoptosis [10]. In addition, PSMC2 was identified to have synthetic lethal interaction with alveolar soft-part sarcoma-associated ASPSCR1 in human cells [11]. Nevertheless, functional validation and mechanistic studies for PSMC2 in cancers have been lacking. Here, we report, for the first time, the effects of PSMC2 on pancreatic cancer cell proliferation and apoptosis.
2. Materials and Methods
2.1 Patients and tumor samples
In this study, stage I-III, formalin-fixed, paraffin-embedded blocks of tumor tissues were obtained from 40 patients with pancreatic adenocarcinoma. All included patients underwent surgical treatment at Wuxi People's Hospital, an affiliate of Nanjing Medical University (Wuxi, China), between January 2013 and December 2015. Patients were enrolled in this study based on the following selection criteria: i) histologically confirmed pancreatic adenocarcinoma other than adenosquamous carcinomas, neuroendocrine carcinomas, mixed subtypes and mucinous neoplasms with associated invasive carcinoma, ii) complete resection performed, iii), sufficient surgically resected tissue was available for formalin-fixed paraffin-embedding. All patients underwent macroscopic curative resection by total pancreatectomy, pancreaticoduodenectomy, or pylorus preserving pancreaticoduodenectomy with lymph node dissection. All resected primary tumors and lymph nodes were examined histologically by hematoxylin and eosin staining according to the TNM classification system [12]. This study was conducted in accordance with recognized ethical guidelines (Declaration of Helsinki) and was approved by the Regional Ethics Committee of the hospital. The written informed consents were from the patients or their living relatives.
2.2 Immunohistochemistry (IHC)
Tissues were fixed in 10% formalin, embedded in paraffin, and dehydrated in 70% ethanol. Five-μm-thick sections were de-paraffinized in xylene, rehydrated in graded ethanol, and subjected to antigen retrieval by steam heating in Citra ™ antigen retrieval solution (BioGenex). PSMC2 primary antibody (Biorbyt Ltd, Cambridge, Cambridgeshire, UK) was applied to tissues at 1:100. It was defined as positive in the presence of ≥ 5% of neoplastic cells, as previously described [10]. It was based on the percentage of neoplastic cells stained positive for PSMC2, with (-) denoting 0.0-5.0% of osteosarcoma cells stained, (+) denoting 5.0-30.0% of osteosarcoma cells stained, (++) denoting 31.0-50.0% of osteosarcoma cells stained and (+++) denoting 51.0-80.0% of osteosarcoma cells stained.
2.3 Oncomine database analysis
Oncomine database (https://www.oncomine.org/resource/login.html), an online database consisting of previously published and open-access microarray data, was performed to identify the transcription level of PSMC2 gene in pancreatic adenocarcinoma [12]. The mRNA expression of PSMC2 in clinical pancreatic adenocarcinoma tissue was compared with that in normal control, using a Student's t-test to generate a p value. The parameters p-value < IE-4, fold change >2, and gene ranking in the top 10% were used to obtain the most significant PSMC2 probes.
2.4 Cell lines and cell culture
Three pancreatic cancer cell lines (SW1990, PANC-1 and AsPC-1 cells) were obtained from American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL Co. Ltd. USA) supplemented with 10% fetal bovine serum(FBS; Gibco), 100U/ml penicillin, and 100mg/ml streptomycin. Cells were cultured in a standard humidified incubator at 37°C in a 5% CO2 atmosphere.
2.5 Lentivirus transduction
Small interference RNA (siRNA) was designed to target the human PSMC2 gene (Gene ID, 5701). The siRNA sequence is as follows: si-PSMC2: 5-GCCAGGGAGATTGGATAGAAA-3. Lentivirus without siRNA insert was used as a control. SiRNA constructs were generated by synthesizing and cloning into the pGCSIL-green fluorescent protein (GFP) plasmid vector GV115 with Age I/EcoRI sites (GeneChem, Shanghai, China). Lentivirus was packaged into 293 T cells using virus titersand Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as soon as cell density reached 70%. The interference efficiency of PSMC2-siRNA in 293 T cells was tested by GFP. On the third day after transfection, we collected lentivirus particles expressing PSMC2-siRNA from cell culture medium. Lentivirus was concentrated using the centrifugal ultrafiltration device (Millipore, Billerica, MA, USA), and then stored at -80°C until use. PANC-1 cells, at 30% confluence in 6-well plates, were infected with a suitable quantity of lentivirus containing PSMC2-siRNA or non-targeting siRNA at 37°C in the presence of 6μg/ml polybrene (Sigma-Aldrich, St Louis, MO, USA). Construction of recombinant lentivirus (LV-PSMC2 (19674-1)) containing PSMC2 (GeneChem, Shanghai, China) was also accomplished by using similar methods as above. After lentiviral construction, PANC-1 cells were transfected with lentivirus to generate PSMC2-overexpressing pancreatic cancer cells. Cells transfected with the empty GFP lentivirus were selected as negative controls.
2.6 Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cells using TrizolReagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Quantity and quality of the extracted RNA were analyzed by spectrophotometer (Nanodrop2000/2000c; Thermo Fisher Scientific, Inc., Wilmington, DE). mRNA (2ug) was reverse-transcribed using M-MLV Reverse Transcriptase (M1705,Promega Corp., Madison, WI) to synthesize cDNA. QRT-PCR was performed using SYBRVR Premix Ex TaqTM (Takara Bio Inc., Dalian, China) and Mx3000P QPCR System (Agilentn Technologies, Santa Clara, CA). Glyceraldehyde 3-phosphatedehydrogenase (GAPDH) was used as an internal control. PCR conditions were strictly controlled as follows: ten minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C. After normalization with controls, fold changes of mRNA levels were calculated via the Δ ΔCt method.
2.7 Western blot analysis
Protein extraction from cells was performed using radioimmunoprecipitation assay (RIPA) buffer. Protein concentrations were determined using the BCA method (P0010S; Beyotime Institute of Biotechnology). Equal amount of protein lysates were electrophoresed by 10% SDS-PAGE and transferred to the polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). The membranes were blocked by TBST (NaCl 500mM, Tris 20 mM, pH 7.5) containing 5% skim milk. Protein expression in tissue samples was probed with rabbit polyclonal anti-PSMC2 (sigma, F1804) and mouse anti-GAPDH (Santa Cruz Biotechnology, CA).Mouse anti-FLAG and anti-GAPDH antibodies (Santa Cruz Biotechnology, CA) were used for analyzing protein expression in SW1990 cells exogenously transfected with a plasmid encoding flag-tagged PSMC2. Blots were visualized with Electro Chemical Luminescence system, and densitometric analysis was measured with image analysis system (Thermo Fisher Scientific, MA). Values for the PSMC2 bands were normalized relative to the GAPDH bands.
2.8 Cell growth assay determined by Celigo imaging cytometer
Celigo Imaging Cytometer (Nexcelom Bioscience, Lawrence, MA) was used to count the number of surviving cells. Seventy-two hours after infection, cells were collected and seeded into 96-well plates at the density of 1500 cells per well. Cell growth curves and fluorescent photomicrographs were taken by measuring cells with green fluorescence with the cytometer in the following 5 days.
2.9 Cell apoptosis analysis
Apoptotic cells were measured using the Annexin V apoptosis kit according to the manufacturer's instructions (88-8007, eBioscience). Briefly, cells were trypsinized, washed once with PBS, and centrifugated at 1300 rpm for 5 min. After that, cells were washed once in 1×binding buffer and resuspended in 1×staining buffer to 106-107cells/ml. Then, 100μl cell (about 5×105 cells) were incubated with 10μlAnnexin V-APC at room temperature for 15 min in the dark. Finally, annexin V-stained cells were analyzed with flow cytometer (Millipore, Billerica, MA) as a measure of cell apoptosis.
2.10 MTT cell proliferation assay
The proliferation viability of SW1990 cells transfected with PSMC2-lentivirus and control lentivirus vectors were measured using MTT assays. Briefly, cells were seeded in 96-well plates at a seeding density of 2000 cells per well. After 1, 2, 3, 4 or 5 days, a total of 20μl MTT solution (5 mg/ml) were added to the cells for 4h. Cell supernatants were then removed and each well was treated with 100μl dimethyl sulfoxide. After 5 min, the optical density (OD) at 490nm was measured by a microplate reader.
2.11 Caspase-3/7 array system
The activity of caspase-3/7, central effector caspases in apoptosis, was measured in SW1990 cells transfected with PSMC2-lentivirusand control lentivirus vectors by using Caspase3/7 assays (G8091, Promega). Briefly, infected cells were seeded in a 96-well plate at a density of 1×104cells per well. After addition of equal volume (100μl) Caspase-Glo 3/7 reagent, each well was constantly shaken for 2h at ambient temperature. Luminescence was measured by using a Tecan Infinite M2009 PR plate reader.
2.12 Statistical analysis
Statistical analysis was performed using SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA). All data were presented as mean ± SD. Each experiment was done in triplicates. Student's t-test, Fisher's exact test and one-way analysis of variance were used to analyze and determine statistical significance. P<0.05 was considered as significant.
3. Results
3.1 The high expression of PSMC2 protein in pancreatic adenocarcinoma
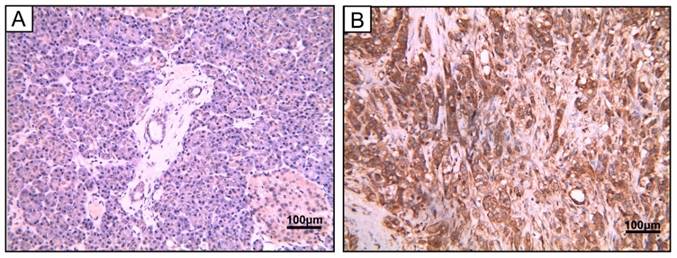
To study the protein expression of PSMC2 in pancreatic adenocarcinoma, 40 pancreatic adenocarcinoma samples and 5 chronic pancreatitis samples were detected by IHC. Of the 40 tumors evaluated in this study, 28 (70%) were strongly cytoplasm positive for PSMC2 expression in cancer cells, while it was negative in all the chronic pancreatitis tissues. The detailed clinical information and the correlation between PSMC2 expression and clinicopathological characteristics were indicated in Table 1, and Figure 1 showed the representative images. The results suggested that PSMC2 expression was significantly associated with vascular invasion (P=0.014) and lymphatic invasion (P=0.008).
PSMC2 expression in pancreatic adenocarcinoma samples. Representative image of pancreatic adenocarcinoma tissue. (A) Hematoxylin and eosin (H&E)-staining (B) PSMC2 positive expression by IHC. Magnification, 400×.
3.2 The transcript expression of PSMC2 in pancreatic adenocarcinoma
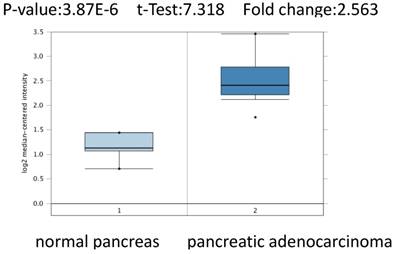
To further confirm the role of PSMC2 in pancreatic adenocarcinoma, we compared the transcription levels of PSMC2 in pancreatic adenocarcinoma tissues with that in normal tissues, using Oncomine database and found that the mRNA expressions of PSMC2 were significantly over-expressed in carcinoma tissues as compared to the normal sample. As show in Figure 2, PSMC2 may work oncogenic function.
Clinicopathological characteristics and the status of PSMC2 expression in pancreatic adenocarcinoma.
| Factors | PSMC2 expression | ||
|---|---|---|---|
| negative | positive | P-value | |
| Age (years) | |||
| ≤60 | 4 | 9 | |
| >60 | 8 | 19 | 0.941 |
| Gender | |||
| Female | 5 | 8 | |
| Male | 7 | 20 | 0.418 |
| Place | |||
| Head | 9 | 21 | |
| Tail | 3 | 7 | 1.000 |
| Vascular invasion | |||
| Yes | 2 | 18 | |
| No | 10 | 10 | 0.014 |
| Lymphatic invasion | |||
| Yes | 5 | 24 | |
| No | 7 | 4 | 0.008 |
| Neruo invasion | |||
| Yes | 7 | 22 | |
| No | 5 | 6 | 0.254 |
| Stats | |||
| I-II | 8 | 4 | |
| III | 4 | 24 | 0.002 |
We conducted cDNA microarray analysis by using the Oncomine database to explore gene expression of PSMC2 in pancreatic adenocarcinoma. The Oncomine database was queried for PSMC2 expression in pancreatic adenocarcinoma tissues and normal tissues [13]. Our analysis revealed that PSMC2 was over-expressed in pancreatic adenocarcinoma, as compared to that in normal tissue (Figure 2).
PSMC2 analysis in pancreatic adenocarcinoma (Oncomine database). The box plot comparing specific PSMC2 expression in normal (left plot) and pancreatic adenocarcinomatissue (right plot) was derived from Oncomine database. The analysis was shown in pancreatic adenocarcinoma relative to normal pancreas.
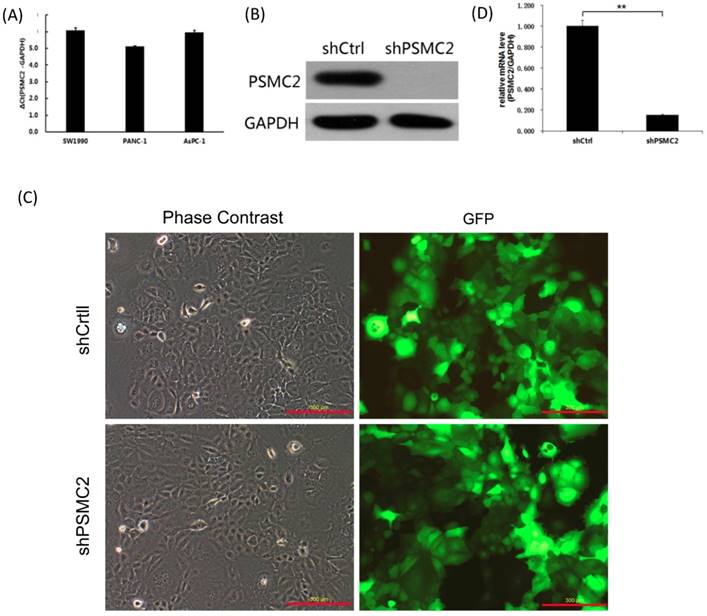
PSMC2 expression was effectively knockdown by shRNA. (A), PSMC2 expression in pancreatic cancer cell lines, as determined by qRT-PCR. (B), Efficiency of PSMC2 silencing in SW1990 cells, as measured by Western blot. (C), Infection efficiency of SW1990 cells with shRNA or shCtrl lentiviral vectors. Cells were assessed by fluorescent microscopy and light microscopy at day 3 post-infection. It is apparent that more than 80% of cells expressed GFP. Magnification, x100. Representative images of the cultures are shown. (**p<0.01). (D), PSMC2 knockdown in SW1990 cells. Levels of PSMC2 in SW1990 cells infected with shRNA or shCtrl lentivirus were determined by using qRT-PCR at day 5 post infection. Note that PSMC2mRNA level was efficiently down-regulated after shRNA infection.
3.3 Expression of PSMC2 in pancreatic cancer cell lines
We first assessed PSMC2 mRNA levels in a panel of different pancreatic cancer cell lines (SW1990, PANC-1 and AsPC-1) using qRT-PCR. Currently, we had no immortalized pancreatic epithelial cell lines. Postoperative specimens of 3 pancreatic cancer patients were selected from the specimen repository, and normal pancreatic epithelial tissues adjacent to cancer were taken. RNA was extracted, and pooled in equal quantities. Reverse transcription of cDNAs were used as a control. As shown in the Figure 3(A), PSMC2 mRNA was highly expressed in the three pancreatic cancer cell lines tested, and the average of ΔCt values were 6.07, 5.12, 5.96 respectively. As PSMC2 is expressed most highly in SW1990 cells, these cells were selected for subsequent PSMC2-knockdown experiments. Effective knockdown of PSMC2 by shRNA was confirmed in SW1990 cells using immunoblotting (Figure 3(B)). After infection with recombinant lentiviruses, >80% of SW1990 cells were shown to express GFP under a fluorescence microscope (Figure 3(C)). Upon lentiviral infection, PSMC2 mRNA was significantly reduced in SW1990 cells compared with cells infected with control shRNA (Figure 3(D)).
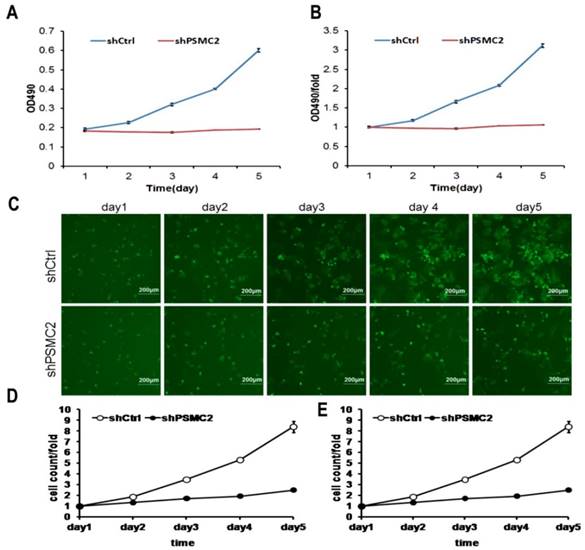
PSMC2 knockdown is associated with reduced proliferation in SW1990 cells. Both Celigo cell counting and MTT absorbance were performed to measure the rate of cell proliferation. (A, B), cell growth curves were plotted using MTT absorbance. (C), fluorescent photomicrographs were taken for cells expressing green fluorescence protein at the indicated times. (D, E), cell growth was depicted every day for 5 days calculated using algorithms and the raw image data (shCtrl vs shPSMC2 , p<0.05).
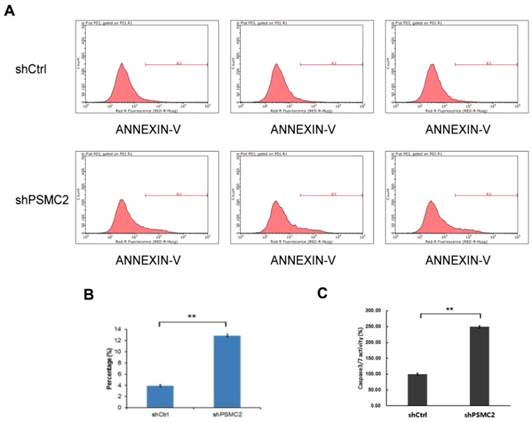
PSMC2 knockdown is associated with elevated apoptosis in SW1990 cells. (A, B), cell death was determined by Annexin V staining followed by flow cytometric analysis.(C), quantification of percentage of apoptotic cells, as measured by caspase 3/7 activity. (**p<0.01).
3.4 Knockdown of PSMC2 inhibited pancreatic cancer cell proliferation
MTT assays were performed to assess proliferation in PSMC2 knockdown SW1990 cells. Growth rate of shPSMC2 SW1990 cells was much slower than that of the shCtrl cells (p<0.01, Figures 4(A,B)). Similarly, the growth curve counted and generated by Nexcelom Celigo Image Cytometer showed that clonogenic survival was significantly decreased following knock down of PSMC2 in SW1990 cells by shRNA (Figures 4C-E). These results suggested that PSMC2 may act as an oncogene that increases proliferation of pancreatic cancer cells.
3.5 Knockdown of PSMC2 increased pancreatic cancer cell apoptosis
Apoptosis of SW1990 cells was measured by FACS analysis of Annexin-V and Caspase 3/7 array. Knockdown of PSMC2 in SW1990 cells increased apoptosis from 4% to 13.12% (Figure 5(A,B)). To provide further evidence for a role of PSMC2 in apoptosis, shPSMC2 cells or shCtrl cells were cultured on a 96-well plate and subjected to Caspase 3/7 activity analysis. Consistent with the results from Annexin V staining, significantly higher levels of caspase3/7 activities were present in cells expressing shPSMC2, compared with those expressing shCtrl (Figure 5(C)). Together, these results showed that PSMC2 was important in the regulation of cell viability.
4. Discussion
The ubiquitin-proteasome system (UPS) is a major mechanism for selective protein degradation in eukaryotic cells [14].The UPS contains two major steps. The first step is the covalent attachment of ubiquitin(s) to a protein substrate, a process called ubiquitination, and the second is the degradation by the 26S proteasome [15]. The 26S proteasome is the major nonlysosomal protease in eukaryotic cells and is responsible for the degradation of all short-lived proteins and 70% to 90% of all long-lived proteins. Thus, it is not surprising that the 26S proteasome plays a critical role in many cellular functions, and, in particular, it is closely involved in many regulatory pathways, including cell proliferation and apoptosis. Indeed, the26S proteasome has been viewed as a potential target for the treatment of human cancers.
Recent studies showed that the proteasome plays an important role in the proliferation and apoptosis of the pancreatic cancer. Two decades ago, Theodore Pet al. had already showed that PSI, a peptide aldehyde inhibitor of the 26S proteasome, effectively induces apoptosis in BxPC-3 human pancreatic cancer cells [16]. Wang et al. showed that quick loss of PSMD11 (26S proteasome non-ATPase regulatory subunit 11) induced rapid or acute apoptosis in pancreatic cancer cells [17]. Furthermore, regulation of an evolutionarily conserved RNA polymerase II-associated factor 1(PAF1) by UPS was involved in pancreatic oncogenesis. In poorly differentiated pancreatic cancer, elevated PAF1 levels were associated with downregulation of proteasomal degradation [18].
PSMC2 is part of the 19S regulatory complex of the 26S proteasome and its expression is essential for 19S and 26S proteasome assembly. In a synthetic lethal screen by comparative genomic approach, PSMC2 was identified to have genetic interaction with ASPSCR1 [11]. This gene pair involves components of the proteasome and is related to chromosome stability. Nijhawan et al. defined PSMC2 as a top one in CYCLOPS genes, which represent a distinct class of cancer-specific liabilities resulting from genome instability [8]. The study indicated that the frequency of partial genomic loss of PSMC2 was 0.10 among 3131 cancers across a wide diversity of cancer types, rendering a high dependence of cancer cells on the remaining PSMC2. This result further suggested PSMC2 could serve as a potential target for cancer treatment. Indeed, the authors showed that PSMC2 suppression decreases ovarian cancer cell proliferation and its expression also correlated with pancreatic cancer cell proliferation [8]. PSMC2 also expressed highly in osteosarcoma samples, as determined by tissue microarrays analysis. Suppression of PSMC2 reduces cell proliferation and increases apoptosis, which is consistent with it acting as an oncogene for osteosarcoma [10]. However, whether PSMC2 carries out the similar functions in pancreatic cancer remains unexplored.
Here, we evaluated PSMC2 expression in pancreatic adenocarcinoma using IHC for the first time and conducted cDNA microarray analysis by using the Oncomine database to explore gene expression of PSMC2 in pancreatic adenocarcinoma tissues. These findings suggested that PSMC2 may work oncogenic function. Then, we demonstrated that PSMC2 expression was critical for pancreatic cancer cell survival. First, we showed that PSMC2 was ubiquitously expressed in SW1990, PANC-1 and AsPC-1pancreatic cancer cell lines. Using RNA interference, we found that PSMC2 was necessary for proliferation and also suppressed apoptosis of pancreatic cancer cells.
The molecular mechanisms underlying PSMC2 in the process of tumorigenesis are not clear. PSMC2 likely drives osteosarcoma via its regulation of cancer-related genes, including ITGA6, FN1, CCND1, CCNE2 and TGFβR2 [10]. Excess PSMC2 in normal cells resides in a complex with PSMC1, PSMD2, and PSMD5, which acts as a reservoir protecting cells from the anti-proliferative effect associated with PSMC2 suppression. Cells harboring partial PSMC2 copy number loss lack this complex and die upon PSMC2 suppression [8]. Besides its proteolytic roles, the 26S proteasome reportedly regulates transcription and promoting sites of active chromatin [19]. In addition, the 19S subunit of the 26S proteasome may also regulate the spreading of heterochromatin [20]. However, these non-canonical functions of the 26S proteasome in transcriptional regulation are still largely debatable. Further studies will be necessary to functionally validate the role of PSMC2 in tumorigenesis in vivo and to uncover the underlying molecular mechanisms.
In conclusion, our study for the first time revealed an important function of PSMC2 in mediating proliferation and apoptosis in pancreatic cancer, and suggested that PSMC2 might serve as a potential therapeutic target for the treatment of pancreatic cancer. Understanding the precise role of PSMC2 in pancreatic cancer pathogenesis will be critical and might facilitate the development of anti-tumor therapies against PSMC2.
Abbreviations
PSMC2: Proteasome 26S subunit ATPase 2; qRT-PCR: quantitative RT-PCR; shRNA: short hairpin RNA; FACS: fluorescence-activated cell sorting; OS: overall survival; CYCLOPS: copy number alterations yielding cancer liabilities owing to partial loss; IHC: Immunohistochemistry; GFP: green fluorescent protein; RIPA: radioimmunoprecipitation assay; PVDF: polyvinylidene difluoride; siRNA: Small interference RNA; H&E: Hematoxylin and eosin; UPS: ubiquitin-proteasome system; PAF1: polymerase II-associated factor 1.
Acknowledgements
This work was supported by National Natural Science Foundation of China (no.81302104 to D.J. and no. 81502038 to F.A.), Zhejiang Province Medical Science Fund Project of China (no.2017194174), Zhejiang Province Traditional Medical Science Project of China (no.2016ZA036) and Wuxi Medical Innovation Team (no. CXTD005).
Ethical approval
This study was approved by the Regional Ethics Committee of Wuxi People's Hospital. All procedures performed in this study were in accordance with the 1964 Declaration of Helsinki and its later amendments.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. et al. Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians. 2016;66:271-89
2. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501
3. da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Molecular cell. 2012;46:54-66
4. Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477-513
5. Grigoreva TA, Tribulovich VG, Garabadzhiu AV, Melino G, Barlev NA. The 26S proteasome is a multifaceted target for anti-cancer therapies. Oncotarget. 2015;6:24733-49
6. Haselbach D, Schrader J, Lambrecht F, Henneberg F, Chari A, Stark H. Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs. Nature communications. 2017;8:15578
7. Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526-38
8. Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE. et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842-54
9. Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T. et al. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914-25
10. Song M, Wang Y, Zhang Z, Wang S. PSMC2 is up-regulated in osteosarcoma and regulates osteosarcoma cell proliferation, apoptosis and migration. Oncotarget. 2017;8:933-53
11. Deshpande R, Asiedu MK, Klebig M, Sutor S, Kuzmin E, Nelson J. et al. A comparative genomic approach for identifying synthetic lethal interactions in human cancer. Cancer research. 2013;73:6128-36
12. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166-80
13. Logsdon C, Simeone D, Binkley C, Arumugam T, Greenson J, Giordano T. et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649-57
14. Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell death and differentiation. 2005;12:1178-90
15. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological reviews. 2002;82:373-428
16. McDade TP, Perugini RA, Vittimberga FJ Jr, Callery MP. Ubiquitin-proteasome inhibition enhances apoptosis of human pancreatic cancer cells. Surgery. 1999;126:371-7
17. Wang L, Zhao L, Wei G, Saur D, Seidler B, Wang J. et al. Homoharringtonine could induce quick protein synthesis of PSMD11 through activating MEK1/ERK1/2 signaling pathway in pancreatic cancer cells. Journal of cellular biochemistry. 2018
18. Ferdoush J, Karmakar S, Barman P, Kaja A, Uprety B, Batra SK. et al. Ubiquitin-Proteasome System Regulation of an Evolutionarily Conserved RNA Polymerase II-Associated Factor 1 Involved in Pancreatic Oncogenesis. Biochemistry. 2017;56:6083-6
19. Bartholomew B. Proteasomes beyond proteolysis: Roles in heterochromatin maintenance. The Journal of biological chemistry. 2017;292:17156-7
20. Seo HD, Choi Y, Kim M, Kang K, Urano T, Lee D. The 19S proteasome is directly involved in the regulation of heterochromatin spreading in fission yeast. The Journal of biological chemistry. 2017;292:17144-55
Author contact
![]() Corresponding author: Junli Ding, Oncology, Wuxi People's Hospital, Affiliated to Nanjing Medical University, No. 299 Qingyang Road, Wuxi, Jiangsu, 214023, P.R. China. Phone: +86510-85350071; Fax: +86510-85350071; E-mail: dingjunliletterscom
Corresponding author: Junli Ding, Oncology, Wuxi People's Hospital, Affiliated to Nanjing Medical University, No. 299 Qingyang Road, Wuxi, Jiangsu, 214023, P.R. China. Phone: +86510-85350071; Fax: +86510-85350071; E-mail: dingjunliletterscom

 Global reach, higher impact
Global reach, higher impact