Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(25):6466-6474. doi:10.7150/jca.33013 This issue Cite
Research Paper
Establishment and characterization of novel human primary endometrial cancer cell line (ZJB-ENC1) and its genomic characteristic
1. Institute of Cancer and Basic Medicine (ICBM), Chinese Academy of Sciences, Hangzhou, 310022, China
2. The Experimental Center, Cancer Hospital of the University of Chinese Academy of Sciences, Hangzhou, 310022, China
3. The Experimental Center, Zhejiang cancer hospital , Hangzhou, 310022, China
4. Medical Support Department, Zhejiang Provincial Hospital of Traditional Chinese Medicine, Hangzhou 310022, China
5. Department of Hematology, The First Affiliated Hospital, Zhejiang University, School of Medicine, Hangzhou, 310022, China
6. Department of Breast Surgery, Cancer Hospital of the University of Chinese Academy of Sciences, Hangzhou, 310022, China
7. Department of Breast Surgery, Zhejiang cancer hospital, Hangzhou, 310022, China
8. The Second Clinical Department, Zhejiang Chinese Medical University, Hangzhou, 310022, China
9. General Surgery Department, Zhejiang Hospital, Hangzhou, 310022, China
10. Department of Breast Thyroid Surgery, Tongde Hospital of Zhejiang Province, Hangzhou, 310022, China
*These authors contributed equally to this work.
Received 2019-1-10; Accepted 2019-8-26; Published 2019-10-20
Abstract
The establishment of human malignant tumor cell lines can provide abundant experimental materials for understanding the biological characteristics of tumors, studying the carcinogenesis, molecular genetics and the mechanism of metastasis and evolution. In this study, a novel cell line designated ZJB-ENC1 has been established from poorly differentiated endometrioid adenocarcinoma. Cytological results showed monolayer-cultured cells were polygonal in shape and a piling-up tendency without contact inhabitation. Immunohistochemistry analysis showed that the cells were negative for ER, PR, c-erbB2, E-CAD, CD117, and OCT3/4, but strongly positive for PTEN and P16. Meanwhile, the tumorigenicity of ZJB-ENC1 was confirmed by subcutaneous transplantation of the cells into a xenograft mouse model. In addition, the results of the whole exome sequencing revealed a unique genomic characteristic of ZJB-ENC1 cells, all common and novel SNPs and InDels were identified. In conclusion, this new stable cell line may promote basic and clinical research on endometrial cancer (EC).
Keywords: Endometrioid adenocarcinoma, Cell line, Tumorigenicity, Immunohistochemisty, Whole exome sequencing
Introduction
Endometrial carcinomas (EC) is a malignant epithelial tumor that arises from the endometrium owing to the precursor lesions such as complex hyperplasia with atypia[1]. It is a common malignancy of female genital tract and the third most common cancer in women[1]. The first sure symptom of EC is often atypical genital bleeding not associated with menstrual period. The most frequent type of EC is endometrioid carcinoma, which counts for more than 80% of cases[2]. The incidence and mortality rate of EC are gradually increasing, according to reports, the incidence rate per 100,000 women is 12.9 in more developed regions[3]. Approximately 52,630 new cases had been diagnosed in 2014 and the annual monitoring of cancer deaths reported almost 8590 events each year in the United States (US)[4]. Only in 2008, in the European countries, 12903 women died from EC, and corrected age-standardized mortality rates have decreased significantly over the past decades in most member states[5]. The increasing incidence of morbidity suggests that the pathogenesis of the disease is an urgent problem to be solved.
Cell lines of endometrial origin may provide useful tools to study the biology of the disease and to develop and test novel therapeutic approaches. A large bank of well-characterized cell lines should reflect the diversity of tumor phenotypes and provide adequate models for the study of tumor heterogeneity. Additionally, disease-orientated drug screening using human tumor cell lines in vitro has some predictive value for the activity of clinical responses[6, 7]. To date, despite a number of endometrial carcinoma cell lines have been reported[8-15], few originated from poorly differentiated endometrioid adenocarcinoma. Therefore, it is absolutely necessary to establish stable and available cell lines of endometrioid adenocarcinoma.
The advent of massively sequencing technologies has markedly expanded knowledge of genome-wide gene abnormalities in various tumors. The whole exome sequencing (WES) is a genomic technique for sequencing all of the protein-coding genes in a genome, which consists of capturing and sequencing of exome. It has been widely used to characterize the mutational spectrum of various cancers[16-18] and provide amount of genetic information for further study.
In this paper, a novel EC cell line ZJB-ENC1 originated from a 58-year-old patient with poorly differentiated endometrioid adenocarcinoma was established and analyzed with respect to the growth property, cellular ultrastructure, neoplastic behavior in SCID nude mice and cell line authentication by short tandem repeat (STR) profiling. Moreover, the mutated genes with known and novel genomic abnormalities were identified by the whole exome sequencing.
Materials and methods
Patient
The cell line was derived from an endometrioid adenocarcinoma patient who was a 58-year-old woman in Zhejiang Cancer Hospital. She was treated with curettage in a local hospital and the symptoms were alleviated subsequently. In May 2015, she underwent surgery for the EC because of recurrence. Laboratory examination results showed CA724 13.36 U/ml, CA125 209.40 U/ml and SCC 2.0 ng/ml. The resected tumor was approximately 2.5×2.3×0.8 cm, pathological results showed moderately poorly differentiated endometrioid adenocarcinoma with chronic inflammation of 18 lymph nodes. The written informed consent was obtained from the patients, which was approved by the Ethical Committees of Zhejiang Cancer Hospital, Hangzhou, China.
Establishment of ZJB-ENC1 cell line
EC tissue was obtained during surgery from the patient and immediately processed. Specimens were washed with RPMI medium supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin and minced into small pieces. Pieces were digested with a mixed enzyme (Vtrypsin-EDTA : Vtype II collagenase = 1:1) for 2 hours and filtered by 40 µm cell strainer to remove large fragment. The flow-through was collected by centrifugation. Cancer cells were resuspended and cultured in growth medium (RPMI medium : DMEM/F12 : DMEM=2:2:1, supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 100 nM hydrocortisone) and incubated at 37 oC in a humidified atmosphere with 5% CO2. The medium was replaced every 3 days. Four days later, the medium was removed and the cells were washed with PBS. Cancer cells were maintained in growth medium till they grew to 80% confluency. The cells were then trypsinized and sub-cultured. Passages 25-40 performed subsequent characterization and testing.
Cell proliferation assays
Suspension of 1×103 logarithmic phase cells was seeded in 96-well plates in triplicate and cultured in the growth medium. The number of cells was counted daily for 8 days using the Cell Counting Kit-8 (Dojindo, Tokyo, Japan) referring to the instructions by measuring the absorbance at 450 nm at the indicated time-points.
Short tandem repeat (STR) analysis
Genomic DNA from ZJB-ENC1 was isolated using genomic extraction kit (Axygen, USA) and amplified by 20-STR amplification protocol. The STR loci and sex gene Amelogenin were detected by an ABI 3730XL Genetic Analyzer. The data were processed using GeneScan and GeneMapperTM ID Software (Invitrgen).
Tumorigenicity in SCID mice
In vivo tumorigenicity of ZJB-ENC1 cell line was assessed based on the ability to form tumors in 50 day-old female nude SCID (severe combined immunodeficiency) (SKXK, China) mice at subcutaneous flank injection sites. A volume of 100 μl was injected in each mouse and consisted of 5×106 cells resuspended in 100 μl of cold phosphate buffered saline (D-PBS) (Thermo Fisher Scientific, Waltham, MA, USA). The animals were housed under sterile conditions in a laminar flow environment with unrestricted access to food and water. Tumor formation was observed on Tuesday and Friday for 35 days. The mice were sacrificed and tumors were removed for H&E staining and pathology examination. All animal studies were performed according to protocols approved by the Institutional Animal Study Committee of Zhejiang University of Traditional Chinese Medicine Animal Testing Center.
Immunohistochemistry
Immunostaining was performed using the Mouse PV Two-Step immunohistochemistry Kit (Beijing ZhongSuanJinQiao Biotechnology Co., LTD., pv-6002). ZJB-ENC1 cells and tumor tissue were processed into paraffin block which could be used for making histological sections. The sections were baked at 60 oC for 1 hour, and incubated with 3% hydrogen peroxide for 10 min after xylol deparaffinization. For all biomarkers, the slides were washed with PBS for 2 × 3 min after incubation with respective primary antibody overnight at 4 °C and incubated for one hour with the secondary antibody at room temperature, washed with PBS for 2 × 3 min again. Image acquisition and section evaluation were performed under a light microscope after DAB coloration.
Whole-exome sequencing (WES)
Genomic DNA was extracted from cells using genomic extraction kit (Axygen, USA) and underwent WES according to the manufacturer's protocols. The exomes were captured using SeqCap EZ Exome V3 (64Mb, Nimblegen, USA) and sequenced on an Illumina HiSeq X Sequencing System (Illumina, San Diego, CA, USA) at Shoudu Technical Service Company (Suzhou, China). All single nucleotide polymorphisms (SNPs) and insertions and deletions (InDels) were identified by Genome Analysis Toolkit v.3.8 (GATK best practices).
Results
Growth characteristics and morphology of ZJB-ENC1 cells
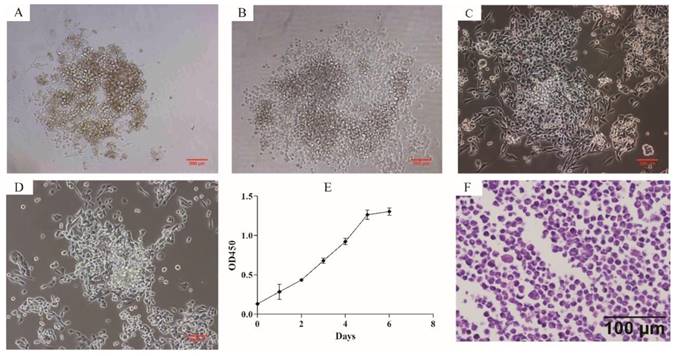
Phase contrast microscope revealed that ZJB-ENC1 cells were anchorage-independent and disorderly grew. Cells tended to formed colonies and pile up upon confluence without obvious touching inhibition, and morphologically most monolayer cells appeared polygonal (Figure 1A-1D). The established cell line was designated as ZJB-ENC1, and cells have been cultured for 2 years (98 passages) with rapid proliferation. The doubling time is approximately 43 hours (Figure 1E). Stained with hematoxylin and eosin, the cells presented polygon-shaped epithelial cells with a high nucleus/cytoplasm ratio and exhibited cellular histomorphology identical to the tumor from the patient (Figure 1F).
Immunohistochemistry
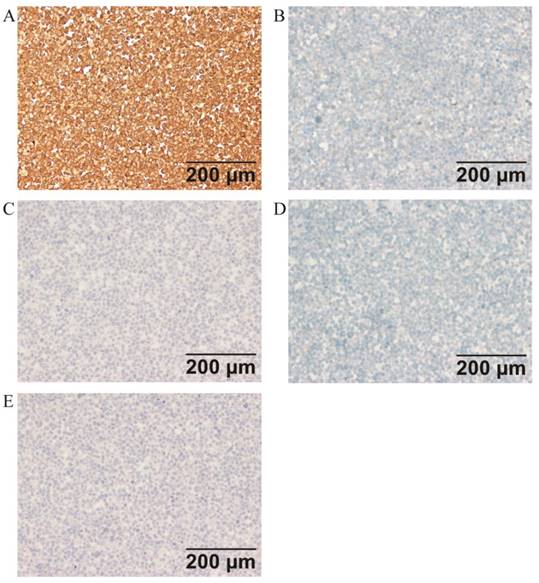
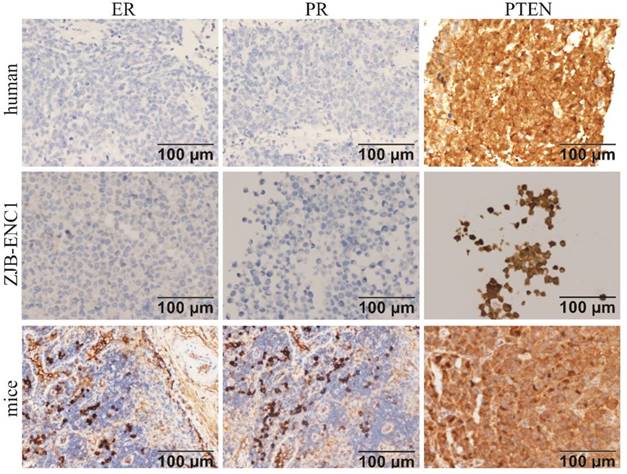
Immunohistochemical analysis of the cell line found that ER, PR, c-erbB2(HER2), E-CAD, CD117, and OCT3/4 were negative, while PTEN and P16 were strongly expressed in ZJB-ENC1 cells (Figure 2). Additionally, immunohistochemical results of tumor tissue from xenograft mice and the clinical patient showed the same results as ZJB-ENC1 cells with the expression pattern of ER (-), PR (-) and PTEN (+), as illustrated in Figure 3.
Cytomorphology and proliferation of cultured ZJB-ENC1 cells. Cells were photographed under microscope at 40 × magnification for passage 0 (A), 7 (B), 17 (C), 50 (D), respectively. (E) The grow curve of ZJB-ENC1 cells. (F) H&E staining of the ZJB-ENC1 cells (Scale bar, 100 µm).
Immunohistochemical characterization of ZJB-ENC1 cells (A) multiple tumor suppressor 1 (P16), (B) KIT proto-oncogene, receptor tyrosine kinase (CD117), (C) cadherin 1 (E-CAD), (D) erb-b2 receptor tyrosine kinase 2 (c-erbB2), (E) POU domain, class 5, transcription factor 1 (OCT3/4).
Immunohistochemical characterization of ZJB-ENC1 cells and tumor tissue of mice model and clinical patient. Estrogen receptor (ER), progesterone receptor (PR), phosphatase and tensin homolog deleted on chromosome ten (PTEN) were immunohistochemically evaluated as indicated (Scale bar, 100 µm).
Short tandem repeat (STR)
We performed STR profiling of ZJB-ENC1 cells to avoid the risk of cross-contamination and to confirm it as a novel human EC cell line. STR examination results showed that neither matched sites in the ATCC, DSMZ, JCRB and RIKEN database nor multiple sites existed, indicating that this is a new cell strain and has not been contaminated, as shown in Table 1.
Short Tandem Repeat (STR) Analysis of ZJB-ENC1 cell lines
| Loci | Allele1 | Allele2 | Loci | Allele1 | Allele2 |
|---|---|---|---|---|---|
| D5S818 | 9 | 10 | FGA | 22 | 26 |
| D13S317 | 10 | 10 | D2S1338 | 20 | 21 |
| D7S820 | 11 | 12 | D21S11 | 30 | 30 |
| D16S539 | 9 | 12 | D18S51 | 12 | 17 |
| VWA | 17 | 20 | D8S1179 | 10 | 10 |
| TH01 | 9 | 9 | D3S1358 | 15 | 15 |
| AMEL | X | X | D6S1043 | 14 | 17 |
| TPOX | 8 | 8 | PENTAE | 14 | 27 |
| CSF1PO | 10 | 11 | D19S433 | 13 | 13 |
| D12S391 | 18 | 19 | PENTAD | 9 | 13 |
The distribution of SNPs and InDels of ZJB-ENC1 cells from the whole exome sequencing
| Term | Number |
|---|---|
| Total | 83926 |
| downstream | 498 |
| exonic | 27745 |
| exonic;splicing | 42 |
| SNPs and InDels in conding region | 27787 |
| synonymous SNV | 11648 |
| nonsynonymous SNV | 14293 |
| stopgain | 710 |
| stoploss | 18 |
| nonframeshift insertion | 121 |
| nonframeshift deletion | 109 |
| frameshift insertion | 108 |
| frameshift deletion | 123 |
| unknown | 657 |
| intergenic | 6522 |
| intronic | 33270 |
| ncRNA_exonic | 3169 |
| ncRNA_exonic;splicing | 6 |
| ncRNA_intronic | 3453 |
| ncRNA_splicing | 25 |
| splicing | 233 |
| upstream | 698 |
| upstream;downstream | 25 |
| UTR3 | 6542 |
| UTR5 | 1692 |
| UTR5;UTR3 | 6 |
Tumorigenicity in vivo
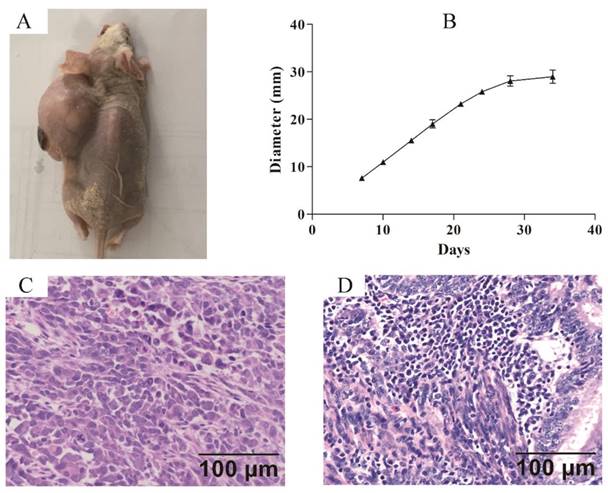
Xenograft model was established to confirm the tumorigenicity of ZJB-ENC1 cells in vivo. Subcutaneous injection of ZJB-ENC1 cells in SCID nude mice all successfully formed tumor at injected sites (Figure 4A). The average tumor diameter of 35 days reached 29 mm (n=6) owing to the efficient proliferation ability of ZJB-ENC1 cells (Figure 4B). As expected, the hematoxylin- and eosin-stained tissue sections of the mice tumor xenografts showed similar histologic features as EC (Figure 4C, 4D).
The whole exome sequencing
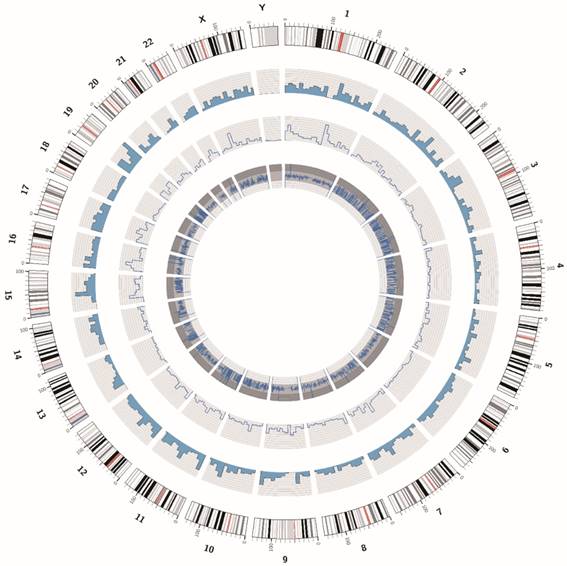
The DNA sample of the ZJB-ENC1 cell line was prepared for the whole exome sequencing. We obtained 129374536 paired raw reads (38.83Gb bases), and the average GC content was 46.44%. The value of Q20 and Q30 were 94.22% and 87.35%, respectively, indicating a high quality of sequencing data. The statistic for the distribution of SNPs and InDels were listed in Table 2. In addition, we showed the circos plot of whole exome sequencing (Figure 5), circles from the outermost to the innermost represented chromosome information, SNV density, depth information and SNV mutation frequency, respectively. Meanwhile, we listed 10 common genes which were proved to be strongly associated with EC, along with their corresponding SNPs and InDels in Table 3.
Mutations of 10 important genes of ZJB-ENC1 cells
| Gene | Ref | Alt | Location | avsnp150 |
|---|---|---|---|---|
| TP53 | G | A | exonic | rs397516436 |
| G | T | intronic | rs17883323 | |
| CCCCAGCCCTCCAGGT | - | intronic | rs59758982 | |
| ARID1A | C | T | intronic | rs767585532 |
| C | T | exonic | rs879255270 | |
| PPP2R1A | C | A | UTR5 | rs535966011 |
| A | G | intronic | rs755531 | |
| C | A | intronic | rs8100600 | |
| C | A | UTR3 | ||
| - | T | intronic | ||
| PIK3R1 | C | T | exonic | rs706713 |
| G | A | intronic | rs171649 | |
| A | C | intronic | ||
| G | A | exonic | rs3730089 | |
| C | T | UTR3 | rs66666989 | |
| A | C | UTR3 | rs78046963 | |
| TG | - | UTR3 | rs58984754 | |
| - | TA | UTR3 | rs531318827 | |
| CTNNB1 | G | A | exonic | rs28931589 |
| G | A | exonic | ||
| C | A | intronic | rs2691680 | |
| - | TAAT | UTR3 | rs16339 | |
| PIK3CA | G | A | exonic | rs1057519929 |
| KRAS | A | T | UTR3 | rs1137189 |
| T | C | UTR3 | rs4597149 | |
| G | A | UTR3 | rs4285970 | |
| A | C | UTR3 | rs712 | |
| C | T | exonic | rs4362222 | |
| - | A | UTR3 | rs71065923 | |
| - | AA | UTR3 | ||
| FGFR2 | G | A | intronic | rs2278202 |
| T | C | exonic | rs1047100 | |
| A | C | intronic | ||
| A | - | intronic | rs796869588 | |
| MLH1 | G | A | intronic | rs41562513 |
| CCNE1 | C | T | exonic | rs7257694 |
| C | A | UTR3 | rs1406 | |
| CTT | - | intronic | rs569537342 | |
| CDKN2A | C | G | UTR3 | rs11515 |
Nude mouse tumorigenicity assay (A) The ZJB-ENC1 cells formed tumors at injected site. (B) The tumor growth curve (n = 6), final diameter reached 29 mm after 35 days, tumor size was measured on Tuesday and Friday for 35 days. H&E staining of the tumor specimens in mice (C) and clinical patient (D) (Scale bar, 100µm).
The circos plot of the whole exome sequencing of ZJB-ENC1 cells. Chromosomes are shown in the color coded of the outermost ring. The second ring shows the SNV density. The inner ring represents depth information and the innermost ring shows SNV mutation frequency.
Discussion
Despite the cell lines may only present a part of the characteristics of the tumor[19], it is still necessary to establish novel and well-characterized EC cell lines to investigate the mechanism of genesis and development, and then so as to develop new therapies against cancer. Many adenocarcinoma cell lines derived from human endometrial tissue have been applied to experimental research[8-15]. In this literature, we established a new human endometrioid adenocarcinoma cell line, designated ZJB-ENC1. The original tumor of ZJB-ENC1 was histologically diagnosed as a poorly differentiated endometrioid adenocarcinoma. The cell line was well characterized with respect to cell morphology, immunohistochemical and histologic characteristics, growth, oncogenicity. ZJB-ENC1 cells grow rapidly in culture and did not express ER, nor PR, except for PTEN. It was confirmed that the cell line was not contaminated with mycoplasma, bacteria and fungi. The tumorigenesis of ZJB-ENC1 cells was confirmed by the formation of solid tumors in nude mice that injected with ZJB-ENC1 cells. The orthotopic implantation model of ZJB-ENC1 cells can be applied to experimental therapeutics for endometrial carcinoma.
To a great extent, progesterone therapy has been considered to be a favorable treatment for patients with EC[20], but poorly differentiated and recurrent endometrial carcinomas always show a poor response to progestin owing to the lack of steroid receptors[21-23]. Notably, endometrioid adenocarcinoma cell lines are mostly negative for hormone receptor because these cell lines mostly source from poorly differentiated tumors[10]. As we expected, ZJB-ENC1 cells did not express ER nor PR in our study. Furthermore, the ZJB-ENC1 cell line is a triple-negative cell line with absence of ER, PR and c-erbB2 protein expression, which indicates this new cell line of EC may have more comprehensive characteristics and potential value for EC research according to the previous studies of the triple-negative phenotype (TNP) breast cancer[24].
Previous studies have found that PTEN gene abnormalities existed in various cancers, such as prostate cancer[25], EC[26], kidney cancer[27], and so on. It is considered to be another tumor suppressor gene that is closely related to tumorigenesis after the found of P53. Unsurprisingly, a high expression level of PTEN was observed in ZJB-ENC1 cells. Meanwhile, several mutations of PTEN were detected by WES, among them, single nucleotide polymorphism (rs701848) carrying the C allele, located on 3-UTR, is associated with increased cancer risk in Asian population[28]. PTEN plays a key role in regulating the PI3K-AKT-mTOR signaling pathway by modulating the phosphatidylinositol 3,4,5-trisphosphate[29, 30], as well as maintaining genetic stability caused by double-strand breaks (DSBs)[31]. It suggests ZJB-ENC1 cells is useful for researching the influence of mutations in PTEN on endometrioid adenocarcinomas. P16, a tumor suppressor protein encoded by the CDKN2A gene, plays an important role in cell cycle regulation by decelerating the cell's progression from G1 phase to S phase[32]. The polymorphism rs11515 with CG phenotype, locate at the 3'UTR of the CDKN2A gene, has a statistically significant correlation with aggressive breast tumors with decreased p16 (INK4a)[33]. On the contrary, the ZJB-ENC1 cells positively expressed P16 protein with CG phenotype at rs11515, further investigation should be conducted to explain this difference.
WES was applied to obtain the genome-wild mutational landscape of ZJB-ENC1 cells, and the total SNPs and InDels were identified. Meanwhile, we listed the common mutation sites of several genes associated with EC, such as TP53, KRAS, FGFR2 and CTNNB1. TP53 encodes the p53 protein, which is involved in cell cycle regulation, apoptosis, senescence, and DNA repair[34]. The p53 gene is highly polymorphic, with at least 13 different polymorphisms described[35, 36]. Among them, p53 intron 3 variant (rs17883323) combined with p73 exon 2 G4A was proved to be associated with a significant increased risk of squamous cell carcinoma of the head and neck (SCCHN)[37]. KRAS is an oncogene which encodes a GDP/GTP-binding protein to regulate cell proliferation and differentiation[38-40], and the relationship between KRAS polymorphisms (rs712) and the risk of cancer in the Chinese population has been revealed[41]. Loss of heterozygosity (LOH) and allele-specific expression (ASE) have frequently observed for tumor suppressor genes. For instance, FGFR2 gene expresses in multiple alternative splicing forms with FGFR2b and FGFR2c as two major transcripts, but it is transcribed as FGFR2b only from one strand (GTA) by mRNA reads at rs1047100[42, 43]. Therefore, these mutations in ZJB-ENC1 show its potential value for further biological research.
Additionally, various novel mutations in these genes were detected, which may reveal some new mechanism in cancer. ARID1A, as a tumor suppressor, regulates CDKN1A and SMAD3 transcription and tumor growth by collaborating with p53[44]. The PPP2R1A gene encodes a constant regulatory subunit of protein phosphatase 2, which is implicated in the negative control of cell growth and division, and its functional mutation (rs11453459) in the promoter contributes to the decreased risk of hepatocellular carcinoma[45]. The catalytic PIK3CA and regulatory PIK3R1 belong to the phosphoinositol-3-kinase (PI3K) family, and the abrogation of these two subunit genes can reduce proliferation, migration, and invasion in cancer cells[46]. MLH1 repairs DNA mismatch by mediating protein-protein interactions, defects in MHL1 will lead to the loss of DNA mismatch repair, which is associated with the microsatellite instability (MSI)[47]. CCNE1, as an oncogene, determines cell division by regulating the transition from G1 to S phase[48]. Based on the WES results, further functional characterization was required for these novel mutations. Thus, ZJB-ENC1 cells also provide an available tool to get a deeper understanding of tumorigenesis and the development of EC.
Conclusion
In summary, ZJB-ENC1 cell line was derived from primary tumor tissue of a patient with poorly differentiated endometrioid adenocarcinoma with specific biological characteristics. It was well-established and serially cultured in vitro for two years. This cell line could be a good model for the study of cancer biology and targeted-therapy of ENC. The whole exome sequencing of the cells also provides deeper insights into genetic abnormalities in ZJB-ENC1 cells and gives the direction for further EC research. However, owing to the deficiencies of the WES method, the whole genome sequencing with high depth will be taken into consideration in future study to obtain a more comprehensive and detailed genomic characteristic of ZJB-ENC1 cells. Moreover, proteomics should be introduced for more tumor-associated markers.
Acknowledgements
The work was partly supported by grants from the National Natural Science Foundation of China (Grant No. 81601899), the Natural Science Foundation of Zhejiang Province (Grant No.17H160013), the Zhejiang Provincial Health Department Foundation (Grant No.2018ky284) and the Zhejiang Provincial Health Department Foundation (Grant No.2015KYB005). The specimens were supported by Zhejiang Cancer Hospital Biospecimen Repository and National Human Genetic Resources Sharing Service Platform (Grant No.YCZYPT[2017]02).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fabjani G, Kucera E, Schuster E, Minai-Pour M, Czerwenka K, Sliutz G. et al. Genetic alterations in endometrial hyperplasia and cancer. Cancer letters. 2002;175:205-11
2. Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, Vergote I. Endometrial cancer. Lancet. 2005;366:491-505
3. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69-90
4. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:104-17
5. Weiderpass E, Antoine J, Bray FI, Oh JK, Arbyn M. Trends in corpus uteri cancer mortality in member states of the European Union. European Journal of cancer. 2014;50:1675-84
6. Dollner R, Granzow C, Werner JA, Dietz A. Is there a role for chemosensitivity tests in head and neck cancer? Onkologie. 2004;27:310-5
7. Newell DR. Flasks, fibres and flanks-pre-clinical tumour models for predicting clinical antitumour activity. British journal of cancer. 2001;84:1289-90
8. Kuramoto H, Tamura S, Notake Y. Establishment of a cell line of human endometrial adenocarcinoma in vitro. American journal of obstetrics and gynecology. 1972;114:1012-9
9. Nishida M, Kasahara K, Kaneko M, Iwasaki H, Hayashi K. Establishment of a new human endometrial adenocarcinoma cell line, Ishikawa cells, containing estrogen and progesterone receptors. Nihon sanka fujinka gakkai zasshi. 1985;37:1103-11
10. Grenman SE, Worsham MJ, Van Dyke DL, England B, McClatchey KD, Babu VR. et al. Establishment and characterization of UM-EC-2, a tamoxifen-sensitive, estrogen receptor-negative human endometrial carcinoma cell line. Gynecologic oncology. 1990;37:188-99
11. Kataoka A, Kojiro M, Yakushiji M, Abe H. Establishment and characterization of unique cell lines (KCC-1a and KCC-1b) from endometrial adenocarcinoma with clear cell carcinoma presenting unusual karyotypes and estrogen secretion. Cancer. 1991;67:1588-98
12. Mobus V, Gerharz CD, Mitze M, Moll R, Pollow K, Kother T. et al. Establishment and characterization of six new human endometrial adenocarcinoma cell lines. Gynecologic oncology. 1993;48:370-83
13. Ohyama A, Nikaido T, Tachibana T, Tominaga N, Toyomura J, Kimura E. et al. Establishment and characterization of a cell line designated Nur-1 derived from human endometrioid adenocarcinoma of uterine corps. Human cell. 2015;28:100-7
14. Isaka K, Nishi H, Sagawa Y, Nakada T, Osakabe Y, Serizawa H. et al. Establishment of a new human cell line (EN) with TP53 mutation derived from endometrial carcinoma. Cancer genetics and cytogenetics. 2003;141:20-5
15. Faruqi SA, Satyaswaroop PG, LiVolsi VA, Deger RB, Noumoff JS. Establishment and characterization of a poorly differentiated lethal human endometrial carcinoma cell line (NOU-1) with karyotype 46,XX. Cancer genetics and cytogenetics. 2002;138:44-9
16. Kim SC, Hong CW, Jang SG, Kim YA, Yoo BC, Shin YK. et al. Establishment and Characterization of Paired Primary and Peritoneal Seeding Human Colorectal Cancer Cell Lines: Identification of Genes That Mediate Metastatic Potential. Translational oncology. 2018;11:1232-43
17. Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K. et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome research. 2012;22:2120-9
18. Kim SI, Lee JW, Lee M, Kim HS, Chung HH, Kim JW. et al. Genomic landscape of ovarian clear cell carcinoma via whole exome sequencing. Gynecologic oncology. 2018;148:375-82
19. Elsasser HP, Lehr U, Agricola B, Kern HF. Structural analysis of a new highly metastatic cell line PaTu 8902 from a primary human pancreatic adenocarcinoma. Virchows archiv. B, cell pathology including molecular pathology. 1993;64:201-7
20. Varga A, Henriksen E. Histologic observations on the effect of 17-alpha-hydroxyprogesterone-17-n-caproate on endometrial carcinoma. Obstetrics and gynecology. 1965;21:656-64
21. Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427-33
22. Thomas DB. Steroid hormones and medications that alter cancer risks. Cancer. 2015;62:1755-67
23. Siiteri PK. Steroid hormones and endometrial cancer. Cancer research. 1978;38:4360
24. Reis-Filho J, Tutt AM. Triple negative tumours: a critical review. Histopathology. 2008;52:108-118
25. Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clinical science. 2017;131:197
26. Zhang HM, Fan TT, Li W, Li XX. Expressions and significances of TTF-1 and PTEN in early endometrial cancer. European review for medical and pharmacological sciences. 2017;21:20-26
27. Que WC, Qiu HQ, Chen Y, Liu MB, Wu CY. PTEN in kidney cancer: A review and meta-analysis. Clinica chimica acta. 2018;480:92-98
28. Song DD, Zhang Q, Li JH, Hao RM, Ma Y, Wang PY. et al. Single nucleotide polymorphisms rs701848 and rs2735343 in PTEN increases cancer risks in an Asian population. Oncotarget. 2017;8:96290-300
29. Mutter GL, Lin MC, Fitzgerald JT, Kum JB, Baak JP, Lees JA. et al. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. Journal of the national cancer institute. 2000;92:924-30
30. Kurose K, Zhou XP, Araki T, Cannistra SA, Maher ER, Eng C. Frequent loss of PTEN expression is linked to elevated phosphorylated Akt levels, but not associated with p27 and cyclin D1 expression, in primary epithelial ovarian carcinomas. American journal of pathology. 2001;158:2097-106
31. Shen W, Balajee AS, Wang JL, Wu H, Charis E, Pier Paolo P. et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157-70
32. Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753-6
33. Royds JA, Pilbrow AP, Ahn A, Morrin HR, Frampton C, Russell IA. et al. The rs11515 polymorphism is more frequent and associated with aggressive breast tumors with increased ANRIL and decreased p16 (INK4a). Expr Front Oncol. 2015;5:306
34. Catherine W, Pharoah PDP, Monica H. p53 polymorphisms: cancer implications. Nature reviews cancer. 2009;9:95-107
35. Matakidou A, Eisen T, Houlston RS. TP53 polymorphisms and lung cancer risk: a systematic review and meta-analysis. Mutagenesis. 2003;18:377
36. Li GJ, Sturgis EM, Wang LE, Chamberlain RM, Amos CI, Spitz MR. et al. Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2004;25:1911
37. Gallì P, Cadoni G, Volante M, Feo ED, Amore R, Giorgio A. et al. A case-control study on the combined effects of p 53 and p 73 polymorphisms on head and neck cancer risk in an Italian population. Bmc cancer. 2009;9:137
38. Bai H, Li HN, Zhang WY, Matkowskyj KA, Liao J, Srivastava SK. et al. Inhibition of chronic pancreatitis and pancreatic intraepithelial neoplasia (PanIN) by capsaicin in LSL-KrasG12D/Pdx1-Cre mice. Carcinogenesis. 2011;32:1689-96
39. Tchernitsa OI, Sers CJ, Hinzmann B, Grips M, Schramme A, Lund P. et al. Transcriptional basis of KRAS oncogene-mediated cellular transformation in ovarian epithelial cells. Oncogene. 2004;23:4536-55
40. Deng M, Tang HL, Zhou YH, Zhou M, Xiong W, Zheng Y. et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. Journal of cell science. 2011;124:2997-3005
41. Zhao WH, Qu XF, Xing ZG, Zhao LQ, Qin L, Lv C. Association of rs712 polymorphism in Kras gene 3'-luntranslated region and cancer risk: a meta-analysis. Journal of B.u.on. Official Journal of the balkan union of oncology. 2015;20:309
42. Katoh M. Cancer genomics and genetics of FGFR2 (Review). International journal of oncology. 2008;33:233
43. Zhao Q, Kirkness E, Caballero O, Galante P, Parmigiani R, Edsall L. et al. Systematic detection of putative tumor suppressor genes through the combined use of exome and transcriptome sequencing. Genome biology. 2010;11:R114
44. Guan B, Wang TL, Iem S. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer research. 2011;71:6718-27
45. Chen HF, Mai JR, Wan JX, Gao YF, Lin LN, Wang SZ. et al. Role of a novel functional variant in the PPP2R1A promoter on the regulation of PP2A-Aalpha and the risk of hepatocellular carcinoma. Plos one. 2013;8:e59574
46. Weber GL, Parat MO, Binder ZA, Gallia GL, Riggins GJ. Abrogation of PIK3CA or PIK3R1 reduces proliferation, migration, and invasion in glioblastoma multiforme cells. Oncotarget. 2011;2:833-49
47. Whelan AJ, Babb S, Mutch DG, Rader J, Herzog TJ, Todd C. et al. MSI in endometrial carcinoma: absence of MLH1 promoter methylation is associated with increased familial risk for cancers. International journal of cancer. 2010;99:697
48. Pils D, Bachmayr-Heyda A, Auer K, Svoboda M, Auner V, Hager G. et al. Cyclin E1 (CCNE1) as independent positive prognostic factor in advanced stage serous ovarian cancer patients - A study of the OVCAD consortium. European journal of cancer. 2014;50:99-110
Author contact
![]() Corresponding authors: Zhiguo Zheng, Tel.: +86-13868015078; E-mail: zhengzhgcom. Xuli Meng, Tel.: +86-13757128803; E-mail: mxlmailcom
Corresponding authors: Zhiguo Zheng, Tel.: +86-13868015078; E-mail: zhengzhgcom. Xuli Meng, Tel.: +86-13757128803; E-mail: mxlmailcom

 Global reach, higher impact
Global reach, higher impact