3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(2):450-459. doi:10.7150/jca.35364 This issue Cite
Research Paper
LINC00511 influences cellular proliferation through cyclin-dependent kinases in papillary thyroid carcinoma
Department of Thyroid & Breast Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, PR China.
Received 2019-3-31; Accepted 2019-9-22; Published 2020-1-1
Abstract
Background: Proverbially, the incidence rate of papillary thyroid carcinoma (PTC) has increased year by year. Many long noncoding RNAs (lncRNAs) have been discovered having a relationship with tumor genesis tightly recently. Thanks to the previous researches, we found long intergenic noncoding RNA 00511 (LINC00511) is overexpressed and acts as an oncogene in non-small-cell lung cancer and breast cancer. However, the biological role and function of LINC00511 are still unclear in PTC.
Methods: We got the expression of LINC00511 in PTC tissues and matched adjacent tissues, as well the cell lines (B-CPAP, KTC-1, and KTC-1) by way of quantitative real-time polymerase chain reaction (qRT-PCR). In vitro, we knocked down the LINC00511 with small interfering RNA in PTC cell lines and demonstrated the function of LINC00511 by Cell Counting Kit-8, cell colony formation, Transwell migration, Transwell invasion, apoptosis assays, and cell cycle assays. Then, we discovered several downstream proteins of LINC00511 using Western blotting.
Results: We proved that LINC00511's expression in PTC tissues and cell lines is higher than the control. LINC00511 promoted cellular proliferation, migration, invasion, G1/S transition and reduced apoptosis in vitro experiment. Knocked-down of LINC00511 resulted in the reduction of histone methyltransferase enhancer of zeste homolog 2 (EZH2), cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase 4 (CDK4).
Conclusions: Our results certified the role and function of LINC00511 in PTC, and it could become a novel tumor therapeutic target.
Keywords: long noncoding RNA 00511, proliferation, cyclin-dependent kinase, papillary thyroid carcinoma.
Introduction
Over the years, the morbidity of thyroid carcinoma is rising all the time and even leaping to the first five in women [1, 2]. Papillary thyroid carcinoma (PTC) accounts for around 80% of all types of thyroid carcinoma, which becomes the commonly histological classification [3]. And PTC usually has a favorable outcome with early diagnosis, which treated by surgery could have a 5-year survival of >97% [4, 5]. However, we shouldn't stop discovering better means such as novel molecular targets to get a faster diagnosis and superior treatment.
Long noncoding RNAs (lncRNAs) are longer than 200 nucleotides in length and have no function in coding protein, which proven to regulate transcription and epigenetic gene [6-8]. There are a large amount of lncRNAs supported closely connecting with tumor genesis and could be therapeutic targets and diagnostic markers [9]. Equally, PVT1, NAMA and H19, and so on all play an important role in thyroid cancer [10-12].
Long noncoding RNA 00511 (LINC00511) is a transcript of 2265 bp. Sun Cheng-Cao et al. found LINC00511 serve as an oncogene and restrain p57 in Non-small-cell Lung Cancer [13]. LINC00511 was discovered up-regulating in breast cancer [14-16]. And Zhao Xiaohui et al. demonstrated that LINC00511 is a competing endogenous RNA in pancreatic ductal adenocarcinoma [17]. But the role and function of LINC00511 in PTC is still unknown. In our study, we illuminated the expression level and functions of LINC00511 in PTC cell lines. These findings all insisted that LINC00511 may be a potential therapeutic target lncRNA in PTC.
Cyclin-dependent kinases (CDKs) are a family associated with cell cycle progression and other complex functions [18]. Among them, CDKs 1, 2, 4 and 6 are directly having contact with cell cycle [19]. CDKs are expressed at a high level in many tumors [20]. CDK2 was found dysregulated in gastric cancer and lung cancer [21, 22]. Zhou Yubing et al. discovered CDK4 was correlated with osteosarcoma [23]. CDK2 and CDK4 could influence breast cancer cell by p27 [24]. Our discovery was sure of the relationship between CDK2, 4 and LINC00511. In the future, we could control the LINC00511 of PTC by CDK2 and CDK4.
Materials and methods
Patient and tissue samples
We gained the information about expression level and clinicopathologic features of LINC00511 from The Cancer Genome Atlas (TCGA). We selected 41 patients suffered from papillary thyroid carcinoma randomly. Major inclusion criteria were: (1) patients with pathologically confirmed thyroid cancer in the primary tumor and without any severe diseases in other organs; (2) patients that had received total/near total thyroidectomy and had not received any radiotherapy; (3) patients with a negative history of any other malignant tumors. Major exclusion criteria were: (1) patients with a positive history of other malignant tumors; (2) patients with severe diseases such as heart failure, stroke, and chronic renal failure; (3) patients with a history of 131I therapy.
In total 41 PTC tissues and matched adjacent tissues were collected after agreement from patients during operation at The First Affiliated Hospital of Wenzhou Medical University in recent two years. All patients had a definite pathological diagnosis. This study got the approval of The Institute Research Medical Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University.
Cell lines
The human thyroid cancer cell lines (B-CPAP, KTC-1, and KTC-1) were obtained from Professor Mingzhao Xing of the Johns Hopkins University School of Medicine, Baltimore, MA, USA. All cell lines were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen). They were lived in a standard cell culture incubator (Thermo, Waltham, MA, USA) at 37℃ with 5% CO2.
Cell transfection
The small interfering RNA (siRNA) targeting LINC00511 was designed by Shanghai Gene Pharma (Shanghai, China). 8*104 (B-CPAP), 8* 104 cells (KTC-1) or 6* 104 cells (TPC-1) were plated the day before transfection. LINC00511 was knocked down using 10μl (TPC-1), 7.5μl (KTC-1) or 5μl (B-CPAP) siRNA and entering the RNAiMAX (Life Technologies, Carlsbad, CA, USA) which to siRNA ratio is 0.4. After 48 hours, the cells could be used further tests. The sequences of LINC00511 were as follows: sense 5'- GACUGAAUGUGGUUCCAGATT-3' and antisense 5'-UCUGGAACCACAUUCAGUCTT-3'.
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was used to test the expression of LINC00511. Total RNA was extracted from PTC cells by Trizol (Invitrogen) and then became cDNA through reverse transcription (Toyobo, Osaka, Japan). cDNA was conducted to run qRT-PCR (Applied Biosystems 7500) with the THUNDERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan). The relative expressions of LINC00511 were analyzed by the 2-ΔΔCt methods comparing to GAPDH. The sequences of the primers used were: LINC00511 Forward: 5′-CGCAAGGACCCTCTGTTAGG-3′, and Reverse: 5′ GAAGGCGGATCGTCTCTCAG-3'; GADPH Forward: 5'-GTCTCCTCTGACTTCAACAGCG-3' and Reverse: 5' ACCACCCTGTTGCTGTAGCCAA-3'.
Colony formation assay
Seeding B-CPAP (1,500 cells/wells), KTC-1 (1,500 cells/wells) and TPC-1 (1,000 cells/wells) in 6-well plates after 48 hours of transfection. 5-7 days later, fixed by 4% paraformaldehyde (PFA) and stained by Crystal violet dye. Then, getting images by camera and performing it in triplicate.
CCK-8 proliferation assay
Cell-counting kit 8 (CCK8) assay is used to assess cell proliferation ability. After cell transfection, putting B-CPAP (1,500 cells/wells), KTC-1 (1,500 cells/wells) and TPC-1 (1,000 cells/wells) in 96-well plates. Adding 10μl CCK8 every well and culturing 2-4 hours at 37℃ in the dark for four consecutive days. And measuring at the 450 nm absorbance in these days.
Cell migration and invasion assays
Putting the digested cells (30,000 cells/well) into the upper chamber with 1640 medium containing 10% FBS. And 600μl medium was filling in the lower chamber without any cells and culturing cells 22-24 hours in the incubator. Then, washing the membrane with PBS, fixing with 4% PFA for 15 min and staining with 0.4% crystal violet solution for 30min. Finally, wiping the crystal violet solution and choosing several views under an inverted microscope at a magnification of x20.
Apoptosis assays
After transfection, resuspending the cells in the 1×binding buffer at 1×106 cells/ml. Using FITC- Annexin V 5μl for 15min and propridium iodide (PI) 5μl for 5 min in the dark at every 100μl suspension. The FACS could measure and analyze the FITC-Annexin V-positive cells as the apoptosis or necrotic cell. PI could distinguish the early and late apoptosis.
Cell cycle assays
The cells were starved 48 hours using serum-free medium before transfection. Cells were fixed by 75% ice-cold ethanol at -20℃ overnight. Next, washed and resuspended by PBS. Adding PI/RNase Staining Buffer (BD, San Diego, CA) 500μl to every Eppendorf tube for 30min in the dark. Samples were run on the C6 flow cytometer and analyzed by Modfit software.
Western blotting
Proteins were isolated by RIPA lysis buffer (Beyotime, Shanghai, China) and degenerated at 100℃ for 10min. In the next moment, proteins were separated by sodium dodecyl sulfatepolyacrylamide gels electrophoresis (BioRad, Berkeley, CA, USA) and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA). Blocking with 5% non-fat milk at 4℃ overnight and probing the membranes with polyclonal antibody overnight with the same conditions. After washed by TBST, the membranes were incubated with the anti-mouse IgG or anti-rabbit IgG (Abcam, Cambridge, MA) at room temperature for 1 hour. Primary antibodies were as follows: EZH2 (Abcam, USA), CDH2 (Abcam, USA), CDH4 (Abcam, USA), β-actin (Abcam, USA).
Statistical analysis
All statistical analyses were using SPSS 22.0 software (SPSS Inc. Chicago, IL, USA). Student's t-test was used between groups. P<0.05 is considered to be statistically significant. The results were presented as mean ± SD.
Results
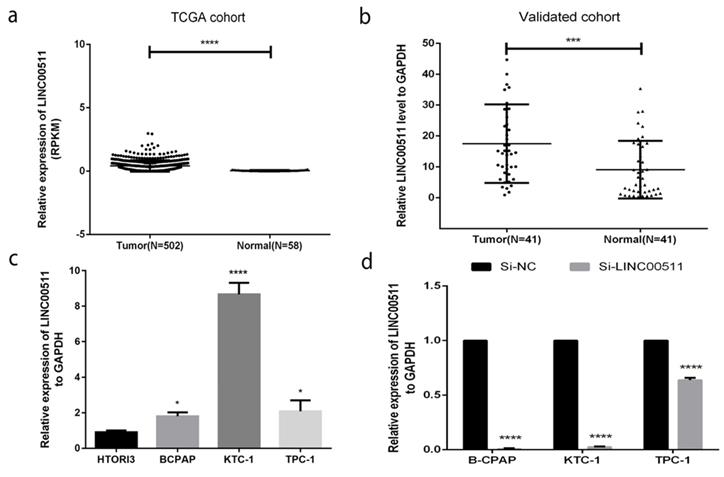
LINC00511 is overexpressed in both PTC tissues and cell lines
To have a rough idea of LINC00511, we got some information from The Cancer Genome Atlas (TCGA) (Fig 1a) and knew that the expression of LINC00511 in PTC tissues is higher than in normal tissues. To further verification, we chose 41 PTC tissues and matched normal tissues to measure their expression level by qRT-PCR. And Fig 1b showed that LINC00511 is up-regulated in PTC tissues (P<0.001). Then, we tested the expression level in cell lines including normal thyroid cell line (HTORI3) and PTC cell lines (B-CPAP, KTC-1, TPC-1) by qRT-PCR. As is shown in Fig.1c, LINC00511 is higher expressed in PTC cell lines. They all supported that LINC00511 is overexpressed in PTC tissues and cell lines.
Next, we used the small interfering RNAs (siRNAs) (Si-NC, Si-LINC00511) to knock down the LINC00511 expression in PTC cell lines for further experiments. Most of the cell lines could be cut to less than 0.1(Fig 1d).
The relationship between LINC00511 expression and clinicopathologic features
To know whether LINC00511 expression is connected with tumorigenesis in PTC, we investigated the relationship between LINC00511 and clinicopathologic features. Firstly, we acquired 41 PTC patients' clinicopathologic information. Then, dividing the patients into two groups according to LINC00511 expression. The low-expression group has 27 patients (T/N<5.9) and the high-expression group has 14 patients (T/N>5.9). The results from the validation cohort showed that the relationship between LINC00511 and tumor size is closer than other features (Table 1). To explore more about this relationship, we divided 502 patients into the low-expression group (T<0.518, n=335) and high-expression group (T>0.518, n=167) according to the expression level of tumor tissues from TCGA. And the patients' clinicopathologic information of TCGA told us that LINC00511 expression is significantly related to tumor size (P=0.019<0.05), T stage (P=0.019<0.05), Lymph node metastasis (P=0.000<0.05), AJCC stage (P=0.001<0.05) and histological type (P=0.000 < 0.05) (Table 2). Generally, LINC00511 expression is most closely to tumor size and it may influence the tumor proliferation.
LINC00511 is overexpressed in human PTC tissues and cell lines. (a) Relationship of expression levels of LINC00511 in thyroid tumor tissues and normal tissues in TCGA. (b) LINC00511 expression level is significantly increasing in 41 human PTC tissues compared to matched adjacent tissues. (c) The relative expression of LINC00511 to GAPDH using qRT-PCR. All cell lines are up-regulated. (d) The efficiency of siRNAs (Si-NC and Si-LINC00511) was measured by qRT-PCR in PTC cell lines. *P < 0.05; ***P < 0.001; ****P < 0.0001 in comparison with the control group using Student's t-test. 2-ΔΔCt is used to represent the fold change in qRT-PCR detection.
The relationship between LINC00511 and clinicopathologic characteristics in the validated cohort.
| Clinicopathologic characteristics | Low expression (%) | High expression (%) | X2 | P |
|---|---|---|---|---|
| Age | 0.579 | 0.447 | ||
| ≤45 | 13(48.1) | 5(35.7) | ||
| >45 | 14(51.9) | 9(64.3) | ||
| Gender | 0.131 | 0.717 | ||
| Female | 17(63) | 8(57.1) | ||
| Male | 10(37) | 6(42.9) | ||
| Tumor size | 2.933 | 0.087 | ||
| ≤10mm | 8(29.6) | 8(57.1) | ||
| >10mm | 19(70.4) | 6(42.9) | ||
| Unilateral or Bilateral | 0.038 | 0.846 | ||
| Unilateral | 17(63) | 10(71.4) | ||
| Bilateral | 10(37) | 4(28.6) | ||
| Extrathyroidal invasion | 0.078 | 0.780 | ||
| YES | 2(7.4) | 0(0) | ||
| NO | 25(92.6) | 14(100) | ||
| Lymph node metastasis | 1.076 | 0.300 | ||
| YES | 9(33.3) | 7(50) | ||
| NO | 18(66.7) | 7(50) | ||
| AJCC | 0.000 | 1.000 | ||
| I+II | 18(66.7) | 9(64.3) | ||
| III+IV | 9(33.3) | 5(35.7) |
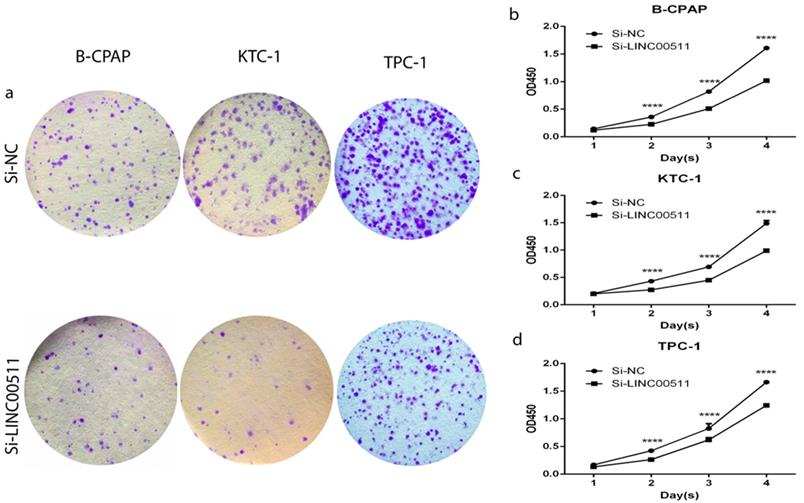
Down-regulation of LINC00511 inhibits proliferation in PTC cell lines
All three PTC cell lines are the high expression of LINC00511, so we chose them to further functional experiments. As is known to us, high expression of LINC00511 could significantly influence tumor size mentioned before. Thanks to the relationship between LINC00511 and tumor size, we did the colony formation assay and CCK-8 assay to prove if knockdown of LINC00511 restrains cellular proliferation. The colony formation assay led to the results that knockdown of LINC00511 inhibits proliferation in B-CPAP, KTC-1 and TPC-1 cell lines compared with the Si-NC group (Fig 2a). And the CCK-8 assay made the results more convincing thanks to the same conclusions (Fig 2b-d).
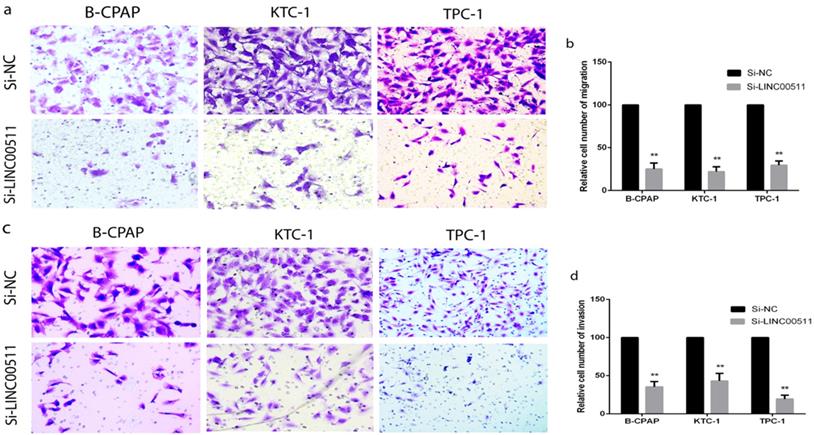
Down-regulation of LINC00511 restrains migration and invasion in PTC cell lines
To explore other potential biological functions of LINC00511, we carried out the Transwell assays to obtain the capacity of migration and invasion using B-CPAP, KTC-1 and TPC-1 cell lines. The migration assays indicated that down-regulated LINC00511 represses cellular migration compared with the negative control in PTC cell lines (Fig 3a, b). The invasion assays also showed that down-regulated LINC00511 represses cellular invasion compared with the Si-NC group (Fig 3c, d). These findings suggested that LINC00511 knockdown suppresses cellular migration and invasion in vitro.
The relationship between LINC00511 and clinicopathologic characteristics in the TCGA cohort.
| Clinicopathologic characteristics | Low expression (%) | High expression (%) | X2 | P |
|---|---|---|---|---|
| Age | 0.08 | 0.777 | ||
| ≤45 | 156(46.6) | 80(47.9) | ||
| >45 | 179(53.4) | 87(52.1) | ||
| Gender | 3.771 | 0.052 | ||
| Female | 254(75.8) | 113(67.7) | ||
| Male | 81(24.2) | 54(32.3) | ||
| Histological type | 44.501 | 0.000* | ||
| Classical | 221(66) | 137(82) | ||
| Follicular | 91(27.2) | 8(4.8) | ||
| Tall Cell | 15(4.5) | 21(12.6) | ||
| other | 8(2.4) | 1(0.6) | ||
| Tumor size | 5.516 | 0.019* | ||
| ≤20mm | 108(32.2) | 37(22.2) | ||
| >20mm | 227(67.8) | 130(77.8) | ||
| T stage | 5.516 | 0.019* | ||
| T1 | 108(32.2) | 37(22.2) | ||
| >T1 | 227(67.8) | 130(77.8) | ||
| Extrathyroidal invasion | 0.306 | 0.58 | ||
| YES | 5(1.5) | 4(2.4) | ||
| NO | 182(54.3) | 100(59.9) | ||
| Lymph node metastasis | 25.472 | 0.000* | ||
| YES | 120(35.8) | 103(61.7) | ||
| NO | 175(52.2) | 54(32.3) | ||
| AJCC | 10.636 | 0.001* | ||
| I+II | 238(71) | 95(56.9) | ||
| III+IV | 95(28.4) | 72(43.1) |
* P-value < 0.05.
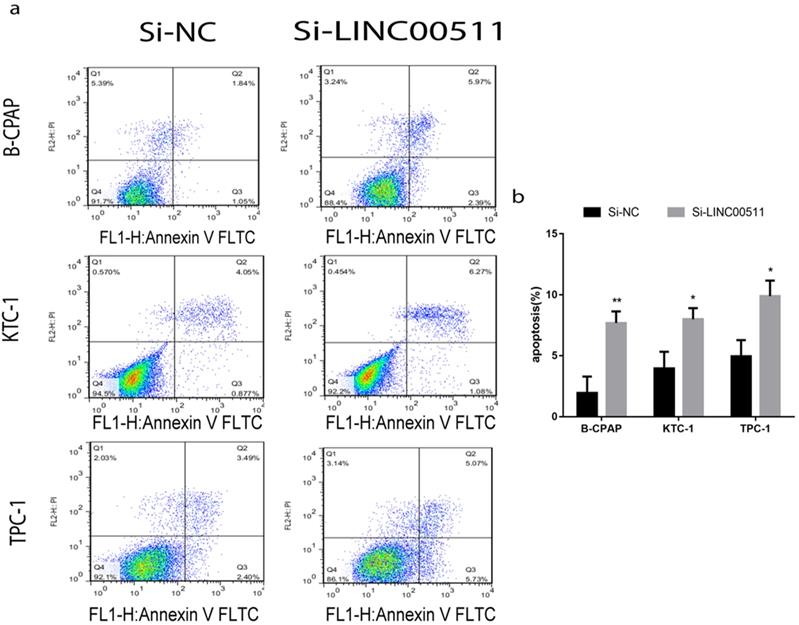
Down-regulation of LINC00511 promotes apoptosis in PTC cell lines
For a closer look at how LINC00511 affects proliferation, we applied flow cytometry to measure cell apoptosis in all three PTC cell lines (B-CPAP, KTC-1 and TPC-1) after transfection. The cells were stained positive by Annexin V/PI on behalf of apoptosis and necrosis. According to the dyeing graph in the person of early and late apoptosis, knockdown of LINC00511 could facilitate apoptosis compared with the control group in vitro (Fig 4a, b).
Down-regulation of LINC00511 decreased G1/S transition in PTC cell lines
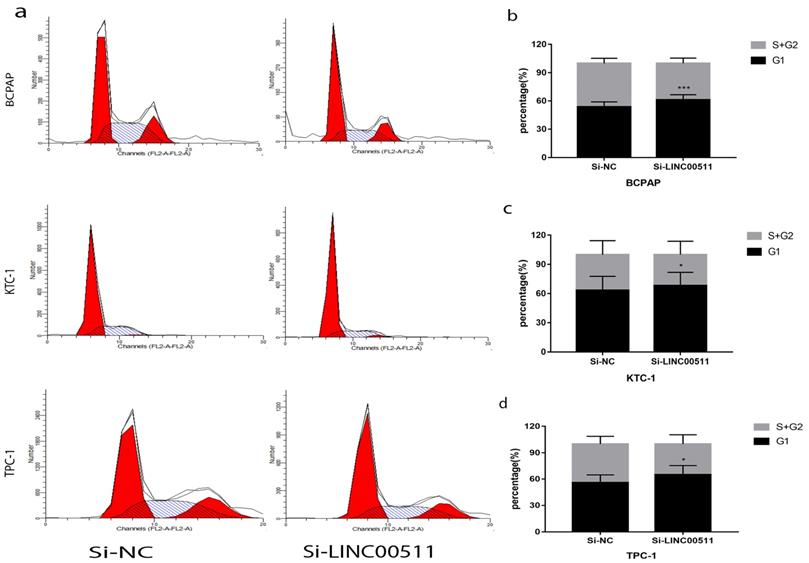
To find other reasons resulting in the reduction of proliferation, we did flow cytometry to study the PTC cell cycle after transfection. And the cells were starved before transfection for cell cycle synchronization. The test manifested that silence of LINC00511 can reduce G1/S transition (Fig 5a). In other words, the number of PTC cells in G1 phase became more and S phase became less. And the histograms showed this experiment is statistically meaningful (Fig 5b-d). These results indicated that knockdown of LINC00511 can lead to PTC cells from G1 phase to S phase.
Effect of LINC00511 on proliferation in PTC cell lines. (a) Colon formation assay indicated that knockdown of LINC00511 inhibits cell proliferation in B-CPAP, KTC-1, and TPC-1 cells. (b -d) CCK-8 assay indicated that knockdown of LINC00511 represses cell proliferation in B-CPAP, KTC-1, and TPC-1 cells. ****P < 0.0001 in comparison with the Si-NC group using Student's t-test.
LINC00511 influences PTC cells migration and invasion. (a, b) Migration assay in PTC cell lines showed that knockdown of LINC00511 decreases the cell migration compared with that in the Si-NC group. (c, d) Knockdown of LINC00511 decreases the ability of cellular invasion compared with that in the Si-NC group. **P < 0.01 in comparison with the Si-NC group using Student's t-test.
LINC00511 facilitates proliferation through CDKs and EZH2 in PTC cell lines
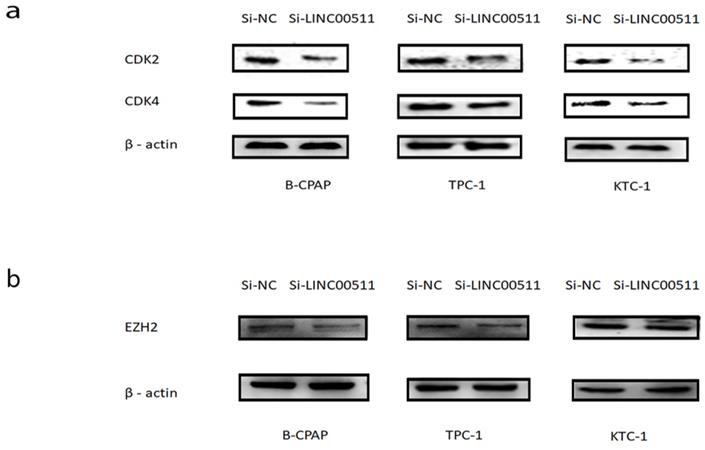
CDKs is a kind of proteins about cell cycle and have a tight relationship with proliferation. In order to seek the potential mechanism of LINC00511 in PTC, we detected some main CDKs about cell cycle by Western blotting. The results suggested that the down regulation of LINC00511 can decrease the expression level of CDK2 and CDK4 (Fig 6a).
Epithelial to mesenchymal transition (EMT) is a crucial character of tumorigenesis [25]. So, we also measured the expression level of several common epithelial markers and mesenchymal markers. And we found that knockdown of LINC00511 can reduce the expression level of EZH2 compared with β-actin (Fig 6b), except N-cadherin, E-cadherin, vimentin, and so on.
From these results, it is clear that LINC00511 can accelerate proliferation by CDKs and EZH2 in PTC cell lines.
Discussion
The incident of papillary thyroid carcinoma is increasing all the time, but the prognosis is not bad under standard treatment such as thyroidectomy [2, 26]. However, PTC is easy to recur so that we need to explore accurate therapeutic method. There are many researchers discovering that gene target therapy is a good manner to cure thyroid cancer. Fang lei et al. found that microRNA-625-3p acts as an oncogene in thyroid cancer by high expression of AEG-1[27]. Han Jiakai et al. detected that anchor protein 4 (AKAP4) promotes the proliferation of thyroid cancer[28].
LncRNAs play a crucial part in transcriptional regulation, epigenetic gene regulation and disease[8]. And many lncRNAs were convinced of having connected with thyroid carcinoma [29-31]. The study shows that LncRNA LINC00511 promotes osteosarcoma through miR-765[32]. And Lu Guanming et al. discovered that LINC00511 leads to breast cancer tumorgenesis via miR-185-3p/E2F1/Nanog axis [33]. But LINC00511'function and mechanism in PTC cell lines are not exactly clear before.
Cyclin-dependent kinases (CDKs) are protein kinases can regulate the cell cycle of tumor cells [18]. It must influence the proliferation of tumor cells so that tumors can be easy to deal with. Recently, CDK inhibitions were deemed to the method of cancer therapy [34]. Epithelial to mesenchymal transition (EMT) is a key feature of cancer progression [35]. EZH2 is one of the EMT features and having important role in cancer progression [36].
Knockdown of LINC00511 promotes cell apoptosis in PTC cell lines. (a, b) Flow cytometry analysis of PTC cell lines indicated that down-regulation of LINC00511 facilitates cell apoptosis in B-CPAP, KTC-1, and TPC-1. *P < 0.05; **P < 0.01 in comparison with the Si-NC group using Student's t-test.
In our study, we caught sight of the high expression of LINC00511 in PTC tissues and cell lines. The relationship between LINC00511 expression and clinicopathologic features certified that LINC00511 influences tumor size from TCGA and our own tissues. It's obviously that LINC00511 expression is related to tumor size, T stage, Lymph node metastasis, AJCC stage and histological type from TCGA. In specification, LINC00511 had tight relations to cell proliferation, tumor metastasis and progression in papillary thyroid carcinoma. But LINC00511 only had connection with tumor size from our tissues data, namely only cell proliferation. The reason for this phenomenon may is the lack of adequate patients of us compared with TCGA. Knockdown LINC00511 repressed the ability of proliferation, migration, invasion and G1/S transition in vitro. But down regulation of LINC00511 could stimulate cellular apoptosis in PTC cell lines. We also found that LINC00511 can affect the expression of CDK2, CDK4, and EZH2. In a word, LINC00511 acted as an oncogene and promoted proliferation through CDKs in PTC.
Nevertheless, our study still exists many inadequacies. Primarily, LINC00511 influence CDK2 and CDK4 directly or through other member remains a question. Secondly, we should validate the function of LINC00511 in vivo to make our study more rigorous. Thirdly, the patients we collected can be more to be receivable.
Overall, we certified that LINC00511 is overexpressed in PTC tissues and cell lines. And clinicopathologic analysis confirmed that tumor size is the most significant feature of LINC00511. Our results described the biological function and potential mechanism of LINC00511 in PTC cell lines. These consequences all indicated that LINC00511 is an oncogene in PTC and promoting proliferation by CDKs. It will be an underlying therapy target in PTC. CDK inhibitors can disturb the CDK thus cure cancer [37]. So maybe the CDK inhibitors can combine with LINC00511 knockdown for the treatment of PTC.
Knockdown of LINC00511 reduces G1/S transition in PTC cell lines. (a) Cell cycle analysis revealed that down-regulation of LINC00511 could repress G1/S transition in PTC cell lines. (b-d) The histograms intuitively showed that knockdown of LINC00511 could influence cell cycle in vitro. *P < 0.05; ***P < 0.001 in comparison with the Si-NC group using Student's t-test.
LINC00511 can affect the expression of CDKs and EZH2. (a) The influence of LINC00511 expression on CDK2 and CDK4 in PTC cell lines by Western blotting. (b) The influence of LINC00511 expression on EZH2 in PTC cell lines by Western blotting.
Acknowledgements
The authors would like to thank all the doctors of Department of Thyroid and Breast Surgery, The First Affiliated Hospital of Wenzhou Medical University, (Wenzhou, China) for providing all the necessary information required for this study.
Ethical Approval and Consent to participate
Ethical approval for this study was obtained from the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University.
Funding
This study was funded by National Natural Science Foundation of China (NO.81572291) and Natural Science Foundation of Zhejiang province (LY17H160053, LGF18H160031, GF18H160071, and LGF18H160032) and the Medical and Health Technology Projects of Zhejiang province (NO.2017187475) and the Science and Technology Project of Wenzhou (Y20170030).
Consent for publication
Written informed consent was issued by the patients for the publication of this research and accompanying images. A copy of the written consent is ready for review by the Editor in Chief of this journal.
Availability of data and materials
The data sets supporting the conclusions of this study are included in this article and its, additional images. Raw data are available on the main electronic data storage system of First Affiliated Hospital of Wenzhou Medical University and access can be provided upon request to the authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988-2005. Cancer. 2009;115(16):3801-3807
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7-30
3. Burns WR, Zeiger MA. Differentiated thyroid cancer. Semin Oncol. 2010;37(6):557-566
4. Lundgren CI, Hall P, Dickman PW, Zedenius J. Clinically significant prognostic factors for differentiated thyroid carcinoma: a population-based, nested case-control study. Cancer. 2006;106(3):524-531
5. Suliburk J, Delbridge L. Surgical management of well-differentiated thyroid cancer: state of the art. Surg Clin North Am. 2009;89(5):1171-1191
6. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298-1307
7. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46-58
8. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629-641
9. Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419-2425
10. Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283(12):2326-2339
11. Yoon H, He H, Nagy R, Davuluri R, Suster S, Schoenberg D, Pellegata N, Chapelle Ade L. Identification of a novel noncoding RNA gene, NAMA, that is downregulated in papillary thyroid carcinoma with BRAF mutation and associated with growth arrest. Int J Cancer. 2007;121(4):767-775
12. Zhou Q, Chen J, Feng J, Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR). Tumour Biol. 2016;37(3):3105-3113
13. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non-small-cell Lung Cancer by Binding to EZH2 and Suppressing p57. Mol Ther Nucleic Acids. 2016;5(11):e385
14. Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016;7(28):43256-43266
15. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16(1):129
16. Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761-772
17. Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y. et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22(1):655-667
18. Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15(6):122
19. Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30(11):630-641
20. Nemunaitis JJ, Small KA, Kirschmeier P, Zhang D, Zhu Y, Jou YM, Statkevich P, Yao SL, Bannerji R. A first-in-human, phase 1, dose-escalation study of dinaciclib, a novel cyclin-dependent kinase inhibitor, administered weekly in subjects with advanced malignancies. J Transl Med. 2013;11:259
21. Tang Z, Li L, Tang Y, Xie D, Wu K, Wei W, Xiao Q. CDK2 positively regulates aerobic glycolysis by suppressing SIRT5 in gastric cancer. Cancer Sci. 2018;109(8):2590-2598
22. Hu S, Danilov AV, Godek K, Orr B, Tafe LJ, Rodriguez-Canales J, Behrens C, Mino B, Moran CA, Memoli VA. et al. CDK2 Inhibition Causes Anaphase Catastrophe in Lung Cancer through the Centrosomal Protein CP110. Cancer Res. 2015;75(10):2029-2038
23. Zhou Y, Shen JK, Yu Z, Hornicek FJ, Kan Q, Duan Z. Expression and therapeutic implications of cyclin-dependent kinase 4 (CDK4) in osteosarcoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt A):1573-1582
24. Patel P, Tsiperson V, Gottesman SRS, Somma J, Blain SW. Dual Inhibition of CDK4 and CDK2 via Targeting p27 Tyrosine Phosphorylation Induces a Potent and Durable Response in Breast Cancer Cells. Mol Cancer Res. 2018;16(3):361-377
25. Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, Kong W, Dan S, Bui MM, Coppola D. et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290(11):6857-6867
26. Markovina S, Grigsby PW, Schwarz JK, DeWees T, Moley JF, Siegel BA, Perkins SM. Treatment approach, surveillance, and outcome of well-differentiated thyroid cancer in childhood and adolescence. Thyroid. 2014;24(7):1121-1126
27. Fang L, Kong D, Xu W. MicroRNA-625-3p promotes the proliferation, migration and invasion of thyroid cancer cells by up-regulating astrocyte elevated gene 1. Biomed Pharmacother. 2018;102:203-211
28. Han J, Gao W, Su D, Liu Y. Silencing of A-Kinase Anchor Protein 4 (AKAP4) Inhibits Proliferation and Progression of Thyroid Cancer. Oncol Res. 2017;25(6):873-878
29. Li X, Zhong W, Xu Y, Yu B, Liu H. Silencing of lncRNA LINC00514 inhibits the malignant behaviors of papillary thyroid cancer through miR-204-3p/CDC23 axis. Biochem Biophys Res Commun. 2019;508(4):1145-1148
30. Liu Y, Yue P, Zhou T, Zhang F, Wang H, Chen X. LncRNA MEG3 enhances (131)I sensitivity in thyroid carcinoma via sponging miR-182. Biomed Pharmacother. 2018;105:1232-1239
31. Zhang D, Liu X, Wei B, Qiao G, Jiang T, Chen Z. Plasma lncRNA GAS8-AS1 as a Potential Biomarker of Papillary Thyroid Carcinoma in Chinese Patients. Int J Endocrinol. 2017;2017:2645904
32. Yan L, Wu X, Liu Y, Xian W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem. 2018
33. Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z. et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37(1):289
34. Heptinstall AB, Adiyasa I, Cano C, Hardcastle IR. Recent advances in CDK inhibitors for cancer therapy. Future Med Chem. 2018;10(11):1369-1388
35. Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119(6):1429-1437
36. Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. 2018;6:10
37. Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2011;137(10):1409-1418
Author contact
![]() Corresponding author: Prof. Dr. Ouchen Wang, Department of Thyroid and Breast Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, People's Republic of China, 325000 Contact number: +86 577 5557 8527; Email: woc863com
Corresponding author: Prof. Dr. Ouchen Wang, Department of Thyroid and Breast Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, People's Republic of China, 325000 Contact number: +86 577 5557 8527; Email: woc863com

 Global reach, higher impact
Global reach, higher impact