3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(4):874-882. doi:10.7150/jca.34723 This issue Cite
Research Paper
Signatures of circulating microRNA in four sarcoma subtypes
1. Department of Soft Tissue, Bone Sarcoma and Melanoma, Maria Sklodowska-Curie Institute - Oncology Centre, Warsaw, Poland
2. Department of Gastroenterology, Hepatology and Clinical Oncology, Centre of Postgraduate Medical Education, Warsaw, Poland
3. Department of Genetics, Maria Sklodowska-Curie Institute - Oncology Centre; 02-781 Warsaw, Poland
*Equal contribution
Received 2019-3-8; Accepted 2019-9-27; Published 2020-1-1
Abstract
Background: Sarcomas are rare malignant tumors of mesenchymal origin. The discovery of circulating biomarkers with high diagnostic value could supplement diagnosis of this heterogenous group of tumors. The aim of this study was to identify the profiles of circulating miRNA (c-miRNAs) in four groups of common bone and soft tissue sarcomas.
Methods: At the time of diagnosis, blood samples were collected from 86 patients: 36 with locally advanced/unresectable/metastatic gastrointestinal stromal tumor (GIST) who received first-line treatment with imatinib; 16 with locally advanced osteosarcoma (OS); 26 with locally advanced synovial sarcoma (SS); and eight with locally advanced Ewing sarcoma (ES). In addition, samples were collected from 30 healthy controls. C-miRNAs were isolated using a miRCURY RNA Isolation Kit, followed by preparation of cDNA libraries and sequencing on the Ion Proton platform.
Results: Pair-wise comparisons identified 156 unique c-miRNAs (adjusted P-value < 0.05) showing significant dysregulation between controls and patients; of these, 24, 36, 42, and 99 differentiated controls from pretherapeutic OS, SS, ES, and GIST, respectively. Ten c-miRNAs were commonly altered in at least three sarcoma types. Receiver operating characteristic curves and area under the curve (ROC-AUC) analyses revealed that a four-miRNA diagnostic classifier was able to differentiate controls from ES, GIST, OS, and SS, with AUC-ROC values of 1, 0.97, 0.95, and 0.94, respectively.
Conclusions: Aberrant miRNA expression signatures were identified in serum from patients with four different sarcoma subtypes. Differences in miRNA expression profiles between sarcoma patients and healthy volunteers suggest that miRNAs may play a role in sarcoma development.
Keywords: miRNA, sarcoma, diagnosis, next generation sequencing
Background
Sarcomas, rare tumors of mesenchymal origin, account for approximately 1% of all adult cancers. Soft tissue sarcomas (STSs) comprise at least 60 histological subtypes, and even more molecular subsets. For example, there are at least six major histological types of bone sarcoma. The majority (close to 75%) of sarcomas arise from soft tissue, with an additional 15% being gastrointestinal stromal tumors (GISTs) and 10% being bone sarcomas (BSs) [1]. The primary treatment for sarcomas is surgery, although (neo)adjuvant chemotherapy is the main treatment for some BS cases [2]. The use of perioperative chemotherapy for STS is uncommon; however, up to date guidelines state that it should be an option for high-risk individuals; this is because approximately half of all STS patients with intermediate or high-grade tumors develop metastatic disease, and the 5 year overall survival is approximately 55% [1,3]. Increased understanding of the molecular mechanisms underlying GIST has led to introduction of targeted therapy with receptor kinase inhibitors. Imatinib mesylate was the first effective systemic therapy for GIST; this treatment shows high efficacy in advanced cases and has improved the outcomes of patients markedly [4]. Making a diagnosis of sarcoma is complicated and is based on adequate imaging and pathological examination, often aided by molecular/cytogenetic diagnostics. As a supplement to traditional diagnostic methods, identifying circulating biomarkers with high diagnostic utility is essential.
Non-coding microRNAs (miRNAs) affect cellular processes by regulating gene expression. As such, over-abundance may play a role in pathogenesis of various diseases, including human neoplastic disorders. It is assumed that cell-free circulating miRNAs are potential cancer biomarkers because they are stable molecules that show expression profiles that correlate specifically with different types of cancer; in addition, they are sampled easily in a relatively non-invasive manner and are detected readily by various techniques [5].
Lawrie et al. were the first to identify circulating miRNAs in serum and to suggest their potential as cancer markers [6]. They found that serum miRNAs (including miR-155, miR-210, and miR-21) in the serum of diffuse large B-cell lymphoma patients were higher than those in in healthy individuals. More interestingly, high miR-21 levels in these patients were associated with relapse-free survival. Mitchell and co-workers [7] found that circulating miRNAs are remarkably stable and are protected from endogenous RNase activity. Following these observations, several studies reported miRNAs differentially expressed in serum or plasma from patients with gastric cancer, non-small-cell lung cancer, hepatocellular carcinoma, rhabdomyosarcoma, head and neck squamous cell carcinoma, breast cancer, bladder cancer, colorectal cancer, epithelial ovarian cancer, renal cancer, pancreatic cancer, esophageal squamous cell carcinoma, chronic lymphocytic leukemia, melanoma, and Hodgkin lymphoma [8-18].
The aim of this study was to identify circulating miRNA (c-miRNA) profiles in serum or plasma from four groups of individuals with common bone and soft tissue sarcomas: osteosarcoma (OS), synovial sarcoma (SS), Ewing sarcoma (ES), and gastrointestinal stromal tumor.
Methods
Patients
Selection of patients and controls was performed at the Department of Soft Tissue/Bone Sarcoma and Melanoma, Maria Sklodowska-Curie Institute - Oncology Center, Warsaw, Poland, which is the reference center for adult sarcomas and is integrated into the European Reference Network EURACAN. Patients were selected according to the following criteria: 1) categorized into homogenous groups of sarcoma patients; and 2) they had a well-established pathological diagnosis performed by expert pathologists specialized in sarcoma diagnostics and confirmed by molecular testing (i.e., cytogenetics results showing translocation of t(X;18) and t(11;22), which is characteristic of SSs and ESs, respectively, and genotyping revealing the presence of oncogenic KIT and PDGFRA driver mutations in GISTs). Computed tomography (CT) or magnetic resonance imaging (MRI) was performed to establish disease staging. The study was approved by the local Bio-Ethics Committee and was conducted in accordance with Good Clinical Practice Guidelines (decision 13/2008). All patients provided written informed consent to participate.
The following patients were included in the analyzed groups: 1) Patients with locally advanced/ unresectable/metastatic GIST who received first-line treatment with imatinib (400 mg OD) until disease progression or unacceptable toxicity, with response to treatment assessed by CT scan every 2-3 months; 2) An OS group comprising patients with localized tumors treated with preoperative chemotherapy (doxorubicin and cisplatin) and 3-6 cycles of post-operative chemotherapy consistent with the same as preoperative scheme; 3) SS patients treated with the internal extensive combined modality treatment protocol, which comprised two cycles of high-dose ifosfamide, preoperative radiotherapy, radical surgery, and post-operative chemotherapy (two cycles of high-dose ifosfamide and two cycles of doxorubicin and cisplatin); and 4) ES patients treated with chemotherapy comprising multi-drug anthracycline- and ifosfamide-based chemotherapy prior to local treatment (surgery and/or radical radiotherapy), followed by continuation of chemotherapy (treatment as a whole lasted 12 months). The characteristics of the four patient groups and controls are listed in Table 1.
Patient characteristics
| Number of patients | Median age at diagnosis (years) | Gender | Location of the primary tumor | Genetic testing | Best response to imatinib treatment |
|---|---|---|---|---|---|
| Gastrointestinal stromal tumor - unresectable/metastatic | |||||
| 36 | 64 | Female: 20 Men: 16 | Stomach: 21 Small intestine: 6 Duodenum: 5 Esophagus: 1 Rectum: 1 Omentum: 1 Retroperitoneum: 1 | KIT/PDGFRA mutational status: KIT exon 11: 14 KIT exon 9: 4 PDGFRA: 2 Wild-type: 2 Not assessed: 14 | Partial response: 12 Stable disease: 24 |
| Synovial sarcoma - localized | |||||
| 26 | 44 | Female: 15 Male: 11 | Upper extremity: 5 Lower extremity: 19 Other: 2 | Yes: 16 No: 3 Not assessable: 7 | Yes: 9 No: 17 |
| Ewing sarcoma - locally advanced | |||||
| 8 | 28 | Female: 0 Male: 8 | Head: 1 Lower limb: 3 Trunk: 4 | Yes: 4 No: 2 Not assessable: 2 | Yes: 3 No: 5 |
| Osteosarcoma - localized | |||||
| 16 | 30 | Female: 6 Male: 10 | Trunk: 2 Upper limb: 3 Lower limb: 10 NA: 1 | Yes: 3 No: 12 NA: 1 | |
| Healthy controls | |||||
| 30 | 31 | Female:16 Male:14 | NA | NA | NA |
Blood sample collection
Blood samples were obtained by venipuncture using Serum Gel S/7.5 ml collection tubes (Sarstedt S-Monovette), allowed to clot for 60 min at room temperature, and then centrifuged at 1300 ×g for 10 min at 4°C. Next, 500 µl of serum was aliquoted into 1.5 ml siliconized polypropylene microtubes (Sigma-Aldrich, T4816) and stored at -80°C until further use. MiRNAs were isolated from 1200 μl of serum mirVana Paris RNA and Native Protein Purification Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The quantity of miRNA was checked using the MyQubit microRNA Assay (Thermo Fisher Scientific). The small RNA fraction was detected using the Small RNA Kit (Agilent) and a Bioanalyzer 2100. Samples that passed the quality check were kept at -80°C until further analyses.
NGS library preparation, sequencing, and data analyses
200ng miRNA was used to construct a sequencing library with an Ion Total RNA‐Seq Kit v2 for Small RNA Libraries and Ion Xpress™ RNA‐Seq Barcode Kit (Thermo Fisher Scientific), according to the manufacturer's protocol. The amount of amplified library and size distribution was assessed on the Bioanalyzer 2100 using a High Sensitivity DNA Kit; the size range for barcoded libraries of miRNA ligation products was 94‐114 bp. The template preparation procedure for clonal amplification of up to 16 miRNA libraries at a concentration of 40 pM, and loading of the Ion PI Chip v3, were performed using The Ion Chef instrument, with a set of reagents from the Ion PI™ Hi-Q™ Chef Kit (Thermo Fisher Scientific). Sequencing was achieved using the Ion Torrent Proton sequencer (Thermo Fisher Scientific) with 500 run flows.
Unmapped bam files were converted into fastq files using a bamToFastq script from bedtools [19]. Read mapping to the human genome (hg19), quantification of known miRNAs (according to miRBase release 18 [20]), and prediction of novel miRNAs was performed using miRDeep2 version 2.0.0.7 [21]. Differential expression of miRNAs was analyzed using edgeR version 3.20.6 [22], using likelihood ratio test. Post-hoc power analysis was conducted with RNASeqPower R package version 1.18.
Statistical analyses
AUC-ROC values were calculated using R in the pROC package version 1.10 [23]. Model variable selection and optimization were conducted using the R package glmulti (Vincent Calcagno (2013); glmulti: Model selection and multi-model inference made easy. R package version 1.0.7. https://CRAN.R-project.org/package=glmulti). Functional analysis was conducted using mirPath version 3 [24], using gene union. P-values were corrected for testing of multiple hypotheses with Benjamini-Hochberg method. Adjusted p-values less or equal to 0.05 were considered significant.
Availability of data
The sequencing datasets generated during the current study are available in the European Nucleotide Archive repository under the PRJEB30542 identifier.
Results
C-miRNA profiles were analyzed by deep sequencing of miRNA transcriptomes isolated from serum obtained from 16, 30, 8, and 36 patients with OS, SS, ES, and GIST, respectively; all were collected at the time of diagnosis. In addition, 30 samples from healthy controls were assessed. There was a significant difference (p-value < 0.05) in age of GIST and SS patients when compared to control group. In 5, 15, and 12 patients with OS, SS, and GIST, respectively, a second blood sample was also collected after treatment. On average, 733051 reads mapped to miRBase were obtained per library, representing 61% of total reads. Altogether, 1419 mature miRNAs were detected, of which 305 generated ten reads on average. Post-hoc power analysis revealed that at power of 0.9,confidence level of 0.05 and coverage of minimum 5x, the minimum detectable fold-change range between 1.6 (GIST group) and 2.1 (ES).
Pair-wise comparisons identified 156 c-miRNAs (adjusted P-value < 0.05) showing significant dysregulation between controls and patients, of which 24, 36, 42, and 99 differentiated controls from pretherapeutic OS, SS, ES, and GIST, respectively; log-fold changes ranged from -5.1 to 6.4, -4.0 to 6.0, -7.6 to 7.2, and -2.5 to 7.2, respectively. Interestingly, three (hsa-miR-483-5p, hsa-miR-96-5p and hsa-miR-150-3p) and seven dysregulated miRNAs (hsa-miR-4772-5p, hsa-miR-582-5p, hsa-miR-450b-5p, hsa-miR-124-3p, and hsa-miR-492, hsa-miR-486-5p and hsa-miR-375) differentiated all and at least three tumor types from healthy controls., respectively. (Figure 1). Of note, another studies [25-45] pointed out 11, 5, 21, and 14 of these c-miRNAs as dysregulated among 291, 58, 150, and 68 c-miRNAs identified in OS, SS, ES, and GIST patients, respectively (Tables S1 and S2). Functional analysis revealed that these miRNAs played roles in regulating 50 pathways, including the p53 signaling pathway (hsa-miR-124-3p, hsa-miR-96-5p, hsa-miR-582-5p, hsa-miR-450b-5p, hsa-miR-375, hsa-miR-483-5p, hsa-miR-486-5p and miR-150-3p) and the TNF signaling pathway (hsa-miR-124-3p, hsa-miR-375, hsa-miR-486-5p, hsa-miR-96-5p, hsa-miR-582-5p, hsa-miR-450b-5p, and miR-150-3p; Table S3).
The diagnostic potential of dysregulated pre-therapeutic c-miRNAs was assessed using ROC curve and AUC analyses. Five (hsa-miR-4772-5p, hsa-miR-582-5p, hsa-miR-424-5p, hsa-miR-223-3p, and hsa-miR-106b-3p), nine (hsa-miR-96-5p, hsa-miR-4746-5p, hsa-miR-424-5p, hsa-miR-323a-3p, hsa-miR-375, hsa-miR-223-3p, hsa-miR-133a, hsa-miR-3173-3p, and hsa-miR-9-5p), and four (hsa-miR-582-5p, hsa-miR-150-5p, hsa-miR-450b-5p, and hsa-miR-450a-5p) c-miRNAs in pre-therapeutic OS, ES, and GIST patients, respectively, showed moderate (good) discriminatory power (AUC-ROC values between 0.8 and 0.9). Only two (hsa-miR-142-3p and hsa-miR-9-3p) miRNAs (both in the ES group) showed high (excellent) discriminatory power (AUC-ROC values > 0.9) (Additional file 1 - Table S1). To determine whether the diagnostic ability of a miRNA signature is higher than that of a single miRNA, we also calculated AUC-ROC values for combinations of the miRNAs showing the best discriminatory power for each tumor type. Using linear models of normalized expression values and a stepwise inclusion approach, we identified four-miRNA diagnostic classifiers that differentiated controls from ES (hsa-miR-424-5p, hsa-miR-3173-3p, hsa-miR-142-3p, and hsa-miR-4746-5p), GIST (hsa-miR-151b, hsa-miR-31-5p, hsa-miR-345-5p, and hsa-miR-486-5p), OS (hsa-miR-133a, hsa-miR-223-3p, hsa-miR-450b-5p, and hsa-miR-548q), and SS (hsa-miR-3613-3p, hsa-miR-450b-5p, hsa-miR-486-5p, and hsa-miR-532-5p) with high discriminatory power (AUC-ROC = 1, 0.97, 0.95, and 0.94, respectively).
Several c-miRNAs differentiated pre-therapeutic patients with particular types of tumor (Table 2 and Table S4). The greatest number of miRNAs differentiated GIST from ES (164 c-miRNAs) and GIST from OS (103 c-miRNAs); other pair-wise comparisons identified smaller numbers of tumor-differentiating c-miRNAs.
Venn diagram with c-miRNA numbers differentiating sarcoma groups from healthy controls. GIST; gastrointestinal stromal tumor.
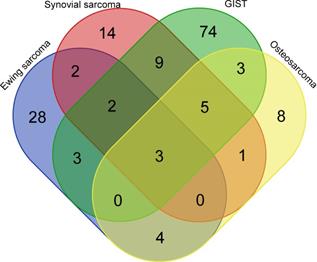
Number of c-miRNAs that differentiate between given types of sarcoma
| GIST vs. OS | GIST vs. SS | GIST vs. ES | ES vs. OS | ES vs. SS | SS vs. OS |
|---|---|---|---|---|---|
| 103 | 28 | 164 | 13 | 23 | 19 |
GIST; gastrointestinal stromal tumor, OS; osteosarcoma, SS; synovial sarcoma, ES; Ewing sarcoma
In five patients with OS, post-therapeutic c-miRNA profiles identified one c-miRNA (hsa-miR-1307-5p, AUC-ROC = 0.83) whose expression differed significantly between patients and healthy controls, and eight c-miRNAs (hsa-miR-24-3p, hsa-miR-223-3p, hsa-miR-487a, hsa-miR-1307-5p, hsa-miR-5583-5p, hsa-miR-33b-5p, hsa-miR-548f, and hsa-miR-124-3p) whose expression differed between pre- and post-therapeutic patients. Of these, the first three on the list showed high discriminatory power (AUC-ROC = 0.97, 0.96, and 0.93, respectively). In 15 patients with SS, 38 c-miRNAs differentiated post-therapeutic patients from healthy controls, and 18 c-miRNAs were differentially expressed between pre-therapeutic and post-therapeutic samples; none of these dysregulated miRNAs showed high discriminatory power (Table S5). Eleven miRNAs (hsa-miR-376b, hsa-miR-134, hsa-miR-376a-3p, hsa-miR-433, hsa-miR-409-3p, hsa-miR-34a-5p, hsa-miR-136-3p, hsa-miR-31-5p, hsa-miR-136-5p, hsa-miR-329, and hsa-miR-127-3p) were identified during both comparisons.
Similar comparisons in GIST patients identified 60 c-miRNAs that differentiated 12 post-therapeutic patients from healthy controls; of these, three (hsa-miR-30c-5p, hsa-miR-125a-5p, and hsa-miR-194-5p) showed high discriminatory power (AUC-ROC = 0.92-0.9). Twenty-two dysregulated c-miRNAs distinguished pre- from post-therapeutic samples (Table S6). Of these, 14 (hsa-miR-485-3p, hsa-miR-194-5p, hsa-miR-144-3p, hsa-miR-543, hsa-miR-125b-5p, hsa-miR-223-3p, hsa-miR-142-5p, hsa-miR-139-5p, hsa-miR-24-2-5p, hsa-let-7d-3p, hsa-miR-125a-5p, hsa-miR-16-5p, hsa-miR-483-3p, and hsa-miR-641) were common to both comparisons. Among GIST patients, 14 showed disease progression under imatinib treatment. In these patients, pre-therapeutic profiles identified 23 c-miRNAs that were dysregulated when compared with healthy controls; however, 52 dysregulated c-miRNAs were identified in non-progressing patients when compared with controls, and 15 c-miRNAs were shared by both subgroups of GIST patients (Table S6). However, none of the differentially expressed c-miRNAs was identified when the pre-treatment miRNA profiles of patients with further observed progression vs. no observed progression under the imatinib treatment were compared.
Discussion
Expression of miRNA is regulated tightly at the translational level. DNA methylation, histone deacetylation, changes in DNA copy number, and gene mutations affect proteins involved in miRNA processing and maturation. In turn, miRNAs regulate cellular proliferation, differentiation, apoptosis, and metabolism. Therefore, dysregulated miRNA expression may play roles in the pathogenesis of numerous human disorders, including cancer development and progression. Indeed, miRNAs act as functional oncogenes or tumor suppressors; increased expression of oncogenic miRNAs and decreased expression of tumor suppressive miRNAs may contribute to tumorigenesis by promoting cellular proliferation and evasion of apoptosis [46,47]. Extracellular miRNAs are highly stable and easily detected in various biofluids, including whole blood, plasma, and serum [25,47]. Although circulating miRNAs are considered to be non-invasive biomarkers for cancer diagnosis and prognosis, the association between circulating miRNA profiles and those in tumor tissues remains unclear [26,48]. Many studies conducted to date have yielded inconsistent results. Thus, the clinical application of miRNA profiling for early detection of cancer requires further study.
Sarcomas are a group of rare heterogeneous tumors that metastasize predominantly via the blood; this makes them particularly attractive to research involving peripheral blood testing. However, assessment of the prognosis of the vast majority of sarcomas is based only on very simple clinical and pathological factors (e.g., tumor size, grade, and localization), which are clearly inadequate in terms of predicting the prognosis of different sarcoma subtypes [1,4,49].
Several studies have examined c-miRNAs in OS [50], but few studies have been conducted in other sarcoma subtypes. Here, we used next generation sequencing to compare c-miRNA profiles of four sarcoma subtypes: OS, SS, ES, and GIST. We found that most c-miRNAs dysregulated in pre-therapeutic serum samples were sarcoma type-specific; only three were found in all sarcoma types.
To date, miRNA profiling studies have detected between tens and hundreds of dysregulated c-miRNAs in sarcomas (Supplementary Table 2); many of these were identified using high-throughput technologies [25-45]. However, we found limited overlap among these findings. Here, we identified only a few c-miRNAs that have been reported by others. We found that 12, 4, 5, and 0 miRNAs had AUC-ROC values > 0.9 in pre-therapeutic samples from EE, GIST, OS, and SS patients, respectively; however, multi-miRNA diagnostic classifiers resulted in higher AUCs for these tumors (1, 0.97, 0.95, and 0.94, respectively).
OS is the most common primary malignant bone tumor. Studies suggest that this tumor is associated with increased levels of circulating miRNAs (including 14q32 miRNAs, miR-95-3p, miR-300, miR-101, miR-542-3p, miR-196a, miR-196b, and miR-491) and with decreased levels of miR-17, miR-497, miR-106a-5p, miR-16-5p, miR-20a-5p, miR-425-5p, miR-451a, miR-25-3p, and miR-139-5p [26-31,47,51,52]. Of these, serum miR-199a-5p concentrations are significantly higher in OS patients than in controls, and are significantly lower in post-operative samples than in preoperative samples [53]. Most aberrantly expressed c-miRNAs discriminated OS patients from healthy volunteers with high specificity and sensitivity; therefore, they were considered promising diagnostic and prognostic biomarkers for patients with advanced tumors [53]. MiR-140 is the first miRNA candidate biomarker associated with drug sensitivity of OS xenografts treated with doxorubicin, cisplatin, and ifosfamide [28].
In addition, TaqMan Low Density Arrays identified 15 miRNAs that are differentially expressed in OS patients compared with healthy controls; of these, miR-215-5p and miR-642a-5p are potential markers for OS diagnosis [32]. Genome-wide profiling identified 56 and 164 c-miRNAs that were upregulated and downregulated, respectively, in OS samples. Of these, expression of three miRNAs (miR-21, miR-221, and miR-106a) was particularly high in patient plasma [25]. In another study, global profiling of c-miRNAs identified 236 serum miRNAs that were expressed at higher levels in OS patients than in controls [33]. The highly dysregulated miR-25-3p correlates with patient prognosis, and might serve as a non-invasive blood-based biomarker for tumor monitoring and prognostic prediction. Correlation between miR-25 expression and bone metabolism was reported previously, with intracellular and extracellular oncogenic functions imputed to miR-25-3p [32,54-56]. Another study conducted a meta-analysis to assess the true diagnostic value of circulating miRNAs for early detection of OS [57]. The authors concluded that c-miRNAs show great promise for the diagnosis of osteosarcoma in Asian populations, particularly when multiple miRNAs are combined to improve diagnostic accuracy. However, the clinical application of miRNA profiling for early detection of OS requires further study.
ESs are highly aggressive sarcomas characterized by oncogenic chromosomal translocations that lead to formation of fusion genes, mainly the EWS gene. Alterations in expression of miRNA in ES involve both EWS/Ets oncogenic fusion-dependent and -independent mechanisms, which contribute to a malignant phenotype. However, some miRNAs with prognostic and therapeutic potential have been identified, including miR-17,92a, miR-106b,25, and miR-106a,363 clusters [34-36,58]. It is hypothesized that miR-125b induces chemoresistance in ES/primitive neuroectodermal tumors [59], and that miR-30a-5p interacts with the 3'UTR region of CD99 to regulate its expression [37]. Serum miR-125b, one of the most consistently dysregulated miRNAs, may serve as a useful non-invasive biomarker for ES [60]; indeed, its expression in ES tumors is lower than that in normal bone tissues [38].
SS shows recurrent alterations in miRNA expression, including that of the miR-143 [39] and miR-183 clusters [40]. In addition, the miR-17-92 cluster (C13orf25) is upregulated in liposarcoma (the most common STS) [41]. Members of the miR-200 family and miR-9/miR-9* are associated with SS and myxoid liposarcomas, respectively. Microarray profiling analysis performed on nine pairs of serum samples obtained from SS patients and healthy individuals identified 49 serum miRNAs that were significantly upregulated in SS patients compared with controls, reaffirming the potential clinical significance of serum miR-92b-3p for identifying SS tumors [42].
GISTs are the most common abdominal sarcoma. They show well-characterized molecular features related to activating mutations in theKIT or PDGFRA genes. Overexpression of miR-196a is associated with a high-risk grade, metastasis, and poor survival [61]; this may allow classification of GIST into various prognostic subtypes [62]. Downregulation of miR-221 and miR-222 is implicated in the pathogenesis of GISTs, and may be involved in secondary resistance to tyrosine kinase inhibitors [43,63]. Expression of miR-221 and miR-222 is significantly lower in KIT-positive GIST cases, and miR-494 correlates inversely with KIT expression [64,65]. Others show that miR-200a-3p correlates negatively with expression of PDGFRA [66]. Downregulation of miR-320a correlates with shorter time to imatinib resistance in patients with GIST, indicating an important role in the mechanism underlying imatinib resistance in these patients [67]. Three other papers show that miRNAs, including miR-218, miR-125a-5p, and miR-518a-5p, play roles regulating sensitivity to imatinib [44,45,68].
The results of the above studies show little overlap: only four miRNAs (hsa-miR-214, hsa-miR-143, hsa-miR-15b, and family hsa-miR-150) are dysregulated in all four tumor types. However, despite the relatively small numbers of samples, particularly in the ES group, we identified ten miRNAs that were dysregulated consistently in at least three tumor types when compared with healthy controls. Eight of these (hsa-miR-124-3p, hsa-miR-96-5p, hsa-miR-582-5p, hsa-miR-450b-5p, hsa-miR-375, hsa-miR-483-5p, hsa-miR-486-5p and miR-150-3p) regulate genes involved in the p53 signaling pathway. A limitation of our study could be a disparity between the ages of GIST and SS patients when compared to control group. The control group was recruited among healthy hospital personnel where young and middle aged individuals dominate. Several studies suggested that the abundance of serum miRNAs can change with age in healthy individuals [69-71], however none of the miRNAs proposed is these studies (Supplementary Table S7) as aging biomarkers differed control from any sarcoma groups.
Conclusions
We identified aberrant expression of miRNA signatures in serum samples from patients with four different sarcoma subtypes. Differences in miRNA expression profiles between sarcoma patients and healthy volunteers suggest that miRNAs play key roles in sarcoma development. Identification of miRNA patterns unique to individual tumor types may yield new diagnostic biomarkers; as such, miRNAs warrant further investigation as therapeutic targets for sarcoma.
Supplementary Material
Supplementary figures and tables.
Abbreviations
c-miRNA: circulating microRNA; STS: soft tissue sarcoma; GIST: gastrointestinal stromal tumor; OS: osteosarcoma; SS: synovial sarcoma; ES: Ewing sarcoma; CT: Computed tomography; ROC-AUC: Receiver operating characteristic curves and area under the curve.
Acknowledgements
This study was supported by the Polish National Science Center (scientific grant 2013/11/B/NZ5/ 03165 [to PR]).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Casali PG, Abecassis N, Bauer S, Biagini R, Bielack S, Bonvalot S, Boukovinas I, Bovee JVMG, Brodowicz T, Broto JM, Buonadonna A, De Álava E, Dei Tos AP. et al. Soft tissue and visceral sarcomas: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv51-67 doi: 10.1093/annonc/mdy096
2. Casali PG, Bielack S, Abecassis N, Aro HT, Bauer S, Biagini R, Bonvalot S, Boukovinas I, Bovee JVMG, Brennan B, Brodowicz T, Broto JM, Brugières L. et al. Bone sarcomas: ESMO-PaedCan-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv79-95 doi: 10.1093/annonc/mdy310
3. Dangoor A, Seddon B, Gerrand C, Grimer R, Whelan J, Judson I. UK guidelines for the management of soft tissue sarcomas. Clin Sarcoma Res. 2016;6:20. doi: 10.1186/s13569-016-0060-4
4. Casali PG, Abecassis N, Aro HT, Bauer S, Biagini R, Bielack S, Bonvalot S, Boukovinas I, Bovee JVMG, Brodowicz T, Broto JM, Buonadonna A, De Álava E. et al. Gastrointestinal stromal tumours: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv267. doi: 10.1093/annonc/mdy320
5. Zen K, Zhang C-Y. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326-48 doi: 10.1002/med.20215
6. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CSR, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672-5 doi: 10.1111/j.1365-2141.2008.07077.x
7. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-8 doi: 10.1073/pnas.0804549105
8. Mo M-H, Chen L, Fu Y, Wang W, Fu SW. Cell-free Circulating miRNA Biomarkers in Cancer. J Cancer. 2012;3:432-48 doi: 10.7150/jca.4919
9. Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174-9 doi: 10.1038/sj.bjc.6605608
10. Moussay E, Wang K, Cho J-H, van Moer K, Pierson S, Paggetti J, Nazarov PV, Palissot V, Hood LE, Berchem G, Galas DJ. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2011;108:6573-8 doi: 10.1073/pnas.1019557108
11. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu C-G, Negrini M, Cavazzini L, Volinia S, Alder H. et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-86 doi: 10.1016/j.ccr.2008.02.013
12. Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55-9 doi: 10.1016/j.ygyno.2008.08.036
13. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118-26 doi: 10.1002/ijc.25007
14. Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-42 doi: 10.1002/mc.20712
15. Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE. 2011;6:e17745. doi: 10.1371/journal.pone.0017745
16. Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2:807-13 doi: 10.1158/1940-6207.CAPR-09-0094
17. Pu X, Huang G, Guo H, Guo C, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin T. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674-80 doi: 10.1111/j.1440-1746.2010.06417.x
18. Wach S, Nolte E, Szczyrba J, Stöhr R, Hartmann A, Ørntoft T, Dyrskjøt L, Eltze E, Wieland W, Keck B, Ekici AB, Grässer F, Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130:611-21 doi: 10.1002/ijc.26064
19. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841-2 doi: 10.1093/bioinformatics/btq033
20. Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-157 doi: 10.1093/nar/gkq1027
21. Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37-52 doi: 10.1093/nar/gkr688
22. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40 doi: 10.1093/bioinformatics/btp616
23. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77
24. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460-466 doi: 10.1093/nar/gkv403
25. Nakka M, Allen-Rhoades W, Li Y, Kelly AJ, Shen J, Taylor AM, Barkauskas DA, Yustein JT, Andrulis IL, Wunder JS, Gorlick R, Meltzer PS, Lau CC. et al. Biomarker significance of plasma and tumor miR-21, miR-221, and miR-106a in osteosarcoma. Oncotarget. 2017;8:96738-52 doi: 10.18632/oncotarget.18236
26. Li S, Gao Y, Wang Y, Wang K, Dai Z-P, Xu D, Liu W, Li Z-L, Zhang Z-D, Yang S-H, Yang C. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol Lett. 2016;12:4905-10 doi: 10.3892/ol.2016.5362
27. Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, Volinia S, Stein GS, Croce CM. et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865-77 doi: 10.1158/0008-5472.CAN-11-2663
28. Kobayashi E, Hornicek FJ, Duan Z. MicroRNA Involvement in Osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739
29. Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, Steer CJ, Modiano JF, Subramanian S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171-81 doi: 10.1016/j.bone.2011.10.012
30. Li H, Zhang K, Liu L-H, Ouyang Y, Guo H-B, Zhang H, Bu J, Xiao T. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol Lett. 2015;10:1662-8 doi: 10.3892/ol.2015.3378
31. Wang S-N, Luo S, Liu C, Piao Z, Gou W, Wang Y, Guan W, Li Q, Zou H, Yang Z-Z, Wang D, Wang Y, Xu M. et al. miR-491 Inhibits Osteosarcoma Lung Metastasis and Chemoresistance by Targeting αB-crystallin. Mol Ther. 2017;25:2140-9 doi: 10.1016/j.ymthe.2017.05.018
32. Monterde-Cruz L, Ramírez-Salazar EG, Rico-Martínez G, Linares-González LM, Guzmán-González R, Delgado-Cedillo E, Estrada-Villaseñor E, Valdés-Flores M, Velázquez-Cruz R, Hidalgo-Bravo A. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum Cell. 2018;31:292-9 doi: 10.1007/s13577-018-0214-1
33. Fujiwara T, Uotani K, Yoshida A, Morita T, Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, Komatsubara T, Takeda K, Kunisada T. et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8:33375-92 doi: 10.18632/oncotarget.16498
34. Dylla L, Jedlicka P. Growth-Promoting Role of the miR-106a∼363 Cluster in Ewing Sarcoma. PLoS ONE. 2013;8:e63032. doi: 10.1371/journal.pone.0063032
35. McKinsey EL, Parrish JK, Irwin AE, Niemeyer BF, Kern HB, Birks DK, Jedlicka P. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene. 2011;30:4910-20 doi: 10.1038/onc.2011.197
36. Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DNT, Schaefer K-L, Nakatani F, Scotlandi K, Reiter M, Strunk D, Speleman F, Vandesompele J. et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing's sarcoma. Oncogene. 2011;30:2173-80 doi: 10.1038/onc.2010.581
37. Franzetti G-A, Laud-Duval K, Bellanger D, Stern M-H, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2013;32:3915-21 doi: 10.1038/onc.2012.403
38. De Vito C, Riggi N, Suvà M-L, Janiszewska M, Horlbeck J, Baumer K, Provero P, Stamenkovic I. Let-7a is a direct EWS-FLI-1 target implicated in Ewing's sarcoma development. PLoS ONE. 2011;6:e23592. doi: 10.1371/journal.pone.0023592
39. Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, Corless CL, Fire AZ, van de Rijn M. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015-26 doi: 10.1038/sj.onc.1210836
40. Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570-80 doi: 10.1158/0008-5472.CAN-10-2074
41. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-69 doi: 10.1038/nrc1840
42. Uotani K, Fujiwara T, Yoshida A, Iwata S, Morita T, Kiyono M, Yokoo S, Kunisada T, Takeda K, Hasei J, Numoto K, Nezu Y, Yonemoto T. et al. Circulating MicroRNA-92b-3p as a Novel Biomarker for Monitoring of Synovial Sarcoma. Sci Rep. 2017;7:14634. doi: 10.1038/s41598-017-12660-5
43. Wang Y, Li J, Kuang D, Wang X, Zhu Y, Xu S, Chen Y, Cheng H, Zhao Q, Duan Y, Wang G. miR-148b-3p functions as a tumor suppressor in GISTs by directly targeting KIT. Cell Commun Signal. 2018;16:16. doi: 10.1186/s12964-018-0228-z
44. Akçakaya P, Caramuta S, Åhlen J, Ghaderi M, Berglund E, Östman A, Bränström R, Larsson C, Lui W-O. microRNA expression signatures of gastrointestinal stromal tumours: associations with imatinib resistance and patient outcome. Br J Cancer. 2014;111:2091-102 doi: 10.1038/bjc.2014.548
45. Shi Y, Gao X, Hu Q, Li X, Xu J, Lu S, Liu Y, Xu C, Jiang D, Lin J, Xue A, Tan Y, Shen K. et al. PIK3C2A is a gene-specific target of microRNA-518a-5p in imatinib mesylate-resistant gastrointestinal stromal tumor. Lab Invest. 2016;96:652-60 doi: 10.1038/labinvest.2015.157
46. Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag Res. 2014;6:15-25 doi: 10.2147/CMAR.S53928
47. Yao Z-S, Li C, Liang D, Jiang X-B, Tang J-J, Ye L-Q, Yuan K, Ren H, Yang Z-D, Jin D-X, Zhang S-C, Ding J-Y, Tang Y-C. et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer Biomark. 2018;22:127-33 doi: 10.3233/CBM-171103
48. Mishra PJ. MicroRNAs as promising biomarkers in cancer diagnostics. Biomark Res. 2014;2:19. doi: 10.1186/2050-7771-2-19
49. Schaefer I-M, Fletcher CDM. Recent advances in the diagnosis of soft tissue tumours. Pathology. 2018;50:37-48 doi: 10.1016/j.pathol.2017.07.007
50. Raimondi L, Luca AD, Costa V, Amodio N, Carina V, Bellavia D, Tassone P, Pagani S, Fini M, Alessandro R, Giavaresi G. Circulating biomarkers in osteosarcoma: new translational tools for diagnosis and treatment. Oncotarget. 2017;8:100831. doi: 10.18632/oncotarget.19852
51. Zhang C, Yao C, Li H, Wang G, He X. Combined elevation of microRNA-196a and microRNA-196b in sera predicts unfavorable prognosis in patients with osteosarcomas. Int J Mol Sci. 2014;15:6544-55 doi: 10.3390/ijms15046544
52. Li Q, Song S, Ni G, Li Y, Wang X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomark. 2018;21:521-6 doi: 10.3233/CBM-170255
53. Zhou G, Lu M, Chen J, Li C, Zhang J, Chen J, Shi X, Wu S. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumour Biol. 2015;36:8845-52 doi: 10.1007/s13277-015-3421-3
54. Gao J, Yang T, Han J, Yan K, Qiu X, Zhou Y, Fan Q, Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem. 2011;112:1844-56 doi: 10.1002/jcb.23106
55. Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325
56. Yoshida A, Fujiwara T, Uotani K, Morita T, Kiyono M, Yokoo S, Hasei J, Nakata E, Kunisada T, Ozaki T. Clinical and Functional Significance of Intracellular and Extracellular microRNA-25-3p in Osteosarcoma. Acta Med Okayama. 2018;72:165-74 doi: 10.18926/AMO/55857
57. Wang X, Ning Y, Yang L, Liu H, Wu C, Wang S, Guo X. Diagnostic value of circulating microRNAs for osteosarcoma in Asian populations: a meta-analysis. Clin Exp Med. 2017;17:175-83 doi: 10.1007/s10238-016-0422-5
58. Dylla L, Moore C, Jedlicka P. MicroRNAs in Ewing Sarcoma. Front Oncol. 2013;3:65. doi: 10.3389/fonc.2013.00065
59. Iida K, Fukushi J-I, Matsumoto Y, Oda Y, Takahashi Y, Fujiwara T, Fujiwara-Okada Y, Hatano M, Nabashima A, Kamura S, Iwamoto Y. miR-125b develops chemoresistance in Ewing sarcoma/primitive neuroectodermal tumor. Cancer Cell Int. 2013;13:21. doi: 10.1186/1475-2867-13-21
60. Nie CL, Ren WH, Ma Y, Xi JS, Han B. Circulating miR-125b as a biomarker of Ewing's sarcoma in Chinese children. Genet Mol Res. 2015;14:19049-56 doi: 10.4238/2015.December.29.12
61. Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T. et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126-36 doi: 10.1158/0008-5472.CAN-11-1803
62. Kupcinskas J. Small Molecules in Rare Tumors: Emerging Role of MicroRNAs in GIST. Int J Mol Sci. 2018:19 doi: 10.3390/ijms19020397
63. Koelz M, Lense J, Wrba F, Scheffler M, Dienes HP, Odenthal M. Down-regulation of miR-221 and miR-222 correlates with pronounced Kit expression in gastrointestinal stromal tumors. Int J Oncol. 2011;38:503-11 doi: 10.3892/ijo.2010.857
64. Liu X, Chu K-M. Molecular biomarkers for prognosis of gastrointestinal stromal tumor. Clin Transl Oncol. 2019;21:145-51 doi: 10.1007/s12094-018-1914-4
65. Kim WK, Park M, Kim Y-K, Tae YK, Yang H-K, Lee JM, Kim H. MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res. 2011;17:7584-94 doi: 10.1158/1078-0432.CCR-11-0166
66. Gyvyte U, Juzenas S, Salteniene V, Kupcinskas J, Poskiene L, Kucinskas L, Jarmalaite S, Stuopelyte K, Steponaitiene R, Hemmrich-Stanisak G, Hübenthal M, Link A, Franke S. et al. MiRNA profiling of gastrointestinal stromal tumors by next-generation sequencing. Oncotarget. 2017;8:37225-38 doi: 10.18632/oncotarget.16664
67. Gao X, Shen K, Wang C, Ling J, Wang H, Fang Y, Shi Y, Hou Y, Qin J, Sun Y, Qin X. MiR-320a downregulation is associated with imatinib resistance in gastrointestinal stromal tumors. Acta Biochim Biophys Sin (Shanghai). 2014;46:72-5 doi: 10.1093/abbs/gmt118
68. Fan R, Zhong J, Zheng S, Wang Z, Xu Y, Li S, Zhou J, Yuan F. microRNA-218 increase the sensitivity of gastrointestinal stromal tumor to imatinib through PI3K/AKT pathway. Clin Exp Med. 2015;15:137-44 doi: 10.1007/s10238-014-0280-y
69. Noren Hooten N, Fitzpatrick M, Wood WH, De S, Ejiogu N, Zhang Y, Mattison JA, Becker KG, Zonderman AB, Evans MK. Age-related changes in microRNA levels in serum. Aging (Albany NY). 2013;5:725-40 doi: 10.18632/aging.100603
70. Smith-Vikos T, Liu Z, Parsons C, Gorospe M, Ferrucci L, Gill TM, Slack FJ. A serum miRNA profile of human longevity: findings from the Baltimore Longitudinal Study of Aging (BLSA). Aging (Albany NY). 2016;8:2971-87 doi: 10.18632/aging.101106
71. Zhang H, Yang H, Zhang C, Jing Y, Wang C, Liu C, Zhang R, Wang J, Zhang J, Zen K, Zhang C, Li D. Investigation of microRNA expression in human serum during the aging process. J Gerontol A Biol Sci Med Sci. 2015;70:102-9 doi: 10.1093/gerona/glu145
Author contact
![]() Corresponding authors: Jerzy Ostrowski, MD, PhD; Maria Sklodowska-Curie Institute - Oncology Centre, Roentgena 5, 02-781 Warsaw, Poland, Tel: +48 225462575, Fax: +48 225462449, E-mail: jostrowcom.pl OR Piotr Rutkowski, MD, PhD; Maria Sklodowska-Curie Institute - Oncology Centre, Roentgena 5, 02-781 Warsaw, Poland, Tel: +48 22 6439375, Fax: +48 22 6439791, E-mail: piotr.rutkowskipl
Corresponding authors: Jerzy Ostrowski, MD, PhD; Maria Sklodowska-Curie Institute - Oncology Centre, Roentgena 5, 02-781 Warsaw, Poland, Tel: +48 225462575, Fax: +48 225462449, E-mail: jostrowcom.pl OR Piotr Rutkowski, MD, PhD; Maria Sklodowska-Curie Institute - Oncology Centre, Roentgena 5, 02-781 Warsaw, Poland, Tel: +48 22 6439375, Fax: +48 22 6439791, E-mail: piotr.rutkowskipl

 Global reach, higher impact
Global reach, higher impact