Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(6):1315-1324. doi:10.7150/jca.39044 This issue Cite
Review
MicroRNA-183 in Cancer Progression
1. College of Animal Sciences, Zhejiang University, Hangzhou, 310058, P. R. China.
2. Sir Run Shaw Hospital, Zhejiang University College of Medicine, Hangzhou, 310058, P. R. China.
Received 2019-8-5; Accepted 2019-11-16; Published 2020-1-1
Abstract
MicroRNA-183(miR-183) is abnormally expressed in many kinds of tumors. It participates in the initiation and development of tumors. There are many pathways regulate the expression of miR-183. The action mechanism of miR-183 in cancer is very extensive, and contradictory conclusions are often drawn. It was upregulated in 18 kinds of cancer, downregulated in 6 kinds of cancer. In addition, there are seven types of cancer, both upregulated and downregulated reports can be found. Evidence showed that miR-183 can not only directly play the role of oncogene or antioncogene, but also regulate the expression of other oncogene or antioncogene in different cancer types. In this review, we discuss the regulator of miR-183 and summarized the expression of miR-183 in different cancers. We also counted the target genes of miR-183 and the functional roles they play. Furthermore, we focused on the roles of miR-183 in cell migration, cell invasion, epithelial-mesenchymal transition (EMT) and microangiogenesis, which play the most important roles in cancer processes. It sheds light on the likely reasons why miR-183 plays different roles in various cancers. In addition, miR-183 and its downstream effectors have the potential to be promising prognostic markers and therapeutic targets in cancer.
Keywords: microRNA-183, cancer progression, EMT, metastasis, microangiogenesis
Introduction
Mature microRNA (miRNA) is a kind of small noncoding single stranded RNA with 18 to 24 nucleotides [1]. The latest release of miRBase (v22) contains miRNA sequences from 271 organisms: 38 589 hairpin precursors and 48 860 mature miRNAs. Human genome contains 1917 annotated hairpin precursors, and 2654 mature sequences [2]. MiRNAs regulate the expression of target genes by completely or incompletely complementing the 3'UTR untranslated region of target miRNAs. A single miRNA can be associated with several or even hundreds of target genes. A gene is regulated by multiple miRNAs at the same time [3]. MiRNAs are ubiquitous in eukaryotic cells and highly conserved among different species. The expression of miRNAs has obvious sequence specificity and tissue specificity [4].
MiR-183 is a member of the microRNA-183-96-182 cluster located at the 7q31-34 locus of human chromosome [5]. It's highly conserved in different organisms. The expression of miR-183 is tissue-specific and highly enriched in visual and sensory organs. It participates in the development of retina and cochlea and plays an important role in maintaining normal function [6].
In recent years, many studies have shown that miR-183 is abnormally expressed in many kinds of tumors, and participates in the initiation and development of tumors. However, in different cancer tissues and cell lines, there are great differences in the expression of miR-183. Even in the same type of cancer, opposite expression results are often reported. While in some studies, the expression of miR-183 was inconsistent in different samples of the same pathological tissue. Therefore, it is urgent to summarize the expression of miR-183 in different types of cancer and function targeted by miR-183. All previous published articles reviewed the three members of miR-183 family together, but the three members of miR-183 family, miR-183, miR-182 and miR-96, differ greatly both in upstream regulators and target genes. It is easy to conceal the function of miR-183 when review the family together. This article is focusing only on miR-183 in cancer progression. The regulatory ways of miR-183 from upstream to downstream regulation are systematically and comprehensively elaborated. In this review, we summarized the expression of miR-183 in different cancers, counted the target genes of miR-183, and focused on the role of miR-183 in cell migration, cell invasion, EMT and microangiogenesis, which play the most important roles in cancer processes. The latest research of miR-183 in lung cancer, ovarian cancer, bladder cancer and prostate cancer has also been fully reflected in this paper. This review provides a basis for elucidating the role of miR-183 in cancer progression.
Regulation of miR-183 expression
The expression of miR-183 varies greatly among different types of cancer. So it is necessary to summarize the upstream regulatory factors of miR-183 in order to clarify the different up-and-down regulation phenomena of miR-183 in different pathological tissues. The upstream regulation mechanism of miR-183 cluster has been reported frequently, but only a part of the factors regulate the expression of miR-183 directly.
The reported literature shows that there are eight regulatory factors that can upregulate miR-183 expression level. They can promote the transcriptional expression of miR-183 by binding to the upstream promoter region. The regulation of Wnt/beta-catenin (CTNNB1) signaling pathway on miR-183 is the most studied. Activation of CTNNB1 can upregulate the expression of miR-183, knockdown of CTNNB1 also showed concordant downregulations of miR-183, suggesting that miR-183 may be an important downstream target gene in the regulation of Wnt signaling pathway [7]. Other studies have shown that expression of GSK3β can inhibit the expression of miR-183 through the β-Catenin/TCF/LEF-1 pathway in human gastric cancer cells, while β-Catenin/TCF/LEF-1 binds to the promoter of miR-183 and thereby activates the transcription of miR-183 [8]. While in non-small cell lung cancer, they found that TFAP2C blocked AKAP12-mediated cyclin D1 inhibition by inducing the overexpression of miR-183, which shows that TFAP2C is one of the upstream regulators of miR-183 [9]. In breast cancer, the transcription of miR-183 was regulated by ZEB1 and HSF2, HSF2 can upregulate miR-183 expression [10]. While the regulatory effect of ZEB1 on miR-183 is more complex. ZEB1 can downregulate the transcriptional expression of miR-183, conversely, ZEB1 can be targeted by miR-183. In addition, there are other regulatory factors can downregulate the transcriptional expression of miR-183 by binding to the upstream promoter region of miR-183. Studies in neuroblastoma show that HDAC(s) involved in miR-183 transcriptional and downregulated it. MYCN was found to recruit HDAC2 in the same complexes to the miR-183 promoter [11]. What's more, inhibition of PI3K/AKT signaling pathway also significantly reduced the expression of miR-183 in esophageal squamous cell carcinoma (ESCC) cells [12].
IncRNA plays an important role in regulating the expression of miRNA. IncRNA MALAT1 function is a sponge competitive endogenous RNA for miR-183 that inhibits the expression and function of miR-183 [13]. The above literature indicates the complexity of the regulation mechanism of miR-183 expression in different cells. We summarize the upstream regulating direction of miR-183 and the binding site location. Details are shown in Table 1 and Fig. 1.
Binding site location of transcriptional regulators. (A) Research in human genome: binding sites of transcriptional factor in upstream of miR-183. (B) Research in mouse genome: binding sites of transcriptional factor in upstream of miR-183.
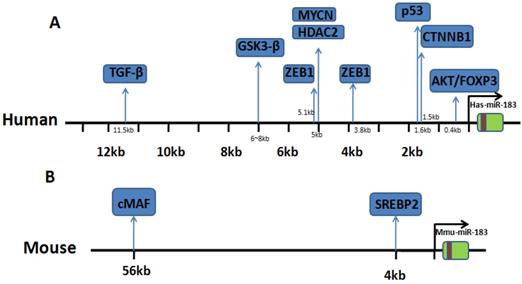
Upstream regulator of miR-183 expression.
| Upstream regulator | Model system | Direction of regulation | Source |
|---|---|---|---|
| IncRNA MALAT1 | Melanoma | ↓ | Sun Y, et al.(2017) [13] |
| ZEB1 | breast cancer | ↓ | Langer EM, et al.(2018) [50] |
| Li P, et al.(2014) [10] | |||
| GSK3β | gastric cancer | ↓ | Tang X, et al.(2013) [8] |
| MYCN, HDAC2 | Neuroblastoma | ↓ | Lodrini M, et al.(2013) [11] |
| CTNNB1 | hepatocellular carcinoma | ↑ | Leung WK, et al.(2015) [7] |
| HSF2 | breast cancer | ↑ | Li P, et al.(2014) [10] |
| TGF-β | lung tumor | ↑ | Trinh TL, et al.(2019) [52] |
| AKT/FOXP3 | leukemia U937 cells | ↑ | Liu W, et al.(2012) [53] |
| TFAP2C | non-small cell lung cancer | ↑ | Kang J, et al.(2017) [54] |
| SREBP2 | Murine liver | ↑ | Jeon T, et al.(2013) [55] |
| cMAF | Murine liver | ↑ | Kelada S, et al.(2013) [56] |
| p53 | Primary human mammary epithelial cells | ↑ | Chang C, et al.(2011) [57] |
Dysregulation of miR-183 in different cancer
Upstream of miR-183 is regulated by a variety of regulatory factors. In different cancers, different transcription factors play a major role and expression of miR-183 varies greatly (Table 2). Results showed that miR-183 was upregulated in 18 kinds of cancer including prostate cancer, ovarian cancer, breast cancer, et al, down regulated in 6 kinds of cancer including osteosarcoma, melanoma, et al. In these cancer cases, miR-183 upregulated and plays an oncogene role while plays as an antioncogene when downregulated. But in 7 kinds of cancer like hepatocellular cancer (HCC), endometrial cancer, some research reported that miR-183 was upregulated in tissues or cell lines, while others reported that miR-183 was downregulated (Table 2). MiR-183 can play a dual role in promoting or inhibiting those cancers. In HCC, Li et al. found that among the 25 samples analyzed, miR-183 was significantly upregulated (twofold to 367-fold) in 17 samples compared with the matching nontumoral liver tissues [14]. Studies in breast cancer have shown similar results [33]. This indicates that miR-183 expression has strong tissue specificity and individual differences. The wide variability in the reported results may be attributable to the differences between blood and tissue or the heterogeneity in cancer cells.
MiR-183 in cancer progression
MiR-183 can play the role of both oncogene and antioncogene through act on different genes among all kinds of cancers. We statistical analyze target genes of miR-183, which have been identified. These target genes are mainly function in EMT, migration, invasion and microangiogenesis. These aspects play a key role in the process of cancer, which directly reflects that miR-183 is a key role in the process of cancer.
Up and down regulation of miR-183 expression in different cancer in published articles.
| Cancer type | Regulated features | Source | |
|---|---|---|---|
| Hepatocellular carcinoma | ↕ | upregulated | Bharali D, et al.(2018) [58] |
| Li ZB, et al.(2015) [59] | |||
| Leung WK, et al.(2015) [7] | |||
| Li J, et al.(2010) [14] | |||
| downregulated | Bian W, et al.(2018) [60] | ||
| Endometrial cancer | ↕ | upregulated | Ruan H, et al.(2017) [61] |
| Xiong H, et al.(2018) [23] | |||
| downregulated | Yan H, et al.(2018) [31] | ||
| Non-small cell lung cancer | ↕ | upregulated | Wang H, et al.(2019) [41] |
| Zhang L, et al.(2015) [62] | |||
| downregulated | Yang C, et al.(2018) [63] | ||
| Yang X, et al.(2018) [64] | |||
| Gastric cancer | ↕ | upregulated | Li C, et al.(2016) [35] |
| Gu W, et al.(2014) [32] | |||
| downregulated | Cao LL, et al.(2014) [65] | ||
| Xu L, et al.(2014) [66] | |||
| Cervical cancer | ↕ | upregulated | Liu SS, et al.(2018) [67] |
| downregulated | Zhang W, et al.(2018) [44] | ||
| Fan D, et al.(2016) [68] | |||
| Lung cancer | ↕ | upregulated | Trinh TL, et al.(2019) [52] |
| downregulated | Meng F, et al.(2019) [69] | ||
| Pancreatic adenocarcinoma | ↕ | upregulated | Miao F, et al.(2016) [70] |
| downregulated | Zhou L, et al.(2014) [71] | ||
| Prostate cancer | ↑ | upregulated | Waseem M, et al.(2019) [72] |
| Ueno K, et al.(2013) [73] | |||
| Ovarian cancer | ↑ | upregulated | Zhou J, et al.(2019) [74] |
| Esophageal squamous cell carcinoma | ↑ | upregulated | Yang M, et al.(2014) [36] |
| Ren L, et al.(2014) [12] | |||
| Mesothelioma | ↑ | upregulated | Suzuki R, et al.(2018) [75] |
| Bladder cancer | ↑ | upregulated | Chen D, et al.(2018) [76] |
| Renal cell carcinoma | ↑ | upregulated | Zhang XL, et al.(2018) [77] |
| Qiu M, et al.(2014) [78] | |||
| Colorectal cancer | ↑ | upregulated | Chen Y, et al.(2018) [79] |
| Yuan D, et al.(2015) [80] | |||
| Tongue carcinoma | ↑ | upregulated | Supic G, et al.(2018) [81] |
| Tongue squamous cell carcinoma | ↑ | upregulated | Yan D, et al.(2016) [82] |
| Pediatric caute myeloid leukemia | ↑ | upregulated | Wang X, et al.(2017) [9] |
| Breast cancer | ↑ | upregulated | Cheng Y, et al.(2016) [37] |
| Li P, et al.(2014) [10] | |||
| Brain glioma | ↑ | upregulated | Ye Z, et al.(2016) [83] |
| Wang Z, et al.(2016) [84] | |||
| Tanaka H, et al.(2013) [109] | |||
| Epithelial ovarian cancer | ↑ | upregulated | Chen H, et al.(2016) [85] |
| Vulvar squamous cell carcinoma | ↑ | upregulated | Yang X, et al.(2016) [86] |
| Papillary thyroid carcinoma | ↑ | upregulated | Wei C, et al.(2015) [38] |
| Synovial sarcoma | ↑ | upregulated | Sarver AL, et al.(2010) [39] |
| Rhabdomyosarcoma (RMS) | ↑ | upregulated | Sarver AL, et al. (2010) [39] |
| Colon cancer | ↑ | upregulated | Sarver AL, et al. (2010) [39] |
| Osteosarcoma | ↓ | downregulated | Sun X, et al.(2018) [24] |
| Zhang J, et al.(2014) [30] | |||
| Nasopharyngeal carcinoma | ↓ | downregulated | Wang G, et al.(2017) [25] |
| Cheung CC, et al.(2016) [87] | |||
| Melanoma | ↓ | downregulated | Sun Y, et al.(2017) [13] |
| Retinoblastoma | ↓ | downregulated | Wang J, et al.(2014) [88] |
| Kidney renal clear cell carcinoma | ↓ | downregulated | Yuan J, et al.(2019) [89] |
| Infected abdominal aortic aneurysm | ↓ | downregulated | Meng C, et al.(2018) [90] |
Roles of miR-183 in EMT
The malignant progression of tumors is mainly manifested in the enhancement of the ability of migration and invasion. While the first step for cell invasion and migration is to change from epithelioid to mesenchymal [15]. EMT refers to the change of epithelial cells, which lose polarity, tight junction and adhesion between cells, acquire the ability of infiltration and migration, and become mesenchymal cells with morphology and characteristics [16]. In recent years, the research of EMT has turned to miRNAs. Results show that miR-183 is involved in many aspects of EMT, including apoptosis, tissue repair, extracellular matrix remodeling, and metastasis of tumors [17].
In malignant epithelial cells, ZEB1 acts as an activator of EMT, triggering the phenotype transformation of epithelial cells to mesenchymal cells [18]. Studies found that miR-183 was able to target ZEB1 [19]. MiR-183 represses EMT through the regulating of ZEB1, overexpression of miR-183 results in the change of cells from mesenchymal to epithelial. E-cadherin is an EMT associated marker. ZEB1 can bind with E-box in E-cadherin promoter to inhibit E-cadherin expression [20]. Several clinical studies have shown that increasing the expression of E-cadherin can significantly improve life quality and prolong the survival time of cancer patients with several types of tumors [21]. Overexpression of miR-183 can downregulate E-cadherin expression and increase N-cadherin expression [22, 23].
Metastasis-associated 1 (MTA1) protein is a prime inducer of EMT and metastatic progression in all solid tumors. MTA1 protein and RNA expression showed opposite trends to miR-183 expression in breast, renal, prostate, and testicular cancer tissues. Sun et al. found that in osteosarcoma tissues and MG63 cells, miR-183 inhibits EMT and apoptosis by targeting MTA1 [24]. In nasopharyngeal carcinoma spheroids, ectopic expression of miR-183 markedly suppressed EMT and strikingly enhanced DDP-induced apoptosis, whereas MTA1 overexpression partially reversed these effects [25]. In human NSCLC cells, miR-183 downregulates MTA1 to inhibit the proliferation and EMT. In pancreatic cancer, Lin et al. found that overexpression miR-183 decreased proliferation and EMT, whereas increased apoptosis by targeting MTA1 [26].
Villin2 (VIL2), the encoding gene of Ezrin protein, is another downstream target gene of miR-183 that regulates EMT. Ezrin is the earliest member of the ERM (ezrin/radixin/moesin) protein family, which is considered to be an important regulator and connector between cell membrane molecules (such as CD44 and ICAM2) and cytoskeleton (such as actin) [27]. It has also been shown that ezrin can coprecipitate with two important adhesion molecules, E-cadherin and beta-catenin, which further illustrates the importance of Ezrin in cell adhesion [28]. Overexpression of miR-183 repressed the expression levels of Ezrin and significantly inhibited the motility of osteosarcoma cells via increased N-cadherin and activating ERK signaling [29, 30]. MiR-183 was decreased in endometrial cancer tissues, while the expression of Ezrin was significantly increased. The downregulation of miR-183 suppressed apoptosis and promoted EMT of human endometrial cancer cells by upregulating Ezrin [31]. In gastric cancer and melanoma, miR-183 acts as a tumor suppressor, the expression was significantly decreased. Overexpression of miR-183 markedly suppressed cell invasion by downregulation of Ezrin expression [32]. In breast cancer, miR-183 targets VIL2 and may play a central role in the regulation of metastasis. The above literature indicates that overexpression of miR-183 inhibits the expression of VIL2 gene, thus ultimately inhibiting EMT of cancer cells.
The above evidences suggest that miR-183 participates in the process of EMT by targeting ZEB1, MTA1 and VIL2. Participates in the process of cancer indirectly and lay foundation for the migration and invasion of cancer cells.
Roles of miR-183 in cell migration and invasion
Numerous studies have shown that miRNAs play an important role in the acquisition of invasive ability and malignant phenotype of cancer cells, including loss of cell adhesion molecules, acquisition of cell motility and proliferation of tumors in target organs. At present, many studies have reported that miR-183 is closely related to the invasion and metastasis of tumors [33]. MiR-183 can regulate about 45 tumor-related genes, which are closely related to the adhesion, migration and invasion of various cancer cells. The most studied genes are programmed cell death factor 4 (PDCD4), early growth response one (EGR1) and integrin beta 1 (ITGB1), which play important roles in the migration and invasion of tumors.
PDCD4 is the most studied target gene of miR-183. It has been found that miR-183 acts as an oncogene by targeting PDCD4 in a variety of tumors. Lu et al. found that miR-183 overexpression could promote cell proliferation, inhibit cell apoptosis and decrease G₀/G₁ arrest in SW1990 pancreatic cancer cells by targeting PDCD4 [34]. In breast cancer, gastric cancer and ESCC, miR-183 is overexpressed, the overexpression significantly enhanced cell proliferation migration, invasion and inhibited cell apoptosis by targeting PDCD4 [35, 36, 37, 38]. Upregulation of miR-183 is associated with advanced clinical stage, positive lymph node, deep stromal invasion, and distant metastasis in cancer patients [32]. Furthermore, knockdown of PDCD4 suppressed expression of p21 and p27 in breast cancer, which shows that miR-183 can inhibit p21 and p27 indirectly [37].
EGR1 plays a very important coupling role in the cascade reaction of information transmission between cells and participates in many life activities such as cell division, proliferation, differentiation and apoptosis. EGR1 can promote apoptosis by activating p53 or binding with transcription factor E-Jun. It has been reported that miR-183 plays an important role in promoting cancer in synovial sarcoma, rhabdomyosarcoma (RMS) and colon cancer by regulating the expression of EGR1 and phosphatase and tensin homolog deleted on chromosome ten (PTEN) [39]. Inhibiting or knocking out miR-183 can lead to a significant increase in the expression of EGR1 and PTEN proteins, as well as the effect of inhibiting cell migration and invasion. Thiel J. et al. found that overexpression of miR-183 in murine CD4+ T cells resulted in decreased EGR1 and PTEN expression, elevated Akt phosphorylation and enhanced proliferation [40]. In NSCLC, Wang et al. found that miR-183-5p could promote lung carcinogenesis by directly targeting PTEN [41].
ITGB1 plays a key role in migration and invasion of tumors [42]. MiR-183 can act as an antioncogene by targeting ITGB1. Many human tumors are related to the increase of ITGB1 expression and activation of ITGB1-related signaling pathways [43]. Sun et al. found a new signaling pathway promoting melanoma development by MALAT1-miR-183-ITGB1 axis [13]. MiR-183 can significantly reduce the expression of ITGB1, thereby inhibiting cell migration and adhesion. MiR-183 is downregulated in cervical cancer cell line HeLa, and miR-183 directly regulates the invasion and metastasis of HeLa cells by acting on its target gene ITGB1 [44].
In addition to the above important genes, there are about thirty miR-183 target genes related to migration and invasion (Table 3). This indicates that miR-183 has a great influence on the invasion and migration of cells. By promoting or inhibiting the migration and invasion of cancer cells, it directly participate the process of cancer.
Genes that have been shown to have a targeting relationship with miR-183 and their functions in the corresponding cancer types or cell lines.
| Model system | Tissue (specimens) | Cell lines | Target gene | Function | Source |
|---|---|---|---|---|---|
| pancreatic cancer | X | SW1990 Pancreatic cancer cell line | PDCD4 | Apoptosis, Proliferation | Lu Y, et al.(2016) [34] |
| breast cancer | √ (n=18) | MCF-7 and MDAMB-231 | PDCD4 | Apoptosis, Proliferation | Cheng Y, et al.(2016) [37] |
| esophageal squamous cell carcinoma | √ (n=81) | EC109 and EC9706 | PDCD4 | Apoptosis, Proliferation | Yang M, et al.(2014) [36] |
| gastric cancer | √ (n=80) | SGC-7901 | PDCD4 | Apoptosis, Proliferation | Gu W, et al.(2014) [32] |
| papillary thyroid carcinoma | √ (n=38) | TPC-1, BCPAP, K1 and NPA PTC | PDCD4 | Migration, Apoptosis | Wei C, et al.(2015) [38] |
| hepatocellular carcinoma | √ (n=25) | HepG2 and Huh7 | PDCD4 | Apoptosis | Li J, et al.(2010) [14] |
| esophageal squamous cell carcinoma | √ (n=32) | Eca109 and TE13 | PDCD4 | Proliferation, Invasion | Ren L, et al.(2014) [12] |
| pediatric caute myeloid leukemia | √ (n=106) | HL60 and K562 | PDCD6 | Apoptosis | Wang X, et al.(2017) [9] |
| melanoma | X | A375 human melanoma cells | MMP-9 | Migration, Invasion | Zhang Y, et al.(2019) [91] |
| endometrial cancer | √ (n=30) | KLE, HEC-1-A and HHUA | MMP-9 | Proliferation, Invasion | Ruan H, et al.(2017) [61] |
| cervical cancer | √ (n=53) | siha and Hela | MMP-9 | Invasion, Metastasis | Fan D, et al.(2016) [68] |
| osteosarcoma | √ (n=25) | hFOB 1.19 and MG63 | MTA1 | Migration, Invasion | Sun X, et al.(2018) [24] |
| nasopharyngeal carcinoma | √ (n=29) | C666-1, CNE1, CNE2, HONE1, and 5-8F | MTA1 | Apoptosis | Wang G, et al.(2017) [25] |
| hepatocellular carcinoma | √ (n=10) | HepG2 | MTA1 | EMT, Metastasis | Bian W, et al.(2018) [60] |
| non-small cell lung cancer | √ (n=194) | H1299, SPC-A-1, 95D and A549 | MTA1 | Migration, Invasion | Yang C, et al.(2018) [63] |
| pancreatic cancer | √ (n=108) | SW1900 Pancreatic cancer cell line | MTA1 | EMT, Migration, Invasion | Lin X, et al.(2017) [26] |
| osteosarcoma | X | F5M2 and F4 | VIL2(Ezrin) | Invasion | Zhao H, et al.(2012) [29] |
| melanoma | X | A375 human melanoma cells | VIL2(Ezrin) | Migration, Invasion | Zhang Y, et al.(2019) [91] |
| endometrial cancer | √ (n=156) | Ishikawa,KLE, JEc, HEc-1-A, and HHUA | VIL2(Ezrin) | EMT, Migration, Invasion | Yan H, et al.(2018) [31] |
| gastric cancer | √ (n=55) | MGC-803, SGC-7901,BGC-823, MKN-45, and MKN-28 | VIL2(Ezrin) | Invasion | Cao LL, et al.(2014) [65] |
| lung cancer cells | X | 801D and 95C | VIL2(Ezrin) | Migration, Invasion | Wang G, et al.(2008) [92] |
| breast cancer | √ (n=70) | MDA-MB-231,T47D,ZR-75-1,and SKBR-3 | VIL2(Ezrin) | Migration | Lowery AJ, et al.(2010) [93] |
| acute myeloid leukemia | √ (n=7) | HL-60, U937 | Erbin | Proliferation | Zheng Z, et al.(2019) [94] |
| pancreatic adenocarcinoma | √ (n=52) | PANC-1 and HPDE6‑C7 | SOCS6 | Proliferation, Invasion, | Miao F, et al.(2016) [70] |
| hepatocellular carcinoma | √ (n=10) | HepG2 and Hep3B | SOCS6 | Growth, Invasion | Li ZB, et al.(2015) [59] |
| cervical cancer | √ (n=43) | SiHa, C-33 A, C-4-I, and CaSki | ITGB1 | Migration, Invasion | Zhang W, et al.(2018) [44] |
| melanoma | √ (n=30) | HBL and SK-MEL-1 | ITGB1 | Tumor progression | Sun Y, et al.(2017) [13] |
| HeLa cells | X | HeLa,HDF and HTM | ITGB1 | Migration, Invasion | Li G, et al.(2010) [95] |
| gastric cancer | X | MKN28 | UVRAG | Autophagosomes ,UVR | Yuan Y, et al.(2018) [96] |
| colorectal cancer | √ (n=14) | HCT116 and HT29 | UVRAG | Autophagosomes, UVR | Huangfu L, et al.(2015) [97] |
| mesothelioma | √ (n=29) | ACC-MESO1 and CRL-5915 | FOXO1 | Migration, Invasion | Suzuki R, et al.(2018) [75] |
| lymphoma | - | L428 | FOXO1 | Proliferation | Xie L, et al.(2012) [98] |
| breast cancer | X | MCF-7 | RAD50 | DNA repair processes | Shirode AB, et al.(2014) [99] |
| prostate cancer | √ (n=49) | DU145, PC3, 22Rv1 and LNCaP-ARhi | KLK3, PSA | Growth | Larne O, et al.(2015) [100] |
| ovarian cancer | √ (n=30) | SKOV-3, OVcAR3 and HOSE | SMAD4 | Apoptosis, Proliferation | Zhou J, et al.(2019) [74] |
| prostate cancer | √ (n=31) | PC-3, DU-145 and LNCaP | SMAD4 | Growth | Ueno K, et al.(2013) [73] |
| lung tumor | √ (n=126) | 293T cells, H1355 and H1299 | MICA,MICB | Immune correlation | Trinh TL, et al.(2019) [52] |
| pancreatic ductal adenocarcinoma | √ (n=91) | ASPC-1, SW1990, BXPC-3, CFPAC-1 and PANC-1 | Bmi-1 | Proliferation | Zhou L, et al.(2014) [71] |
| gastric cancer | √ (n=65) | AGS, SGC7901, MKN28, MGC803 and HGC27 | Bmi-1 | Proliferation, Invasion | Xu L, et al.(2014) [66] |
| osteosarcoma | X | MG63 and U20S | LRP-6 | Invasion | Yang X, et al.(2018) [64] |
| retinoblastoma | √ (n=6) | Y79, SO-RB50 and WERI-RB1 | LRP-6 | Growth, Invasion | Wang J, et al.(2014) [88] |
| synovial sarcoma | √ (n>300) | SYO-1 and FUJI | EGR1 | Migration | Sarver AL, et al.(2010)[39] |
| colon cancer | √ (n=80) | HCT116 and DLD1 | |||
| rhabdomyosarcoma | √ (n=42) | Rh30 and JR1 | |||
| non-small cell lung cancer | √ (n=55) | A549, SPCA-1, PC-9, and 95-D | PTEN | Tumor progression | Wang H, et al.(2019) [41] |
| leukemia | X | leukemia U937 cells | ADAM17, NFK-β | -- | Liu W, et al.(2012) [53] |
| renal cell carcinoma | X | ACHN, 786-O, Caki-1 and Caki-2 | DKK-3 | Proliferation, Metastasis | Zhang XL, et al.(2018) [77] |
| prostate cancer | √ (n=31) | PC-3, DU-145 and LNCaP | DKK-3 | Growth | Ueno K, et al.(2013) [73] |
| gastric cancer | √ (n=876) | AGS and MKN-45 | TCF12 | Tumor progression | Wang X, et al.(2019) [101] |
| lung cancer | √ (n=15) | A549, 95D, H1299 and H1650 | PIK3CA | Migration, Invasion | Meng F, et al.(2019) [69] |
| bladder cancer | √ (n=23) | 5637 and T24 | AXIN2 | Growth, Apoptosis | Chen D, et al.(2018) [76] |
| endometrial cancer | √ (n=208) | Ishikawa and RL95-2 | CPEB1 | EMT, Migration, Invasion | Xiong H, et al.(2018) [23] |
| glioblastoma | √ (n=39) | U251R | LRIG1 | Radioresistance | Fan H, et al.(2018) [102] |
| breast cancer cell line | X | MDA-MB-231 and MDA-MB-468 | RB1 | Metastasis | Macedo T, et al.(2017) [103] |
| lung adenocarcinoma | √ | CSLCs | PTPN | Migration, Invasion | Zhu C, et al.(2016) [104] |
| colon cancer | √ (n=10) | DLD1, Caco-2, HCT116, DiFi, and HCA7 | ABCA1 | Apoptosis, Proliferation | Bi DP, et al.(2016) [105] |
| glioma | √ (n=44) | U87MG, U251, and LN229 | NEFL | Proliferation, Invasion | Wang Z, et al.(2016) [84] |
| lung cancer | X | 344SQ, 344LN, 531LN1 and 531LN2 | FOXF2 | EMT, invasion | Kundu ST, et al.(2016) [22] |
| breast cancer | √ (n=2) | MCF-7,MDA-MB-231 and MCF-10A et al. | RAB40B ,RAB21 | Proliferation, Migration | Li P, et al.(2014) [10] |
| renal cancer cells | √ (n=16) | ACHN and A498 | PP2A-Cα, PP2A-Cβ, PP2A-B56-γ | Migration, Invasion | Qiu M, et al.(2014) [78] |
| glioma | √ (n=88) | U251, U87MG, T98G, A172 and SF126 | IDH2 | Angiogenesis | Tanaka H, et al.(2013) [109] |
| ovarian cancer | √ (n=17) | SKOV-3ip and HO-8910PM | TIAM1 | Migration, Invasion | Li J, et al.(2012) [106] |
| HeLa cells | X | HeLa | KIF2A | Migration, Invasion | Li G, et al.(2010) [95] |
| HUVECs | X | HUVECs | IRS1 | Angiogenesis | Zhang Y, et al.(2019) [47] |
| diabetic retinopahy | X | BTG1 | Angiogenesis | Zhang ZZ, et al.(2019) [107] | |
| NF-KB1 | -- | Sha F, et al.(2014) [108] |
X: No relevant data or use of pathological tissue for testing. √: Pathological tissue tests were carried out.
Roles of miR-183 in microangiogenesis
Experimental and clinical studies have confirmed that the growth rate of vascularized tumors is significantly faster than that of non-vascularized tumors [45]. Hypoxia is a major factor in neovascularization of tumors. It stimulates tumour cells to produce a variety of angiogenic factors, such as vascular endothelial growth factor (VEGF). Fibronectin 1 and serine protease inhibitor family E member 2 (SERPINE2) are essential for vascular mimicry in this system. Low-density lipoprotein receptor-related protein 1 (LRP1) and ITGB1 are receptors for these secreted proteins. Evidence shows that miR-183 regulates those autocrine signal and VM formation [51]. In addition, hypoxia-induced transcription factors (HIFs) are upregulated before angiogenesis due to hypoxia. Hypoxia in microenvironment can result in upregulation of growth factor receptors, angiogenic cytokines and proteolytic enzymes [46]. Studies in glioma suggest that miR-183 upregulation in malignant gliomas induces HIF-1α expression by targeting IDH2 [110]. Reducing the translation of miR-183-inhibited HIF-1α and other hypoxic markers can increase angiogenesis and improve oxygen transport in hypoxic tissues.
Effect of miR-183 on human umbilical vascular endothelial cells (HUVECs) and vascular endothelial cells, indirectly showed the role of 183 in angiogenesis. Inhibition miR-183 expression in HUVECs injury could enhance cell activity, inhibit inflammatory level, and thus resist cell injury by upregulating expression of IRS1 [47]. Overexpression miR-183 promoted cell growth and tube formation while it suppressed cell apoptosis of vascular endothelial cells [48]. In addition, miR-183 also participates in microangiogenesis during tumor growth through some signaling pathways. PI3K/AKT/VEGF signaling pathway plays an important role in angiogenesis. Research in diabetic retinopathy revealed the involvement of miR-183 via the PI3K/AKT/VEGF signaling pathway by targeting BTG1. MiR-183 overexpression activated the PI3K/AKT/VEGF signaling pathway and inhibited BTG1 [48]. These evidences suggest that miR-183 is involved in microangiogenesis through multiple regulations. Angiogenesis plays a decisive role in the growth of tumors and metastasis of cancer cells, which further demonstrates that miR-183 is closely involved in the progression of cancer.
miR-183 in cancer diagnosis and treatment
In recent decades, the rapid rise of posttranscriptional mechanistic studies, especially miRNA studies, has brought a new perspective to early diagnosis and precise treatment of tumors. Because the expression of miRNA in blood is relatively stable, the content of miRNA in serum can provide important information for the prediction and diagnosis of cancer. Compared with the level of expression of mRNA and protein, miRNA will not face the fate of being regulated and degraded, so its detection results are relatively more reliable. MiR-183 is highly abnormally expressed in cancer patients and easy to detect. Therefore, on the basis of understanding miR-183 and its mechanism, the expression of miR-183 in blood can be used to diagnose cancer and determine the appropriate treatment strategies.
Targeting drug brings some diagnostic advantages. However, it does not work for a considerable number of patients, or some of the active patients develop drug resistance after a period of treatment, leading to further deterioration of the condition and increasing the difficulty of diagnosis. A study found that miR-183 may be involved in paclitaxel-induced chemotherapy when studying the miRNA related to paclitaxel-induced apoptosis of hepatocellular carcinoma [110]. Abnormal expression of miR-183 in ovarian cancer cells also results in chemical resistance to paclitaxel [111]. It has been reported that miR-183 regulates multidrug resistance (MDR) in hepatocellular carcinoma through a feedback loop of miR-183 idh2/SOCS6-HIF-1α axis [112]. MiR-183 has been up-regulated in MDR Ehrlich ascites tumor cells and regulated MDR in hepatocellular carcinoma cells. Kouri et al. synthesized spherical nucleic acid based on functionalized gold nanoparticles with mature miR-182 double chains could reduce the burden of tumors and increase the survival rate of animals through intravenous application [113], which can also use anti-miR-183 as a possible treatment in cancer. Notably, miR-183 might affect the prediction for PSA-dependent diagnosis and prognosis via regulating PSA expression [100]. However, there are many kinds of miRNA involved in this process. Therefore, further research is needed to thoroughly investigate how complex miRNA regulatory networks contribute to the MDR process [114]. Study of the specific regulatory mechanisms of miR-183 in these tumors can further elucidate the molecular mechanisms of tumorigenesis and development from the perspective of genes, which will form a complete molecular signaling pathway and functional gene network system related to miR-183. This provides an important guiding value for the treatment of clinical tumors, and also makes the early diagnosis and precise treatment of tumors possible.
Discussion
The recognized mechanism of miRNAs is to influence the stability of the target gene by complementary or incomplete pairing, inhibit its translation and ultimately achieve the regulation of protein expression [49]. Previous studies have suggested that miRNAs play a role only in complete or incomplete pairing with the 3'-UTR region of mRNA. Studies have shown that it can also play a role in combination with the coding region of mRNAs and the 5'UTR region [50]. Therefore, miR-183 may have more target genes and play more important roles in the biological process, especially in cell proliferation, migration, invasion and EMT (Fig. 2). Many biological processes, such as growth, development, embryo implantation and embryo development, all involve cell proliferation, migration, invasion, and EMT. This indicates that miR-183 may play a much more important role in biological physiological regulation.
Mechanisms of miR-183 in cancer progression at cellular and molecular level.
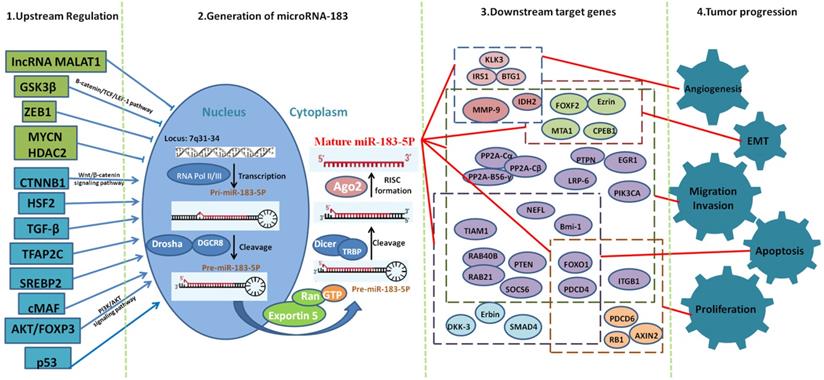
While in cancer, the change pattern of miR-183 varies greatly among different types of cancer. Different upstream regulators play a key role in the expression of miR-183 in different cancer types. We can conclude that miR-183 is upregulated in most cancer types. Meanwhile, up to now, 8 upstream regulators can upregulate the expression of miR-183 and 5 upstream regulators can downregulate the expression of miR-183. We speculate that miR-183 is upregulated in most cancer types because of the fact that there are more upregulating effect transcription factors than downregulating effect transcription factors.
It has been reported that miR-183 plays different roles in cell migration and invasion in different tissues. We speculate that the main reason for this result is that a miRNA can have hundreds of target genes. Through network miRNAs target gene prediction system, we found that the potential downstream targets of miR-183 include oncogenes, antioncogenes, cell signal transduction molecules, cell cycle regulation genes and molecules related to invasion and metastasis. As far as the mechanism of miRNAs is concerned, miR-183 can play the role of antioncogene by inhibiting the expression of oncogene, inhibiting cell migration and invasion. Of course, it can also play the role of oncogene by inhibiting the expression of antioncogene, promoting cell migration and invasion. This suggests that miR-183 plays different roles in different tumors, and its molecular mechanisms are complex and diverse, which need to be further explored. In addition, the abnormal expression of miR-183 in cancer progression can be used as a biological molecule for cancer diagnosis and treatment; it has great significance for cancer diagnosis and development of new therapeutic drugs.
Acknowledgements
This research was supported by grants from Department of Science Technology of Tongxiang City, Team Technology Commissioner Project (Tongxiang) and Leading Innovation Team Project of South Taihu Elite Program.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Taylor MA, Schiemann WP. Therapeutic opportunities for targeting microRNAs in cancer. Mol Cell Ther. 2014;2:1-13
2. Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155-62
3. Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215-33
4. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
5. Lagos-quintana M, Rauhut R, Meyer J. et al. New microRNAs from mouse and human. RNA. 2003;9:175-9
6. Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30:3254-63
7. Leung WK, He M, Chan AW. et al. Wnt/β-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015;362:97-105
8. Tang X, Zheng D, Hu P. et al. Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the β-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res. 2014;42:2988-98
9. Wang X, Zuo D, Yuan Y. et al. MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J Cancer Res Clin. 2017;143:169-80
10. Li P, Sheng C, Huang L. et al. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014;16:473
11. Lodrini M, Oehme I, Schroeder C. et al. MYCN and HDAC2 cooperate to repress miR-183 signaling in neuroblastoma. Nucleic Acids Res. 2013;41:6018-33
12. Ren L, Chen W, Li S. et al. MicroRNA-183 promotes proliferation and invasion in oesophageal squamous cell carcinoma by targeting programmed cell death 4. Brit J Cancer. 2014;111:2003-13
13. Sun Y, Cheng H, Wang G. et al. Deregulation of miR-183 promotes melanoma development via lncRNA MALAT1 regulation and ITGB1 signal activation. Oncotarget. 2017;8:3509-18
14. Li J, Fu H, Xu C. et al. MiR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010;10:354-64
15. Yoon SK. The biology of cancer stem cells and its clinical implication in hepatocellular carcinoma. Gut Liver. 2012;6:29-40
16. Sánchez-Tilló E, Lázaro A, Torrent R. et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490-500
17. Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142-65
18. De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97-110
19. Karamitopoulou E, Haemmig S, Baumgartner U. et al. MicroRNA dysregulation in the tumor microenvironment influences the phenotype of pancreatic cancer. Mod Pathol. 2017;30:1116-25
20. Peinado H, Olmeda D, Cano A. Snail, zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415-28
21. Peña C, García JM, Silva J. et al. E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: clinicopathological correlations. Hum Mol Genet. 2005;14:3361-70
22. Kundu ST, Byers LA, Peng DH. et al. The miR-200 family and the miR-183-96-182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173-86
23. Xiong H, Chen R, Liu S. et al. MicroRNA-183 induces epithelial-mesenchymal transition and promotes endometrial cancer cell migration and invasion in by targeting CPEB1. J Cell Biochem. 2018;119:8123-37
24. Sun X, Xu Y, Zhang S. et al. MicroRNA-183 suppresses the vitality, invasion and migration of human osteosarcoma cells by targeting metastasis-associated protein 1. Exp Ther Med. 2018;15:5058-64
25. Wang G, Wang S, Li C. MiR-183 overexpression inhibits tumorigenesis and enhances DDP-induced cytotoxicity by targeting MTA1 in nasopharyngeal carcinoma. Tumour Biol. 2017;39:1-11
26. Lin X, Zheng L, Song H. et al. Effects of microRNA-183 on epithelial-mesenchymal transition, proliferation, migration, invasion and apoptosis in human pancreatic cancer SW1900 cells by targeting MTA1. Exp Mol Pathol. 2017;102:522-32
27. Algrain M, Turunen O, Vaheri A. et al. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol. 1993;120:129-39
28. Bruce B, Khanna G, Ren L. et al. Expression of the cytoskeleton linker protein ezrin in human cancers. Clin Exp Metastas. 2007;24:69-78
29. Zhao H, Guo M, Zhao G. et al. MiR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int J Mol Med. 2012;30:1013-20
30. Zhang J, Zuo J, Lei M. et al. Ezrin promotes invasion and migration of the MG63 osteosarcoma cell. Chin Med J (Engl). 2014;127:1954-9
31. Yan H, Sun BM, Zhang YY. et al. Upregulation of miR-183-5p is responsible for the promotion of apoptosis and inhibition of the epithelial-mesenchymal transition, proliferation, invasion and migration of human endometrial cancer cells by downregulating Ezrin. Int J Mol Med. 2018;42:2469-80
32. Gu W, Gao T, Shen J. et al. MicroRNA-183 inhibits apoptosis and promotes proliferation and invasion of gastric cancer cells by targeting PDCD4. Int J Clin Exp Med. 2014;7:2519-29
33. Li P, Sheng C, Huang L. et al. MiR-183/-96/-182 cluster is upregulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014;16:473-90
34. Lu Y, Zheng J, Liu J. et al. MiR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed Pharmacother. 2015;70:151-7
35. Li C, Deng L, Zhi Q. et al. MicroRNA-183 functions as an oncogene by regulating PDCD4 in gastric cancer. Anticancer Agents Med Chem. 2016;16:447-55
36. Yang M, Liu R, Li X. et al. MiRNA-183 suppresses apoptosis and promotes proliferation in esophageal cancer by targeting PDCD4. Mol Cells. 2014;37:873-80
37. Cheng Y, Xiang G, Meng Y. et al. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod Biol. 2016;16:225-33
38. Wei C, Song H, Sun X. et al. MiR-183 regulates biological behavior in papillary thyroid carcinoma by targeting the programmed cell death 4. Oncol Rep. 2015;34:211-20
39. Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570-80
40. Thiel J, Alter C, Luppus S. et al. MicroRNA-183 and microRNA-96 are associated with autoimmune responses by regulating T cell activation. J Autoimmun. 2019;96:94-103
41. Wang H, Ma Z, Liu X. et al. MiR-183-5p is required for non-small cell lung cancer progression by repressing PTEN. Biomed Pharmacother. 2019;111:1103-11
42. Ramsay AG, Marshall JF, Hart IR. Integrin trafficking and its role in cancer metastasis. Cancer Metastasis Rev. 2007;26:567-78
43. Marotta A, Tan C, Gray V. et al. Dysregulation of integrin-linked kinase (ILK) signaling in colonic polyposis. Oncogene. 2001;20:6250-7
44. Zhang W, Zhang M, Liu L. et al. MicroRNA-183-5p inhibits aggressiveness of cervical cancer cells by targeting integrin subunit beta1 (ITGB1). Med Sci Monitor. 2018;24:7137-45
45. Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011-27
46. Steeg PS. Tumor metastasis mechanistic insights and clinical challenges. Nat Med. 2006;12:895-904
47. Zhang Y, Zhan Y, Liu D. et al. Inhibition of microRNA-183 expression resists human umbilical vascular endothelial cells injury by upregulating expression of IRS1. Drug Deliv. 2019;26:612-21
48. Wang Z, Zhang J, Chen L. et al. Glycine suppresses AGE/RAGE signaling pathway and subsequent oxidative stress by restoring Glo1 function in the aorta of diabetic rats and in HUVECs. Oxid Med Cell Longev. 2019;2019:1-14
49. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5
50. Tay Y, Zhang J, Thomson AM. et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124-8
51. Langer EM, Kendsersky ND, Daniel CJ. et al. ZEB1-repressed microRNAs inhibit autocrine signaling that promotes vascular mimicry of breast cancer cells. Oncogene. 2018;37:1005-19
52. Trinh TL, Kandell WM, Donatelli SS. et al. Immune evasion by TGFβ-induced miR-183 repression of MICA/B expression in human lung tumor cells. Oncoimmunology. 2019;8:e1557372
53. Liu W, Chang L. Suppression of Akt/Foxp3-mediated miR-183 expression blocks Sp1-mediated ADAM17 expression and TNFα-mediated NFκB activation in piceatannol-treated human leukemia U937 cells. Biochem Pharmacol. 2012;84:670-80
54. Kang J, Kim W, Lee S. et al. TFAP2C promotes lung tumorigenesis and aggressiveness through miR-183 and miR-33a mediated cell cycle regulation. Oncogene. 2017;36:1585-96
55. Jeon TI, Esquejo RM, Roqueta-Rivera M. et al. An SREBP responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18:51-61
56. Kelada S, Sethupathy P, Okoye IS. et al. MiR-182 and miR-10a are key regulators of treg specialisation and stability during schistosome and leishmania-associated inflammation. Plos Pathog. 2013;9:e1003451
57. Chang C, Chao C, Xia W. et al. P53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317-23
58. Bharali D, Jebur HB, Baishya D. et al. Expression analysis of serum microRNA-34a and microRNA-183 in hepatocellular carcinoma. Asian Pac J Cancer Prev. 2018;19:2561-68
59. Li ZB, Li ZZ, Li L. et al. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur Rev Med Pharmacol Sci. 2015;19:3208-17
60. Bian W, Zhang H, Tang M. et al. Potential role of microRNA-183 as a tumor suppressor in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51:2065-72
61. Ruan H, Liang X, Zhao W. et al. The effects of microRNA-183 promots cell proliferation and invasion by targeting MMP-9 in endometrial cancer. Biomed Pharmacother. 2017;89:812-8
62. Zhang L, Quan H, Wang S. et al. MiR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol. 2015;36:8121-6
63. Yang C, Zheng X, Ye K. et al. MicroRNA-183 acts as a tumor suppressor in human non-small cell lung cancer by downregulating MTA1. Cell Physiol Biochem. 2018;46:93-106
64. Yang X, Wang L, Wang Q. et al. MiR-183 inhibits osteosarcoma cell growth and invasion by regulating LRP6-Wnt/β-catenin signaling pathway. Biochem Bioph Res Co. 2018;496:1197-203
65. Cao LL, Xie JW, Lin Y. et al. MiR-183 inhibits invasion of gastric cancer by targeting Ezrin. Int J Clin Exp Pathol. 2014;7:5582-94
66. Xu L, Li Y, Yan D. et al. MicroRNA-183 inhibits gastric cancer proliferation and invasion via directly targeting Bmi-1. Oncol Lett. 2014;8:2345-51
67. Liu SS, Chan K, Chu D. et al. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Mol Oncol. 2018;12:2009-22
68. Fan D, Wang Y, Qi P. et al. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol. 2016;141:166-74
69. Meng F, Zhang L. MiR-183-5p functions as a tumor suppressor in lung cancer through PIK3CA inhibition. Exp Cell Res. 2019;374:315-22
70. Miao F, Zhu J, Chen Y. et al. MicroRNA-183-5p promotes the proliferation, invasion and metastasis of human pancreatic adenocarcinoma cells. Oncol Lett. 2016;11:134-40
71. Zhou L, Zhang WG, Wang DS. et al. MicroRNA-183 is involved in cell proliferation, survival and poor prognosis in pancreatic ductal adenocarcinoma by regulating Bmi-1. Oncol Rep. 2014;32:1734-40
72. Waseem M, Ahmad MK, Serajuddin M. et al. MicroRNA-183-5p: a new potential marker for prostate cancer. Indian J Clin Biochem. 2019;34:207-12
73. Ueno K, Hirata H, Shahryari V. et al. MicroRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Brit J Cancer. 2013;108:1659-67
74. Zhou J, Zhang C, Zhou B. et al. MiR-183 modulated cell proliferation and apoptosis in ovarian cancer through the TGF-β/Smad4 signaling pathway. Int J Mol Med. 2019;43:1734-46
75. Suzuki R, Amatya VJ, Kushitani K. et al. MiR-182 and miR-183 promote cell proliferation and invasion by targeting FOXO1 in mesothelioma. Front Oncol. 2018;8:446-55
76. Chen D, Li S, Chen J. et al. MiR-183 maintains canonical Wnt signaling activity and regulates growth and apoptosis in bladder cancer via targeting AXIN2. Eur Rev Med Pharmaco. 2018;22:4828-36
77. Zhang XL, Xu G, Zhou Y. et al. MicroRNA-183 promotes the proliferation and metastasis of renal cell carcinoma through targeting Dickkopf-related protein 3. Oncol Lett. 2018;15:6003-8
78. Qiu M, Liu L, Chen L. et al. MicroRNA-183 plays as oncogenes by increasing cell proliferation, migration and invasion via targeting protein phosphatase 2A in renal cancer cells. Biochem Bioph Res Commun. 2014;452:163-9
79. Chen Y, Song W. Wnt/catenin beta1/microRNA 183 predicts recurrence and prognosis of patients with colorectal cancer. Oncol Lett. 2018;15:4451-6
80. Yuan D, Li K, Zhu K. et al. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol Ther. 2015;16:268-75
81. Supic G, Zeljic K, Rankov AD. et al. MiR-183 and miR-21 expression as biomarkers of progression and survival in tongue carcinoma patients. Clin Oral Invest. 2018;22:401-9
82. Yan D, Cai X, Feng Y. MiR-183 modulates cell apoptosis and proliferation in tongue squamous cell carcinoma SCC25 cell line. Oncol Res. 2016;24:399-404
83. Ye Z, Zhang Z, Wu L. et al. Upregulation of miR-183 expression and its clinical significance in human brain glioma. Neurol Sci. 2016;37:1341-7
84. Wang Z, Xiong J, Zhang S. et al. Upregulation of microRNA-183 promotes cell proliferation and invasion in glioma by directly targeting NEFL. Cell Mol Neurobiol. 2016;36:1303-10
85. Chen H, Zhang L, Zhang L. et al. MicroRNA-183 correlates cancer prognosis, regulates cancer proliferation and bufalin sensitivity in epithelial ovarian cancer. Am J Transl Res. 2016;8:1748-55
86. Yang X, Wu X. MiRNA expression profile of vulvar squamous cell carcinoma and identification of the oncogenic role of miR-590-5p. Oncol Rep. 2016;35:398-408
87. Cheung CC, Lun SW, Chung GT. et al. MicroRNA-183 suppresses cancer stem-like cell properties in EBV-associated nasopharyngeal carcinoma. BMC Cancer. 2016;16:495-504
88. Wang J, Wang X, Li Z. et al. MicroRNA-183 suppresses retinoblastoma cell growth, invasion and migration by targeting LRP6. FEBS J. 2014;281:1355-65
89. Yuan J, Dong R, Liu F. et al. The miR-183/182/96 cluster functions as a potential carcinogenic factor and prognostic factor in kidney renal clear cell carcinoma. Exp Ther Med. 2019;17:2457-64
90. Meng C, Guo Z, Li D. et al. MiR-183 and miR-141 in lesion tissues are potential risk factors for poor prognosis in patients with infected abdominal aortic aneurysm. Exp Ther Med. 2018;16:4695-9
91. Zhang Y, Wang G. MicroRNA-183 inhibits A375 human melanoma cell migration and invasion by targeting Ezrin and MMP-9. Oncol Lett. 2019;17:548-54
92. Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582:3663-8
93. Lowery AJ, Miller N, Dwyer RM. et al. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502-14
94. Zheng X, Zheng Z, Zhu Y. et al. MiR-183-5p inhibits occurrence and progression of acute myeloid leukemia via targeting Erbin. Mol Ther. 2019;27:542-58
95. Li G, Luna C, Qiu J. et al. Targeting of integrin β1 and kinesin 2α by microRNA-183. J Biol Chem. 2010;285:5461-71
96. Yuan Y, Zhang Y, Han L. et al. MiR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int J Mol Med. 2018;42:3562-70
97. Huangfu L, Liang H, Wang G. et al. MiR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget. 2016;7:4735-45
98. Xie L, Ushmorov A, Leithauser F. et al. FOXO1 is a tumor suppressor in classical hodgkin lymphoma. Blood. 2012;119:3503-11
99. Shirode AB, Kovvuru P, Chittur SV. et al. Antiproliferative effects of pomegranate extract in MCF-7 breast cancer cells are associated with reduced DNA repair gene expression and induction of double strand breaks. Mol Carcinog. 2014;53:458-70
100. Larne O, Östling P, Haflidadóttir BS. et al. MiR-183 in prostate cancer cells positively regulates synthesis and serum levels of prostate-specific antigen. Eur Urol. 2015;68:581-8
101. Wang X, Gao S, Xie F. et al. High expression of TCF12 contributes to gastric cancer development via being target regulated by miR-183 and activating PI3K/AKT pathway. J Cell Biochem. 2019;120:13903-11
102. Fan H, Yuan R, Cheng S. et al. Overexpressed miR-183 promoted glioblastoma radioresistance via down-regulating LRIG1. Biomed Pharmacother. 2018;97:1554-63
103. Macedo T, Silva-Oliveira RJ, Silva VAO. et al. Overexpression of mir-183 and mir-494 promotes proliferation and migration in human breast cancer cell lines. Oncol Lett. 2017;14:1054-60
104. Zhu C, Deng X, Wu J. et al. MicroRNA-183 promotes migration and invasion of CD133(+)/CD326(+) lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol. 2016;37:11289-97
105. Bi DP, Yin CH, Zhang XY. et al. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep. 2016;35:2873-9
106. Li J, Liang S, Jin H. et al. Tiam1, negatively regulated by miR-22, miR-183 and miR-31, is involved in migration, invasion and viability of ovarian cancer cells. Oncol Rep. 2012;27:1835-42
107. Zhang ZZ, Qin XH, Zhang J. MicroRNA-183 inhibition exerts suppressive effects on diabetic retinopathy by inactivating BTG1-mediated PI3K/Akt/VEGF signaling pathway. Am J Physiol Endocrinol Metab. 2019;316:E1050-60
108. Sha F, Wu S, Zhang H. et al. MiR-183 potentially inhibits NF-κB1 expression by directly targeting its 3'-untranslated region. Acta Bioch Bioph Sin. 2014;46:991-6
109. Tanaka H, Sasayama T, Tanaka K. et al. MicroRNA-183 upregulates HIF-1α by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J Neuro-Oncol. 2013;111:273-83
110. Yan H, Wang S, Yu H. et al. Molecular pathways and functional analysis of miRNA expression associated with paclitaxel-induced apoptosis in hepatocellular carcinoma cells. Pharmacology. 2013;92:167-74
111. Wang YQ, Guo RD, Guo RM. et al. MicroRNA-182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J Cell Biochem. 2013;114:1464-73
112. Wang XJ, Zhang DL, Fu C. et al. MiR-183 modulates multi-drug resistance in hepatocellular cancer (HCC) cells via miR-183-IDH2/SOCS6-HIF-1α feedback loop. Eur Rev Med Pharmacol Sci. 2016;20:2020-7
113. Kouri FM, Hurley LA, Daniel WL. et al. MiR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732-45
114. Husted S, Søkilde R, Rask L. et al. MicroRNA expression profiles associated with development of drug resistance in Ehrlich ascites tumor cells. Mol Pharm. 2011;8:2055-62
Author contact
![]() Corresponding author: Zhengguang Wang, College of Animal Sciences, Zhejiang University, 866 Yuhangtang Road, Hangzhou, 310058 P. R. China. E-mail: wzhguang68edu.cn.
Corresponding author: Zhengguang Wang, College of Animal Sciences, Zhejiang University, 866 Yuhangtang Road, Hangzhou, 310058 P. R. China. E-mail: wzhguang68edu.cn.

 Global reach, higher impact
Global reach, higher impact