Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(13):3893-3902. doi:10.7150/jca.42944 This issue Cite
Research Paper
Self-enforcing HMGB1/NF-κB/HIF-1α Feedback Loop Promotes Cisplatin Resistance in Hepatocellular Carcinoma Cells
1. Hepatology Unit and Department of Infectious Diseases, Nanfang Hospital, Southern Medical University, Guangzhou 510515, China
2. Department of Radiation Oncology, Nanfang Hospital, Southern Medical University, Guangzhou, PR China.
Received 2019-12-10; Accepted 2020-3-27; Published 2020-4-6
Abstract
Hepatocellular carcinoma (HCC) is ranked the sixth most common cancer and the fourth leading cause of cancer-related death worldwide, and its incidence is expected to increase in the future. Cisplatin has been widely used in chemotherapy and transarterial chemoembolization in treatment for HCC. However, the main obstacle to the clinical use of cisplatin is the development of resistance, the mechanisms of which are poorly defined. Therefore, it is imperative to investigate the cellular mechanisms mediating cisplatin resistance in HCC. Here, we demonstrated that high mobility group box 1 (HMGB1) is upregulated in patients with cancer, and implicated in a tumor-supportive role. Further, we showed that HMGB1 has an important role in mediating cisplatin resistance via an HMGB1/ nuclear factor kappa-B (NF-κB)/ hypoxia inducible factor-1α (HIF-1α) feedback loop. The study findings reveal an unappreciated molecular mechanism of HMGB1-mediated cisplatin resistance and may provide a new clue in cancer therapy.
Keywords: HMGB1, Hepatocellular carcinoma, Cisplatin resistance, HIF-1α, NF-κB, Self-enforcing
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer, and the fourth leading cause of cancer-related death worldwide, with more than 840,000 new cases diagnosed and 781,000 deaths in 2018 [1]. Although there is a wide range of available therapeutic options for HCC, chemotherapy is nonetheless still often adopted in patients with highly advanced HCC [2]. Chemotherapeutic agents such as cisplatin are widely used in transarterial chemoembolization [3]. However, chemotherapy often fails due to multidrug resistance (MDR), leading to a poor prognosis in patients with advanced disease [3, 4]. Therefore, it is imperative to investigate the cellular mechanisms mediating cisplatin resistance in HCC.
High mobility group box 1 (HMGB1) is a nonhistone chromatin-binding protein that shows high electrophoretic mobility when run on polyacrylamide gels [5]. HMGB1 has multiple functions depending on its location [6]. In the nucleus, HMGB1 takes part in DNA replication, repair, recombination, transcription, and genomic stability [7, 8]. Cytoplasmic HMGB1 expression has a role in autophagy regulation in cancer cells [9, 10]. In addition, extracellular HMGB1, after either active release from cells or passive release upon lytic cell death, functions as a pro-inflammatory cytokine and perpetuates inflammatory responses in a wide variety of pathologies [11]. Owing to its versatile role in cancer, HMGB1 has been proposed as a potential cancer biomarker of survival and a target for cancer therapy [12]. Although the role of HMGB1 in tumorigenesis, progression, invasion, metastasis, and prognosis in different malignancies has been frequently discussed, its function in cisplatin resistance in HCC is not fully understood [13-15]. Understanding the role of HMGB1 is necessary to address the current problem of cisplatin resistance.
In the present study, we found that HMGB1 expression was prominently induced in HCC cells treated with cisplatin and significantly upregulated in HCC tissues than in matched adjacent liver tissues. Functionally, HMGB1 silencing decreased cancer cell proliferation. HMGB1 markedly encouraged resistance to cisplatin by protecting HCC cells from apoptosis. Mechanistically, we report the identification of a positive HMGB1/NF-κB/HIF-1α feedback loop and define its critical role in cisplatin resistance.
Materials and Methods
Cell culture
The human hepatocarcinoma cell lines SK-Hep-1, SMMC-7721, and Huh7, and the hepatocyte cell line HL-7702 were purchased from the Cell Bank of Type Culture Collection (Shanghai, China). SK-Hep-1 and Huh7 cells were cultured in Dulbecco's modified eagle medium (DMEM, Gibco, MA USA); SMMC-7721 cells were incubated in RPMI 1640 (Thermo Fisher, MA USA), supplemented with 10% fetal bovine serum (FBS, Biological Industries, CT USA) in a moist incubator with 5% CO2.
Western blot analysis
The extraction of HCC cell total proteins was acquired using RIPA lysis buffer (Cell Signaling Technology, MA USA) containing protease and phosphatase inhibitors (Roche, Basel, Switzerland). Protein concentration was determined by the BCA Protein Assays kit (ThermoFisher, MA USA). Lysates were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and electro-transferred to a polyvinylidene difluoride membrane (Bio-Rad, CA USA) that was blocked with 5% skimmed milk for 1 hour at room temperature and cultured overnight at 4°C with primary antibodies followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Abcam, MA USA). Immunodetection was performed using chemiluminescence detection system (Bio-Rad, CA USA). Antibodies include HMGB1, (1:1000, from Proteintech, IL USA), PARP, cleaved-PARP (1:1000, from Abcam, MA USA), NF-κB-p65, phosphor-NF-κB-p65 (1:1000, from CST, MA USA), and HIF-1α (1:1000, from BD Biosciences, CA USA); β-actin was an internal control.
Reagents
Cisplatin (Sigma, MA USA), stable in sterile solutions in 0.9% sodium chloride protected from light for 28 days at room temperature. TNF-α (PeproTech, NJ USA), dissolved in phosphate-buffered saline (PBS) to 10μg/μL stock at -20°C. Cobalt (II) chloride hexahydrate (Sigma, MA USA), dissolved in deionized water to 20mM, freshly prepared solution and used at room temperature.
Clinical human tissue samples
Six pairs of tumor tissues and adjacent normal tissues were obtained from Nanfang Hospital, Southern Medical University, Guangzhou, China from June 2017 to July 2018, and the informed consent was signed by each participant. This experiment was permitted by the ethics committee of the Nanfang Hospital. Patients who met the following inclusion criteria were enrolled: (I) received liver resection for primary HCCs for the first time; (II) no evidence of extrahepatic metastasis at the initial stage; (III) Child- Pugh class A of liver function at the initial stage. Patients who received radiotherapy or chemotherapy before surgery were excluded. The resulting tissue samples were frozen in liquid nitrogen immediately.
Cancer biostatistical analysis
Relative expression levels of HMGB1 in hepatocellular carcinoma patients compared to normal samples were analyzed using previously published microarray database (GEO: GSE25097, IGCG: IGCG-LIRI-JP). Patients were divided into low-HMGB1 and high-HMGB1 groups based on HMGB1 expression for overall survival analysis.
The linear regression was analyzed using data from the GEO: GSE364 dataset.
Cell small interfering RNA transfection
Specific siRNAs against HMGB1 (si-HMGB1#1 and si-HMGB1#2) or HIF-1α (si-HIF-1α) and their corresponding NCs (si-NC) were acquired from RiboBio (Guangzhou, China). Cells were transfected with these siRNAs through Lipofectamine RNAiMAX (Invitrogen, MA USA) for 48h.
Cell plasmid transfection
The HMGB1 and P65 overexpression plasmid was purchased from Genechem (Shanghai, China). The plasmid was used to transfect well-growing state cells by using Lipofectamine 3000 transfection reagent (Invitrogen, MA USA) for 48h.
5-ethynyl-20-deoxyuridine (EdU) assays
Transfected SK-Hep-1 or SMMC-7721 cells were incubated for 2 h by use of 50 μM EdU (RiboBio, Guangzhou China), followed by Apollo staining (RiboBio, Guangzhou China) and then staining with DAPI (ZSGB-Bio, Beijing China). At length, EdU-positive cells were revealed via a fluorescence microscope (Oympus, Tokyo Japan) in eight random fields. Four independent experiments were performed.
CCK8 assay
The CCK8 assay was carried out following the manufacturer's instructions (Dojindo Laboratories, Osaka Japan). Absorbance of each well was quantified at 450 nm by an enzyme-labeled instrument, and cell growth was assayed every day for five days.
Real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted from cell lines and isolated by TRIzol reagent (Invitrogen, MA USA) according to the manufacturer's protocol. qRT-PCR was performed using the SYBR Green PCR kit (Takara Biotechnology, Tokyo Japan). β-actin was used as internal control for NF-κB and HIF-1α. The specificity of amplification products was confirmed by melting curve analysis. Triplicate samples were analyzed in three independent experiments.
Statistical analysis
The quantitative data are presented as the means ± SD of at least three independent experiments. The statistical analyses were performed using SPSS V.19.0 software and GraphPad Prism 8. Student's t test was applied to compare the significant differences between two groups, and analysis of variance (ANOVA) was used for multiple comparisons. All tests were two-sided, and differences were considered statistically significant at a p-value below 0.05.
Results
HMGB1 is upregulated in cancer cells treated with cisplatin and human cancer tissues
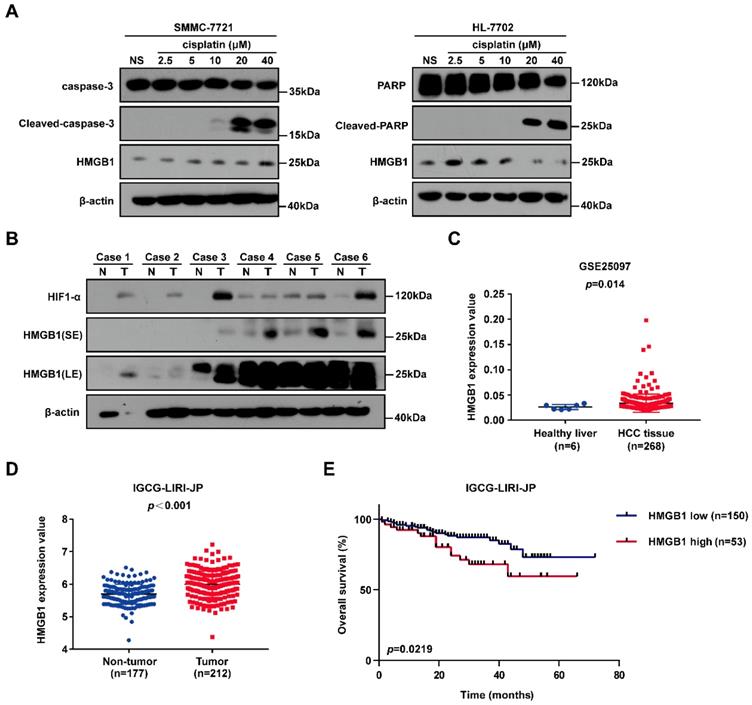
To identify the gene involved in cisplatin resistance of liver cancer cells, we first treated SMMC-7721 cancer cells and normal human HL-7702 hepatocytes with increasing doses of cisplatin. Immunoblotting showed that HMGB1 protein levels were significantly higher in SMMC-7721 cells than in HL-7702 cells, suggesting that HMGB1 may be related to cisplatin resistance (Fig. 1A). Next, we explored the expression of HMGB1 in tumors. Immunoblotting showed the upregulation of HMGB1 in HCC tumor samples of the patients. Moreover, HIF-1α upregulation was observed (Fig. 1B). For further verification, we analyzed data from the GSE25097 and IGCG-LIRI-JP datasets; both of them showed that HMGB1 expression was increased in HCC tissues (Figs. 1C and 1D). More importantly, to investigate the clinical importance of HMGB1, we performed a survival analysis based on data from the IGCG-LIRI-JP dataset. The survivorship curve showed that patients with high HMGB1 expression exhibited poorer overall survival (OS) (Fig. 1E). Multivariate analysis results showed that OS in the IGCG-LIRI-JP cohort could be predicted based on significant prognostic factors, such as portal vein invasion, alcohol consumption, and HMGB1 expression (Table 1 and Table 2). Taken together, these results suggested that HMGB1 may be related to cisplatin resistance and that in a substantial proportion of HCCs, HMGB1 is upregulated and associated with poor clinical outcomes.
The associations of HMGB1 expression with clinicopathological characteristics in ICGC-LIRI-JP (n=203) cohort
| Feature | Low expression of HMGB1 (n=150) | High expression of HMGB1 (n=53) |
|---|---|---|
| Age(years) | ||
| ≥50 | 139 | 47 |
| <50 | 11 | 6 |
| Gender | ||
| Male | 109 | 44 |
| Female | 41 | 9 |
| Virus | ||
| HBV | 35 | 18 |
| HCV | 88 | 29 |
| HBV and HCV | 2 | 2 |
| Negative | 25 | 4 |
| TNM stage | ||
| I+II | 101 | 28 |
| III+IV | 49 | 25 |
| Portal vein invasion | ||
| Positive | 35 | 14 |
| Negative | 115 | 39 |
| Vein invasion | ||
| Positive | 25 | 12 |
| Negative | 125 | 41 |
| Bile duct invasion | ||
| Positive | 9 | 4 |
| Negative | 141 | 49 |
| Fibrosis | ||
| Yes | 142 | 52 |
| No | 8 | 1 |
| Alcohol | ||
| Yes | 58 | 32 |
| No | 84 | 20 |
| NA | 8 | 1 |
| Smoking | ||
| Yes | 78 | 36 |
| No | 65 | 16 |
| NA | 7 | 1 |
Abbreviations: NA, not available.
HMGB1 silencing decreases cancer cell proliferation
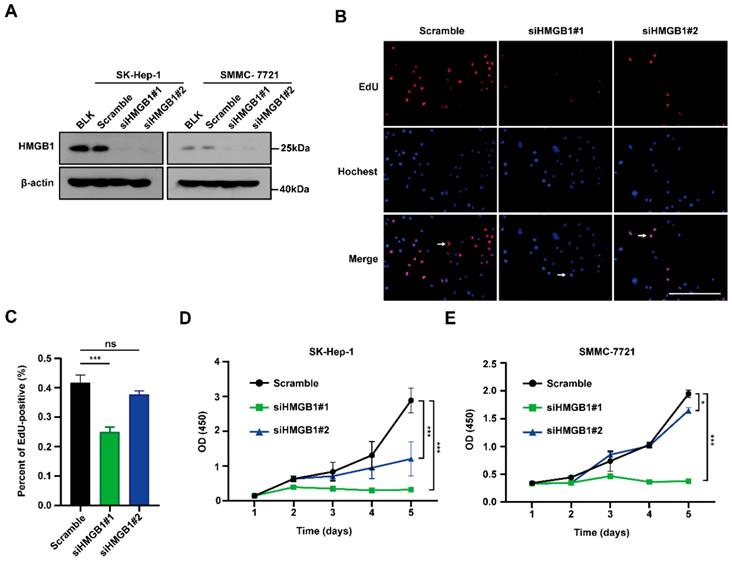
To examine the biological functions of HMGB1 in tumors, first SMMC-7721 and SK-Hep-1 cells were transfected with HMGB1 siRNA; HMGB1 expression levels were successfully reduced in both systems by immunoblotting (Fig. 2A). We then conducted EdU incorporation assays to assess HCC cell proliferations. The percentage of EdU-positive cells was significantly reduced in the HMGB1-silenced group than in the control group (Figs. 2B and 2C). In addition, we obtained the same result using CCK8 assays in SK-Hep-1 and SMMC-7721 cells (Fig. 2D and 2E). The above results show that HMGB1 silencing decreased cancer cell proliferation.
Univariate and multivariate analyses of OS in ICGC-LIRI-JP (n=203) cohort by Cox regression analysis
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | CI (95%) | P value | HR | CI (95%) | P value | ||
| Age (years) | 0.922 | 0.325-2.616 | 0.878 | ||||
| Gender | 0.536 | 0.265-1.083 | 0.082 | ||||
| Virus (HBV or/and HCV) | 1.654 | 0.506-5.408 | 0.406 | ||||
| TNM stage | 2.830 | 1.446-5.540 | 0.002* | ||||
| Portal vein invasion | 3.135 | 1.595-6.160 | 0.001* | 2.395 | 1.109-5.169 | 0.026* | |
| Vein invasion Bile duct invasion | 2.394 0.531 | 1.172-4.891 0.072-3.890 | 0.017* 0.533 | ||||
| Fibrosis | 1.158 | 0.276-4.853 | 0.841 | ||||
| Alcohol | 0.476 | 0.242-0.936 | 0.031* | 0.390 | 0.193-0.786 | 0.009* | |
| Smoking | 0.758 | 0.391-1.471 | 0.413 | ||||
| HMGB1 | 2.162 | 1.098-4.255 | 0.026* | 2.192 | 1.102-4.363 | 0.025* | |
Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus; CI, confidence interval; HR, hazard radio; *The values had statistically significant differences.
HMGB1 is upregulated in cancer cells treated with cisplatin and human cancer tissues. (A) Lysates from SMMC-7721 cells and HL-7702 cells treated with graduate increased dosage of cisplatin were subjected to immunoblotting. NS: normal saline. (B) the expression of HIF-1α and HMGB1 in 6 pairs of HCC tissues (T) and their matched normal-appearing tissues (N). SE: short exposure, LE: long exposure. (C) HMGB1 expression in healthy liver and HCC tissue using the GSE25097 (n=6 or 268) cohort. p Value was determined by unpaired two-sample Student's t test. (D) HMGB1 expression in non-tumor tissue (n=177) and tumor tissue (n=212) using the IGCG-LIRI-JP cohort. p Value was determined by Student's t test. (E) Analysis of overall survival (OS) in hepatocellular carcinoma patients with high or low HMGB1 expression using the IGCG-LIRI-JP (n=53 or 150) cohort. p Value was determined by log-rank (Mantel-Cox) test.
Silencing of HMGB1 decreases cancer cells proliferation. (A) Validation of two independent HMGB1 siRNAs by immunoblotting. (B) The percentage of S phase cell in HCC cells transfected with siRNAs against HMGB1 was detected by the EdU incorporation assays. Scale bars, 100μm. Arrows: EdU-positive cells. (C) Quantification of the number of EdU-positive cells is shown in the right. n=8, p Value was determined by Student's t test (*p < 0.05, ** p < 0.01, *** p < 0.001, ns: not significant). (D-E) The proliferation ability of with or without knockdown HMGB1 in SK-Hep-1 and SMMC-7721 cells evaluated by CCK8 assay at various time points. The experiments were performed by triplicate. p Value was determined by factorial design ANOVA. (*p < 0.05, ** p < 0.01, *** p < 0.001, ns: not significant).
HMGB1 confers cisplatin resistance to HCC cells via HIF-1α
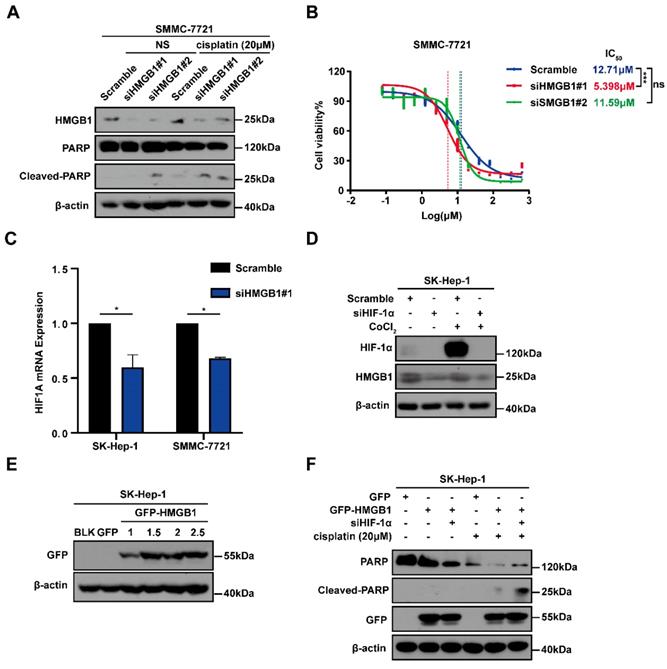
To explore the potential involvement of HMGB1 in cisplatin resistance in HCC cells, we first treated SMMC-7721 cells with or without HMGB1 knockdown and subsequently cultured them with normal saline (NS) or cisplatin (20μM) for 24 hours. Using immunoblotting, we detected the apoptosis-related protein cleaved-PARP expression, which showed that HMGB1 silencing promoted cell apoptosis in cisplatin treatment (Fig. 3A). We then examined cisplatin IC50 in HCC cells with or without HMGB1 knockdown after culturing them with the indicated cisplatin and using CCK8 assays to verify cell viability. We observed that HMGB1 silencing significantly reduced IC50 values as compared with normal controls (Fig. 3B). In the previous experiment, we found that HIF-1α was also upregulated in tumor tissues synchronized with HMGB1 (Fig. 1B). We, therefore, explored the relationship between HMGB1 and HIF-1α. RT-qPCR was used to examine mRNA expression of HIF-1α in HMGB1-silenced HCC cells. The mRNA expression of HIF-1α was downregulated by HMGB1 knockdown in both SK-Hep-1 and SMMC-7721 cells (Fig. 3C). It has been reported that HIF-1α levels were elevated in response to cobalt chloride (CoCl2) [16], so we treated cells with CoCl2 to mimic overexpression of HIF-1α. Immunoblotting analysis showed that HMGB1 was significantly upregulated under CoCl2, and deficiency of HIF-1α suppressed the CoCl2-induced increase in HMGB1 expression levels (Fig. 3D). The above results suggested that there is a positive regulatory loop between HMGB1 and HIF-1α. Because HMGB1 induced cisplatin resistance in HCC cells, we speculated that HIF-1α is involved in this process. To validate this hypothesis, we first transfected SK-Hep-1 cells with plasmid to obtain cells with overexpressed HMGB1, verified using immunoblotting (Fig. 3E). We then silenced HIF-1α in overexpressed HMGB1 HCC cells, cultured them with cisplatin (20μM) for 24 hours, and collected the lysates for immunoblotting. The results showed that the protective influence of HMGB1 overexpression on cisplatin-induced cell apoptosis was significantly reversed by HIF-1α deficiency (Fig. 3F). These findings demonstrate that HMGB1 contributed to cisplatin resistance in HCC cells via HIF-1α.
HMGB1 conferred cisplatin resistance to HCC cells via HIF-1α. (A) Lysates from SMMC-7721 cells with or without HMGB1 knockdown after normal saline (NS) or cisplatin treatment (20μM, 24h) were subjected to immunoblotting. (B) Dose response of the cisplatin treatment was assessed by optical density in SMMC-7721 cell lines with or without HMGB1 knockdown. The experiments were performed by triplicate. (C) Total RNAs from SK-Hep-1 and SMMC-7721 cells with or without HMGB1 knockdown were subjected to RT-qPCR. p Value was determined by Student's t test (*p < 0.05, ** p < 0.01, *** p < 0.001, ns: not significant). (D) Lysates from SK-Hep-1 cells with or without HIF-1α knockdown and treated with CoCl2 (100mM, 24h) were subjected to immunoblotting. (E) Validation of the transfection efficiency of HMGB1 plasmid by immunoblotting. (F) Lysates from SK-Hep-1 cells transfected as indicated and treated with NS or cisplatin (20μM, 24 hours) were subjected to immunoblotting.
Positive feedback regulatory loop between HMGB1 and HIF-1α is mediated by NF-κB
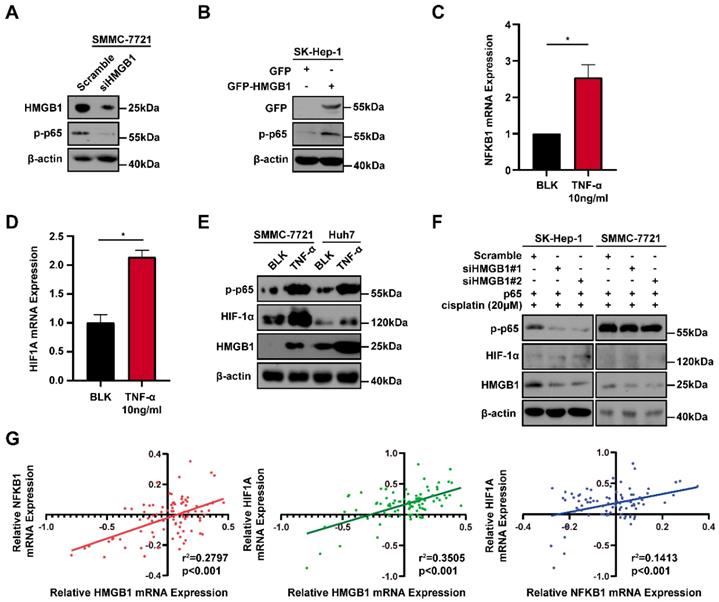
Because the positive regulatory loop between HMGB1 and HIF-1α induced cisplatin resistance, we sought to further explore the mechanism of this loop. The phosphorylated p65 protein level, which is an essential contributor to NF-κB activation was detected using immunoblotting [17]. HMGB1 knockdown reduced the phosphorylated p65 level and HMGB1 overexpression increased the phosphorylated p65 level (Figs. 4A and 4B). These results suggest that HMGB1 could activate NF-κB signaling. It has been reported that tumor necrosis factor-α (TNF-α) can activate receptors and trigger potent and persistent activation of NF-қB signaling [18]. Therefore, we tested the effect of NF-κB activation on HIF-1α using TNF-α. RT-qPCR showed that the mRNA expressions of NFKB1 and HIF1A were significantly upregulated by TNF-α (10ng/mL) treatment (Figs. 4C and 4D). Furthermore, we detected HIF-1α and HMGB1 protein levels in SMMC-7721 and Huh7 human liver cancer cells after TNF-α (10ng/mL) treatment. Immunoblotting showed that both HIF-1α and HMGB1 were increased in the TNF-α treatment groups (Fig. 4E). We then silenced HMGB1 in p65-overexpressed HCC cells, cultured them with cisplatin (20μM) for 24 hours, and collected the lysates. Immunoblotting showed that the supportive regulation of p65 overexpression on phosphorylated p65 level was reversed when combined with HMGB1 knockdown in both SK-Hep-1 and SMMC-7721 cells. In addition, suppressive regulation of HMGB1 silencing on HIF-1α was rescued in NF-κB-p65 overexpression (Fig. 4F). To confirm their association, we analyzed data from the GSE364 dataset, the linear regression results showed that HMGB1 mRNA expression positively correlated with NF-κB and HIF-1α mRNA levels, NF-κB mRNA expression also positively correlated with HIF-1α mRNA levels (Fig. 4G). Taken together, these results suggest that the HMGB1/NF-κB/HIF-1α positive feedback loop makes HMGB1 self-enforcing in HCC cells (Fig. 5).
Discussion
Our findings led us to reach several relevant conclusions.
First, we identified that HMGB1 was significantly up-regulated in both cisplatin-treated liver cancer cells and tumor tissues; this, also correlated with poor prognosis in patients with HCC. The results of multivariate analysis results showed that portal vein invasion, alcohol consumption, and HMGB1 expression were significant and independent prognostic factors that could be associated with OS in patients with HCC.
Subsequently, to investigate the potential biological functions of HMGB1 in tumors, we generated HMGB1 knockdown HCC cells. EdU and CCK8 assays showed that reduced HMGB1 expression can inhibit proliferation and promote apoptosis of HCC cells.
Furthermore, we found that HMGB1 was significantly elevated after cisplatin treatment, and HMGB1 deficiency increased the expression of apoptosis-related protein. In addition, our results revealed that the protective influence of HMGB1 overexpression on cisplatin-induced cell apoptosis was significantly reversed by HIF-1α deficiency. These findings indicate that HMGB1 promoted cisplatin resistance in HCC cells via HIF-1α.
A positive feedback regulatory loop between HMGB1 and HIF-1α is mediated by NF-κB. (A) Lysates from SMMC-7721 cells with or without HMGB1 knockdown were subjected to immunoblotting. p-p65: phosphorylated p65 (B) Lysates from SK-Hep-1 cells transfected with HMGB1 plasmid were subjected to immunoblotting. (C-D) Total RNAs from SMMC-7721 cells treated with TNF-α (10ng/mL, 24h) were subjected to RT-qPCR. p Value was determined by Student's t test (*p < 0.05, ** p < 0.01, *** p < 0.001, ns: not significant). (E) Lysates from SMMC-7721 cells and Huh7 cells treated with TNF-α (10ng/mL, 24h) were subjected to immunoblotting. (F) Lysates from SK-Hep-1 and SMMC-7721 cells transfected as indicated and treated with NS or cisplatin (20μM, 24 hours) were subjected to immunoblotting. (G) Correlation analysis among HMGB1, NF-κB and HIF-1α using the GSE364 dataset.
The HMGB1/NF-κB/HIF-1α positive feedback loop makes HMGB1 self-enforcing in HCC cells.
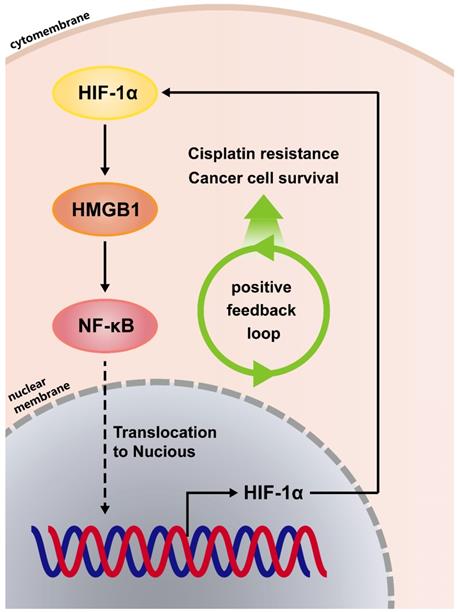
Finally, we further explored the mechanism between HIF-1α and HMGB1. HMGB1 could activate NF-κB signaling, and both HIF-1α mRNA and protein levels were increased in the follow-up step. In addition, reciprocally positive connections were found among HMGB1, NF-κB and HIF-1α. As shown in Fig. 5, we found that HMGB1 self-enforces via the HMGB1/NF-κB/HIF-1α positive feedback loop to promote cisplatin resistance in HCC cells.
The role of HMGB1 has been reported by research groups worldwide in the genesis of different cancers, such as lung and breast cancer. Many investigations have suggested that HMGB1 gene polymorphisms are correlated with risk factors of lung cancer development [19]. A recent report showed that HMGB1 may be an important factor in the development of chemoresistance in cervical cancer [20]. Studies have also reported that chemoresistance of cancer cells is regulated via HMGB1-mediated cell autophagy [21, 22]. This study confirmed that HMGB1 promotes cancer and takes part in cisplatin resistance. What is different to the classical HMGB1/RAGE/autophagy pathway, however, is that we verified hepatocellular carcinoma cells resistance to cisplatin treatment by HMGB1 via HMGB1/NF-κB/HIF-1α feedback loop; HMGB1 not only plays the role of cisplatin resistance through this positive feedback loop, but also promotes its high expression in tumor cells.
The NFκB proteins are key regulators of cell proliferation, inhibit apoptosis, promote cell migration and invasion, and stimulate angiogenesis and metastasis [23]. NF-κB also regulates the survival and proliferation of tumor cells following cancer chemotherapy [24]. When NF-κB was activated, it translocated to the nucleus, where it acted as a nuclear transcription factor [25]. It has been shown that NF‐κB is a direct modulator of HIF-1α expression [26]. In our study, we verified through RT-qPCR and immunoblotting that TNF-α activated NF‐κB and followed by stimulating HIF-1α.
The nuclear transcription factor, HIF-1α, is a crucial biomarker of the hypoxia tumor microenvironment, which is due to the rapid proliferation of tumor cells [27]. Studies have reported that HMGB1 recruited macrophages into HCC tissue via HIF-1α, or HMGB1-regulated HIF-1α to promote the tumor malignancy [28, 29]. It can be seen that HMGB1 and HIF-1α act as partners in promoting cancer. Here we reported that the protective influence of HMGB1 overexpression on cisplatin-induced cell apoptosis was significantly reversed by HIF-1α deficiency. Our findings verified that HMGB1 induced cisplatin resistance in HCC cells via HIF-1α, enriched the studies on the role of HMGB1 and HIF-1α in promoting cancer, and it will be interesting to further determine their synergism to induce cisplatin resistance in vivo.
It has been demonstrated that NF-κB and HIF-1α play a role in the development of many cancers, such as LMP1-induced HIF-1α transcription via ERK1/2/NF-κB pathway in nasopharyngeal carcinoma cells to promote angiogenesis[30]; metformin-sensitized response of oral squamous cell carcinoma to cisplatin treatment through inhibition of the NF-κB/HIF-1α signal axis[31], and our study finding that show HMGB1-involvement in the NF-κB/HIF-1α pathway, which caused cisplatin resistance in HCC cells. It has been shown that extracellular HMGB1 induces angiogenesis and promotes rheumatoid arthritis via NF-κB/HIF-1α activation, and cilostazol inhibits NF-κB-mediated transcription leading to the inhibition of synovial angiogenesis [32]. In our study, we provide new evidence of a cytoplasmic HMGB1 regulation mechanism whereby a HMGB1/NF-κB/HIF-1α feedback loop regulates HMGB1 expression and function in HCC cells, thus suggesting that suppressing the link in this loop may stop the vicious circle and achieve therapeutic effects.
In summary, for the first time, we confirmed a reciprocal regulatory interaction loop between HMGB1, NF-κB, and HIF-1α, which implements HMGB1 self-enforcing, leading to cisplatin resistance in HCC. The inhibition of this positive regulatory loop could be a valuable pharmaceutical target to disrupt HMGB1 self-enforcing and reversing cisplatin resistance in clinical antitumor therapeutics [33].
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81773008, 81672756), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015), and Guangdong Natural Science Foundation (Grant No. 2017A030311023). Guangzhou Science and Technology Program projects (No.201804010044). We gratefully recognize the patients who participated in this study. We thank Wenwen Li and Editage (www.editage.cn) for English language editing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Ikeda M, Morizane C, Ueno M, Okusaka T, Ishii H, Furuse J. Chemotherapy for hepatocellular carcinoma: current status and future perspectives. Jpn J Clin Oncol. 2018;48:103-114
3. Song JE, Kim DY. Conventional vs drug-eluting beads transarterial chemoembolization for hepatocellular carcinoma. World J Hepatol. 2017;9:808-814
4. Buschauer S, Koch A, Wiggermann P, Muller M, Hellerbrand C. Hepatocellular carcinoma cells surviving doxorubicin treatment exhibit increased migratory potential and resistance to doxorubicin re-treatment in vitro. Oncol Lett. 2018;15:4635-4640
5. Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L. et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1-116
6. Deng M, Scott MJ, Fan J, Billiar TR. Location is the key to function: HMGB1 in sepsis and trauma-induced inflammation. J Leukoc Biol. 2019;106:161-169
7. Mandke P, Vasquez KM. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: Implications in DNA repair and immune responses. DNA Repair (Amst). 2019;83:102701
8. Liu Y, Prasad R, Wilson SH. HMGB1: roles in base excision repair and related function. Biochim Biophys Acta. 2010;1799:119-130
9. Xu T, Jiang L, Wang Z. The progression of HMGB1-induced autophagy in cancer biology. Onco Targets Ther. 2019;12:365-377
10. Khambu B, Huda N, Chen X, Antoine DJ, Li Y, Dai G. et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J Clin Invest. 2018;128:2419-2435
11. Bianchi ME, Crippa MP, Manfredi AA, Mezzapelle R, Rovere Querini P, Venereau E. High-mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol Rev. 2017;280:74-82
12. Lv DJ, Song XL, Huang B, Yu YZ, Shu FP, Wang C. et al. HMGB1 Promotes Prostate Cancer Development and Metastasis by Interacting with Brahma-Related Gene 1 and Activating the Akt Signaling Pathway. Theranostics. 2019;9:5166-5182
13. Tripathi A, Shrinet K, Kumar A. HMGB1 protein as a novel target for cancer. Toxicol Rep. 2019;6:253-261
14. Wu L, Yang L. The function and mechanism of HMGB1 in lung cancer and its potential therapeutic implications. Oncol Lett. 2018;15:6799-6805
15. Wang X, Xiang L, Li H, Chen P, Feng Y, Zhang J. et al. The Role of HMGB1 Signaling Pathway in the Development and Progression of Hepatocellular Carcinoma: A Review. Int J Mol Sci. 2015;16:22527-22540
16. Masoud GN, Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378-389
17. Pradere JP, Hernandez C, Koppe C, Friedman RA, Luedde T, Schwabe RF. Negative regulation of NF-kappaB p65 activity by serine 536 phosphorylation. Sci Signal. 2016;9:ra85
18. Webb LV, Ley SC, Seddon B. TNF activation of NF-kappaB is essential for development of single-positive thymocytes. J Exp Med. 2016;213:1399-1407
19. Hu W, Liu PY, Yang YC, Chen PC, Su CM, Chao CC. et al. Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects. Int J Med Sci. 2017;14:1197-1202
20. Xia J, Yu X, Song X, Li G, Mao X, Zhang Y. Inhibiting the cytoplasmic location of HMGB1 reverses cisplatin resistance in human cervical cancer cells. Mol Med Rep. 2017;15:488-494
21. Shi Y, Gong W, Lu L, Wang Y, Ren J. Upregulation of miR-129-5p increases the sensitivity to Taxol through inhibiting HMGB1-mediated cell autophagy in breast cancer MCF-7 cells. Braz J Med Biol Res. 2019;52:e8657
22. Pan B, Chen D, Huang J, Wang R, Feng B, Song H. et al. HMGB1-mediated autophagy promotes docetaxel resistance in human lung adenocarcinoma. Mol Cancer. 2014;13:165
23. Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18:309-324
24. Tian M, Tian D, Qiao X, Li J, Zhang L. Modulation of Myb-induced NF-kB -STAT3 signaling and resulting cisplatin resistance in ovarian cancer by dietary factors. J Cell Physiol. 2019;234:21126-21134
25. Dolcet X, Llobet D, Pallares J, Matias-Guiu X. NF-kB in development and progression of human cancer. Virchows Arch. 2005;446:475-482
26. van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem J. 2008;412:477-484
27. Singh D, Arora R, Kaur P, Singh B, Mannan R, Arora S. Overexpression of hypoxia-inducible factor and metabolic pathways: possible targets of cancer. Cell Biosci. 2017;7:62
28. He H, Wang X, Chen J, Sun L, Sun H, Xie K. High-Mobility Group Box 1 (HMGB1) Promotes Angiogenesis and Tumor Migration by Regulating Hypoxia-Inducible Factor 1 (HIF-1alpha) Expression via the Phosphatidylinositol 3-Kinase (PI3K)/AKT Signaling Pathway in Breast Cancer Cells. Med Sci Monit. 2019;25:2352-2360
29. Tian Z, Hou X, Liu W, Han Z, Wei L. Macrophages and hepatocellular carcinoma. Cell Biosci. 2019;9:79
30. Sung WW, Chu YC, Chen PR, Liao MH, Lee JW. Positive regulation of HIF-1A expression by EBV oncoprotein LMP1 in nasopharyngeal carcinoma cells. Cancer Lett. 2016;382:21-31
31. Qi X, Xu W, Xie J, Wang Y, Han S, Wei Z. et al. Metformin sensitizes the response of oral squamous cell carcinoma to cisplatin treatment through inhibition of NF-kappaB/HIF-1alpha signal axis. Sci Rep. 2016;6:35788
32. Kim HY, Park SY, Lee SW, Lee HR, Lee WS, Rhim BY. et al. Inhibition of HMGB1-induced angiogenesis by cilostazol via SIRT1 activation in synovial fibroblasts from rheumatoid arthritis. PLoS One. 2014;9:e104743
33. Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell. 2017;168:37-57
Author contact
![]() Corresponding author: Prof. Li Liu, Hepatology Unit and Department of Infectious Diseases, Nanfang Hospital, Southern Medical University, Guangzhou 510515, China. E-mail: liulismu.edu.cn
Corresponding author: Prof. Li Liu, Hepatology Unit and Department of Infectious Diseases, Nanfang Hospital, Southern Medical University, Guangzhou 510515, China. E-mail: liulismu.edu.cn

 Global reach, higher impact
Global reach, higher impact