Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(14):4007-4014. doi:10.7150/jca.37164 This issue Cite
Research Paper
Role of the E3 Ubiquitin Ligase TRIM4 in Predicting the Prognosis of Hepatocellular Carcinoma
Department of General Surgery, Qilu Hospital, Shandong University, Jinan 250012, China
*Those authors contributed equally to this work.
Received 2019-5-30; Accepted 2020-3-3; Published 2020-4-6
Abstract
The E3 ubiquitin ligase TRIM4 has been reported to regulate the assembly of the antiviral signalling complex, induce mitochondrial aggregation and sensitize cells to H2O2-induced death. However, the relationship between TRIM4 and human malignancies, including hepatocellular carcinoma (HCC), is unclear. In this study, we detected the expression of TRIM4 in 134 pairs of HCC tissues and peritumoural tissues and investigated the association of TRIM4 expression with the prognosis of HCC. We found that the TRIM4 expression was much lower in HCC tissues than in peritumoural tissues and was significantly associated with vascular invasion, tumour capsule and Hong Kong Liver Cancer (HKLC) stage. Univariate and multivariate analyses revealed that the TRIM4 expression was an independent prognostic factor for overall survival (OS) and recurrence-free survival (RFS) in our HCC cohort. Patients with higher TRIM4 expression had a lower incidence of intrahepatic recurrence and a higher OS rate (p<0.001 and p<0.01, respectively). These results were further validated in another independent cohort of 200 HCC patients. In conclusion, the TRIM4 level in HCC tissues is an independent prognostic factor for HCC patients. Close clinical monitoring is recommended for patients with low TRIM4 expression.
Keywords: E3 ubiquitin ligase, TRIM4, hepatocellular carcinoma, progression
Introduction
Ubiquitination is a multistep enzymatic process that results in the attachment of ubiquitin, or chains of ubiquitin, to the target protein. Previous studies have confirmed that ubiquitination is an important posttranslational modification and is involved in the regulation of a number of intracellular events, including transcription, the cell cycle, apoptosis, tumorigenesis and development [1-4]. The biochemical reaction of ubiquitination requires several essential enzymes, including E1, E2 and E3 enzymes. Among these enzymes, E3 ubiquitin ligases are regarded as receptors for recognizing specific target proteins [1-5].
Tripartite motif (TRIM)-containing proteins serve an important function as E3 ubiquitin ligases. Generally, TRIM proteins are defined by the presence of an N-terminal RING finger, one or two B-boxes and a coiled-coil domain. A RING domain catalyses ubiquitin chain formation on its substrate via binding to a ubiquitin-conjugating enzyme (E2). A B-box domain may also exhibit E3 ligase activity in certain TRIM proteins that lack a functional RING domain. TRIM genes have been subclassified based on differences in their C-terminal domains [6-8]. Recent studies have shown that several TRIM proteins are involved in the tumorigenesis and development of solid malignancies, including hepatocellular carcinoma (HCC) [9-10]. For example, TRIM24 can function as a liver-specific tumour suppressor in mice and co-regulate hepatocarcinogenesis in an antagonistic manner with Rara [9]. In addition, TRIM16 has been proven to inhibit cell migration, invasion and epithelial-mesenchymal transition in HCC to suppress tumour progression [10].
TRIM4 is a member of the TRIM family and its cellular function remains unclear. TRIM4 has been shown to regulate the assembly of the mitochondrial antiviral signalling complex and K63-linked ubiquitination of RIG-1 [11]. Recently, TRIM4 has also been reported to transiently interact with mitochondria, induce mitochondrial aggregation and sensitize HSK293T cells to H2O2-induced cell death [12]. However, little is known about the biological effects of TRIM4 on HCC. In this study, we detected the expression of TRIM4 in HCC tissues and corresponding peritumoural tissues and investigated the association of TRIM4 expression with HCC patient's prognosis. Our data demonstrate that decreased TRIM4 expression is significantly associated with HCC progression and worse patient survival and is an independent prognostic factor for postoperative recurrence.
Material and Methods
Patients and Tissue Samples
This study was conducted with the approval of the Ethics Committee of Qilu Hospital, Shandong University (Jinan, China). Informed consent was obtained from each patient. Eight paired frozen HCC and adjacent non-tumour liver tissue samples were collected from patients who underwent curative resection at Qilu Hospital, Shandong University, between January 2010 and December 2012. Archival specimens used for tissue microarrays (TMAs) were obtained from HCC patients who underwent curative resection. The inclusion and exclusion criteria of the patient cohorts included (a) having a distinctive pathologic diagnosis of HCC, (b) having no anticancer treatment before liver resection, (c) having curative liver resection, (d) having suitable formalin-fixed, paraffin-embedded tissues and (e) having complete clinicopathologic and follow-up data. All patients were followed regularly in the outpatient clinic and were monitored prospectively for recurrence according to a standard protocol as previously described [13-15].
Western Blot Analysis
Proteins extracted from tissues were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride membranes, and incubated with a TRIM4 antibody and a secondary antibody. Proteins were visualized using HRP-conjugated IgG and Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Construction of Tissue Microarrays and Immunohistochemistry
Tissue microarray (TMA) was constructed as described in our previous studies [13-15]. Immunohistochemical staining for TRIM4 was performed on formalin-fixed sample sections from the TMA. The slides were dewaxed and washed three times with xylene. Tissues were then rehydrated, and endogenous peroxidase activity was blocked. After antigen retrieval, the TMA slides were incubated in 5% normal goat serum at room temperature to reduce nonspecific reactions. Subsequently, the slides were incubated overnight at 4°C with rabbit monoclonal anti-TRIM4 antibody, incubated with secondary antibody, and then stained with diaminobenzidine (DAB)-H2O2. Finally, the TMA slides were counterstained with haematoxylin, dehydrated, and mounted with a coverslip using standard medium. Phosphate buffer solution was used instead of the primary antibody as a negative control.
Evaluation of Immunostaining Intensity
TRIM4 was immunohistochemically stained in the cell cytoplasm. Cases were scored independently based on the intensity of cellular staining and the proportion of stained tumour cells. The product of the intensity and proportion scores was used to determine the level of TRIM4, as described in our previous studies [13, 14].
Statistical Analysis
Fisher's exact test or χ2 test was used to evaluate relationships between TRIM4 expression and clinicopathological parameters of patients with HCC. The relative prognostic significance of variables for predicting OS and RFS were analysed using Cox proportional hazards regression models. All p-values were two-sided, and p<0.05 was regarded as statistically significant. All statistical analyses were performed using SPSS 21.0 for Windows (SPSS, Chicago, IL).
Results
TRIM4 Expression in Human HCC Tissues
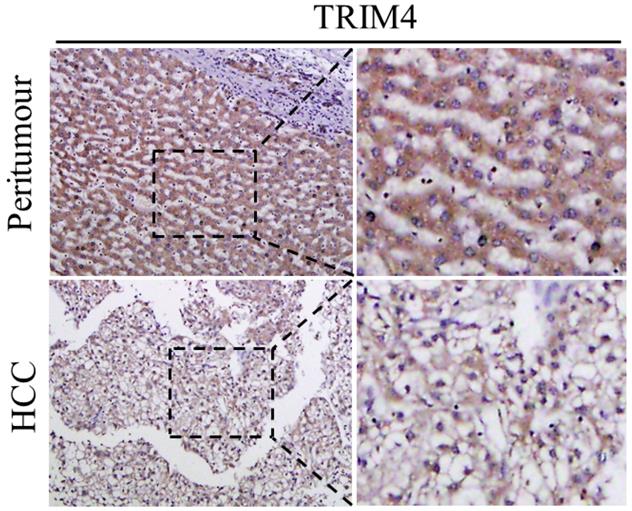
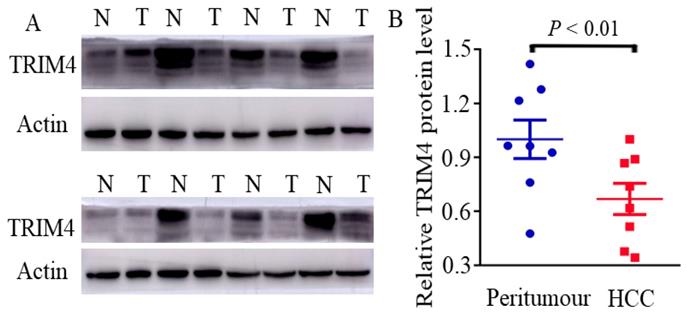
First, immunohistochemical staining was used to detect TRIM4 expression in eight human HCC tissues and corresponding peritumoural tissues. Our results showed that TRIM4 expression was much lower in human HCC tissues than in corresponding peritumoural tissues (Figure 1). Second, western blotting was performed to determine TRIM4 protein levels in these samples. These findings, in accordance with the immunohistochemical staining results, also revealed that TRIM4 levels were significantly lower in human HCC tissues than in peritumoural tissues (p<0.01, Figure 2A and B).
Clinicopathological Features of TMAs
We further used immunohistochemistry and the TMA technique to evaluate TRIM4 levels in human HCC tissues. In our present study, samples from 134 primary HCC cases were stained for TRIM4. Individuals in the patient cohort, which consisted of 115 males and 19 females, had a median age of 57 years (range: 26 to 74 years) at the time of surgery. There were 109 HBsAg-positive patients and 25 HBsAg-negative patients; all of the patients were negative for HCV. Patients were histopathologically categorized according to the Hong Kong Liver Cancer (HKLC) staging system. An early-stage tumour was defined as a tumour with diameter ≤5 cm, ≤3 tumour nodules and no intrahepatic venous invasion [16]. Tumours beyond the early stage were categorized as advanced-stage tumours. There were 57 early-stage tumours and 77 advanced-stage tumours based on the HKLC staging system.
Immunohistochemical analysis of TRIM4 expression in HCC and peritumoural tissues.
Immunoblotting analysis of TRIM4 levels in HCC and peritumoural tissues. (N: peritumoural tissue; T: HCC tissue)
Relationship between TRIM4 Expression and the Clinicopathological Parameters of HCC Patients
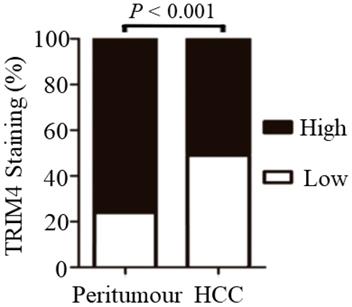
Significant differences in TRIM4 staining patterns were generally observed between tumour-adjacent tissues and primary HCC tissues. Statistical analysis revealed that TRIM4 expression was much lower in HCC tissues than in matched peritumoural tissues (p<0.001, Figure 3).
Immunohistochemical analysis revealed that TRIM4 expression was much lower in HCC tissues than in matched peritumoural tissues
We then further investigated the correlations between TRIM4 expression and HCC patients' clinicopathological parameters. Based on TRIM4 staining scores (Figure 4), we divided our HCC cohort into the high TRIM4 cohort and the low TRIM4 cohort. Our results suggested that the TRIM4 level was associated with vascular invasion (p=0.029), tumour capsule (p<0.01) and HKLC stage (p=0.034); however, no significant associations were detected between TRIM4 expression and age, sex, tumour size, hepatitis B virus (HBV) infection, tumour number, cirrhosis, grade of tumour differentiation or serum alpha-fetoprotein (AFP) concentration (Table 1).
Correlation of TRIM4 Expression with the RFS and OS of HCC Patients
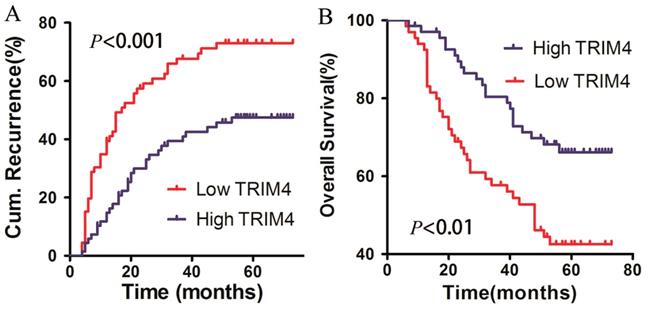
To determine the association between TRIM4 protein levels in HCC tissues and patient prognosis, all patients in our cohort were followed up to determine OS and RFS after surgery. The 1-, 2-, and 5-year cumulative incidences of intrahepatic recurrence in the high TRIM4 cohort were 14.78%, 29.99%, and 47.45%, respectively, while the 1-, 2-, and 5-year cumulative incidences of intrahepatic recurrence in the low TRIM4 cohort were 39.39%, 59.12%, and 73.03%, respectively. The high and low TRIM4 cohorts had significantly different cumulative incidences of intrahepatic recurrence (log-rank test, χ2=12.190, p<0.001, Figure 5A). The 1-, 2-, and 5-year OS rates of the high TRIM4 cohort were 97.04%, 87.94%, and 66.16%, respectively, significantly better than those of the low TRIM4 cohort (92.42%, 67.32%, and 42.52%, respectively) (χ2=8.874, p<0.01, Figure 5B).
Association of TRIM4 level with the Clinicopathological Parameters of 134 HCC Patients
| Clinicopathological Parameters | Total | High TRIM4 expression | ||
|---|---|---|---|---|
| N (%) | χ2 | p value | ||
| Sex | 134 | |||
| Male | 115 | 59(51.30%) | 0.101 | 0.751 |
| Female | 19 | 9(47.37%) | ||
| Age range, yr | 2.971 | 0.085 | ||
| ≤ 57 | 69 | 40(57.97%) | ||
| >57 | 65 | 28(43.08%) | ||
| Tumor size, cm | 1.883 | 0.170 | ||
| ≤5 | 73 | 41(56.16%) | ||
| >5 | 61 | 27(44.26%) | ||
| Tumor number | 0.170 | 0.680 | ||
| 1 | 114 | 57(50.00%) | ||
| ≥2 | 20 | 11(55.00%) | ||
| Grade of differentiation | 0.088 | 0.767 | ||
| I-II | 100 | 50(50.00%) | ||
| III-IV | 34 | 18(52.94%) | ||
| Vascular invasion | 4.740 | 0.029 | ||
| Yes | 41 | 15(36.59%) | ||
| No | 93 | 53(56.99%) | ||
| Tumor capsule | 9.655 | <0.01 | ||
| None | 61 | 22 (36.07%) | ||
| Yes | 73 | 46 (63.01%) | ||
| HBsAg | 0.093 | 0.761 | ||
| Positive | 109 | 56(51.38%) | ||
| Negative | 25 | 12(48.00%) | ||
| Cirrhosis | 2.502 | 0.114 | ||
| Positive | 74 | 33(44.60%) | ||
| Negative | 60 | 35(58.33%) | ||
| AFP, ng/ml | 0.448 | 0.503 | ||
| ≤400 | 85 | 45(52.94%) | ||
| >400 | 49 | 23(46.94%) | ||
| HKLC stage | 4.507 | 0.034 | ||
| Early | 57 | 35(61.40%) | ||
| Advanced | 77 | 33(42.86%) | ||
Multivariate Analyses of Predictive Factors for Intrahepatic Recurrence
Univariate analysis revealed that TRIM4 expression, tumour diameter, vascular invasion and tumour capsule were predictive factors for intrahepatic recurrence. Multivariate analysis revealed that TRIM4 expression (hazard ratio, 0.566; 95% CI, 0.345-0.930; p=0.025), tumour diameter (hazard ratio, 1.804; 95% CI, 1.131-2.876; p=0.013) and vascular invasion (hazard ratio, 1.802; 95% CI, 1.115-2.912; p=0.016) were independent predictive factors for RFS (Table 2).
Multivariate Analyses of Predictive Factors for the OS of HCC Patients
Univariate analyse revealed that TRIM4 expression, tumour diameter, and vascular invasion were the factors that significantly influenced the OS of HCC patients. Subsequently, the multivariate analyses further revealed that the TRIM4 level (hazard ratio, 0.550; 95% CI, 0.321-0.944; p=0.030), tumour diameter (hazard ratio, 2.191; 95% CI, 1.264-3.798; p<0.01), and vascular invasion (hazard ratio, 2.249; 95% CI, 1.322-3.826; p<0.01) were independent predictive factors for OS of HCC patients (Table 3).
Univariate and multivariate analysis of prognostic factors of RFS in 134 HCC patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| χ2 | p value | HR (95%Cl) | p value | |
| TRIM4 expression (high vs low) | 12.190 | <0.001 | 0.345-0.930 | 0.025 |
| Tumor diameter (>5 cm vs ≤5 cm) | 11.162 | 0.001 | 1.131-2.876 | 0.013 |
| Vascular invasion (yes vs no) | 14.189 | <0.001 | 1.115-2.912 | 0.016 |
| Tumor capsule (none vs yes) | 4.726 | 0.030 | 0.798-2.091 | 0.298 |
| Tumor number (≥2 vs 1) | 0.991 | 0.320 | - | n.a. |
| Sex (male vs female) | 1.000 | 0.317 | - | n.a. |
| Age (years) (≤57 vs>57) | 0.110 | 0.740 | - | n.a. |
| AFP (ng/ml) (≤400 vs >400) | 2.264 | 0.132 | - | n.a. |
| Tumor differentiation (III-IV vs I-II) | 0.859 | 0.354 | - | n.a. |
| Liver cirrhosis (yes vs none) | 0.007 | 0.932 | - | n.a. |
| HKLC stage (advanced vs early) | 16.275 | <0.001 | - | n.a. |
n.a., not applicable.
Univariate and multivariate analysis of prognostic factors of OS in 134 HCC patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| χ2 | p value | HR (95%Cl) | p value | |
| TRIM4 expression (high vs low) | 8.874 | <0.01 | 0.321-0.944 | 0.030 |
| Tumor diameter (cm) (>5 vs ≤5) | 14.497 | <0.001 | 1.264-3.798 | <0.01 |
| Vascular invasion (yes vs no) | 15.798 | <0.001 | 1.322-3.826 | <0.01 |
| Tumor capsule (none vs yes) | 0.385 | 0.535 | - | n.a. |
| Tumor number (≥2 vs 1) | 0.559 | 0.455 | - | n.a. |
| Sex (male vs female) | 0.308 | 0.579 | - | n.a. |
| Age (years) (≤57 vs >57) | 0.003 | 0.955 | - | n.a. |
| AFP (ng/ml) (≤400 vs >400) | 2.150 | 0.143 | - | n.a. |
| Tumor differentiation (III-IV vs I-II) | 1.411 | 0.235 | - | n.a. |
| Liver cirrhosis (positive vs negative) | 0.499 | 0.480 | - | n.a. |
| HKLC stage (advanced vs early) | 13.465 | <0.001 | - | n.a. |
n.a., not applicable.
Validation
We next validated the prognostic value of TRIM4 in another independent cohort of 200 HCC patients who underwent hepatectomy and had a median age of 50 years. The results we obtained were similar to those acquired for the training set. Patients with lower TRIM4 levels had a higher cumulative incidence of intrahepatic recurrence and a lower OS rate in this cohort of HCC patients. Multivariate analyses also confirmed that TRIM4 expression was an independent predictor of RFS and OS (Tables 4 and 5).
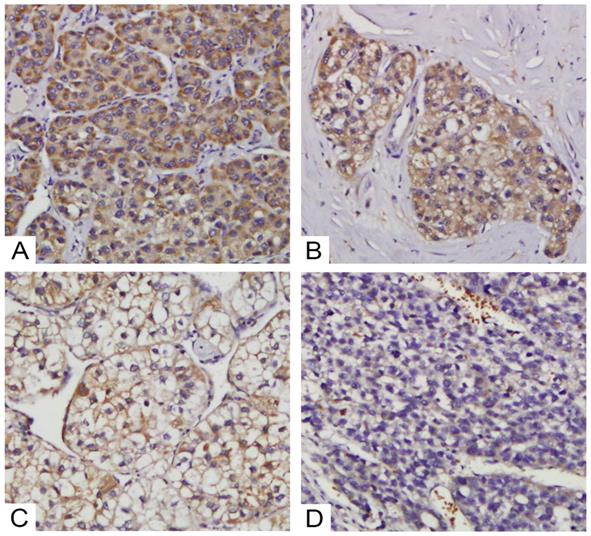
Various levels of TRIM4 staining were observed in HCC tissues. Representative images of staining in tumour samples: high expression (A and B) and low expression (C and D).
Kaplan-Meier curves for intrahepatic recurrence (A) and overall survival (B), comparing HCC patients with high and low TRIM4 levels.
Univariate and multivariate analysis of prognostic factors of RFS in 200 HCC patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| χ2 | p value | HR (95%Cl) | p value | |
| TRIM4 expression (high vs low) | 10.795 | 0.001 | 0.371-0.795 | 0.002 |
| Tumor diameter (cm) (>5 vs ≤5) | 0.130 | 0.719 | - | n.a. |
| Vascular invasion (yes vs no) | 2.707 | 0.100 | - | n.a. |
| Tumor capsule (none vs yes) | 1.744 | 0.187 | - | n.a. |
| Tumor number (≥2 vs 1) | 20.094 | <0.001 | 1.620-3.713 | <0.001 |
| Sex (male vs female) | 0.002 | 0.968 | - | n.a. |
| Age (years) (≤50 vs >50) | 1.020 | 0.313 | - | n.a. |
| AFP (ng/ml) (≤400 vs >400) | 0.163 | 0.686 | - | n.a. |
| Tumor differentiation (III-IV vs I-II) | 0.008 | 0.929 | - | n.a. |
| Liver cirrhosis (positive vs negative) | 3.090 | 0.079 | - | n.a. |
n.a., not applicable.
Univariate and multivariate analysis of prognostic factors of OS in 200 HCC patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| χ2 | p value | HR (95%Cl) | p value | |
| TRIM4 expression (high vs low) | 9.004 | <0.01 | 0.386-0.951 | 0.029 |
| Tumor diameter (cm) (>5 vs ≤5) | 1.338 | 0.247 | - | n.a. |
| Vascular invasion (yes vs no) | 8.596 | <0.01 | 0.916-2.477 | 0.106 |
| Tumor capsule (none vs yes) | 1.866 | 0.172 | - | n.a. |
| Tumor number (≥2 vs 1) | 12.678 | <0.001 | 1.240-3.185 | <0.01 |
| Sex (male vs female) | 0.148 | 0.701 | - | n.a. |
| Age (years) (≤50 vs >50) | 0.722 | 0.395 | - | n.a. |
| AFP (ng/ml) (≤400 vs >400) | 5.018 | 0.025 | 0.699-1.948 | 0.556 |
| Tumor differentiation (III-IV vs I-II) | 0.434 | 0.510 | - | n.a. |
| Liver cirrhosis (positive vs negative) | 4.463 | 0.035 | 0.904-3.633 | 0.094 |
n.a., not applicable.
Discussion
As the third leading cause of cancer mortality worldwide, HCC is one of the most prevalent malignancies and remains a major public health challenge. Partial hepatectomy is the most common curative treatment for HCC [17, 18], but the high incidence of intrahepatic recurrence and metastasis is the major challenge that must be overcome in order to improve the prognosis of HCC patients undergoing surgery. Therefore, it is important to identify predictive factors for HCC recurrence and metastasis to ensure close monitoring for patients with high recurrence risk and then utilize individualized therapy strategies.
The role of E3 ubiquitin ligases is to mediate the chemical transfer of the C-terminus of ubiquitin from an E2 ubiquitin-conjugating enzyme to a protein substrate, and ubiquitinated substrates undergo different fates depending on the site and type of ubiquitination. Traditionally, E3 ubiquitin ligases have been categorized into two classes of RING/U-box ligases and HECT ligases. Both types of ligases have been reported to lead to positive or negative regulation of multiple cancer-related signal pathways and maybe potential novel targets for anticancer therapies [13, 19].
TRIM4, a member of the TRIM family with a unique domain organization that belongs to the RING family of ubiquitin ligases [20, 21], is a cytoplasmic E3 ubiquitin ligase; this localization differs from that of certain other TRIM family members, such as TRIM24, which has been verified to be a nuclear protein. Evidence has demonstrated that TRIM4 can sensitize HEK293 cells to H2O2-induced cell death. However, the role of TRIM4 in hepatocarcinogenesis and HCC development remains to be elucidated. This study indicated that TRIM4 expression was much lower in human HCC tissues than in peritumoural tissues. The OS rate was significantly higher for HCC patients with high TRIM4 expression than for HCC patients with low TRIM4 expression. These results reveal that TRIM4 can be a valuable biomarker for evaluating HCC prognosis and that the loss of the TRIM4 protein in HCC tissues consistently suggests a worse prognosis.
Numerous studies have revealed that vascular invasion is an independent risk factor for the prognosis of HCC patients who underwent surgical treatment [22-24]. The presence of vascular invasion of the portal or hepatic veins is associated with a high risk of tumour recurrence and is regarded as a prerequisite for systemic tumour dissemination [25, 26]. Our study revealed that TRIM4 expression levels in HCC tissues were negatively associated with vascular invasion and the absence of a tumour capsule, indicating that TRIM4 may be a potential prognostic marker for HCC.
For HBV-related HCC, the HKLC staging system is thought to exhibit good prognostic accuracy, and the use of this system is associated with high therapeutic efficacy [16, 27]. In our cohort, 81.34% (109/134) of patients were positive for HBsAg. Therefore, our patients were categorized according to HKLC stage. We found that low TRIM4 expression in HCC is correlated with advanced HKLC stage. Therefore, we can propose that HCC cells with low TRIM4 protein levels may have higher invasive and metastatic potential, which promotes HCC progression and leads to a worse prognosis for HCC patients.
However, the molecular mechanisms by which TRIM4 regulates HCC progression must be further illuminated. Previous studies have revealed the role of some other TRIM family members in carcinogenesis or tumour progression. For example, TRIM29, which appears to be localized primarily to the cytoplasm, can bind to and stabilize Dvl-2; this interaction results in the release of β-catenin from the destruction complex and the activation of downstream β-catenin/TCF-regulated target genes and subsequently contributes to the development of pancreatic cancer [28]. TRIM25, which plays a dual role in regulating the p53/Mdm2 circuit, can not only increase the abundance of p53 and Mdm2 by inhibiting their ubiquitination and degradation by 26S proteasomes but also inhibit the activity of the p53 protein [29, 30]. Further research will provide evidence regarding whether an aberrant TRIM4 level can influence the Wnt and/or p53 signalling pathways.
In conclusion, our study revealed that TRIM4 expression levels in HCC tissues were an independent prognostic factor for OS and RFS in HCC patients. Close clinical monitoring is recommended for patients with low TRIM4 expression, and the biological functions of TRIM4 in regulating HCC progression require further elucidation.
Abbreviations
HCC: hepatocellular carcinoma; OS: overall survival; RFS: recurrence-free survival; TMAs: tissue microarrays; HBV: hepatitis B virus; AFP: alpha-fetoprotein.
Acknowledgements
This work was supported by the Taishan Scholars Program for Young Expert of Shandong Province (tsqn20161064), the National Natural Science Foundation of China (81602548, 81572328, 81874178), the Natural Science Foundation of Shandong Province (ZR2014HZ002, ZR2016HM06) and the Key Research and Development Project of Shandong Province (2018GSF118162).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li H, Li J, Chen L. et al. HERC3-Mediated SMAD7 Ubiquitination Degradation Promotes Autophagy-Induced EMT and Chemoresistance in Glioblastoma. Clin Cancer Res. 2019;25(12):3602-16
2. Bai L, Chen MM, Chen ZD. et al. F-box/WD Repeat-Containing Protein 5 Mediates the Ubiquitination of Apoptosis Signal-Regulating Kinase 1 and Exacerbates Nonalcoholic Steatohepatitis in Mice. Hepatology. 2019;70(6):1942-57
3. Jin Q, Martinez CA, Arcipowski KM. et al. USP7 Cooperates with NOTCH1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clin Cancer Res. 2019;25(1):222-39
4. Liu Y, Meng F, Wang J. et al. A Novel Oxoglutarate Dehydrogenase-Like Mediated miR-214/TWIST1 Negative Feedback Loop Inhibits Pancreatic Cancer Growth and Metastasis. Clin Cancer Res. 2019;25(17):5407-21
5. Metzger MB, Pruneda JN, Klevit RE. et al. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843(1):47-60
6. Grütter MG, Luban J. TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr Opin Virol. 2012;2(2):142-50
7. Streich FC Jr, Ronchi VP, Connick JP. et al. Tripartite motif ligases catalyse polyubiquitin chain formation through a cooperative allosteric mechanism. J Biol Chem. 2013;288(12):8209-21
8. Bell JL, Malyukova A, Holien JK. et al. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS One. 2012;7(5):e37470
9. Khetchoumian K, Teletin M, Tisserand J. et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39(12):1500-06
10. Li L, Dong L, Qu X. et al. Tripartite motif 16 inhibits hepatocellular carcinoma cell migration and invasion. Int J Oncol. 2016;48(4):1639-49
11. Yan J, Li Q, Mao AP. et al. TRIM4 modulates typeI interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J Mol Cell Biol. 2014;6(2):154-63
12. Tomar D, Prajapati P, Lavie J. et al. TRIM4: a novel mitochondrial interacting RING E3 ligase, sensitizes the cells to hydrogen peroxide (H2O2) induced cell death. Free RadicBiol Med. 2015;89:1036-48
13. Dong ZR, Sun D, Yang YF. et al. TMPRSS4 Drives Angiogenesis in Hepatocellular Carcinoma by Promoting HB-EGF Expression and Proteolytic Cleavage. Hepatology. 2019 Dec 22. doi: 10.1002/hep.31076. [Epub ahead of print]
14. Dong XF, Liu TQ, Zhi XT. et al. COX-2/PGE2 Axis Regulates HIF2α Activity to Promote Hepatocellular Carcinoma Hypoxic Response and Reduce the Sensitivity of Sorafenib Treatment. Clin Cancer Res. 2018;24(13):3204-16
15. Wang CH, Guo ZY, Chen ZT. et al. TMPRSS4 facilitates epithelial-mesenchymal transition of hepatocellular carcinoma and is a predictive marker for poor prognosis of patients after curative resection. Sci Rep. 2015;5:12366
16. Yau T, Tang VY, Yao TJ. et al. Development of Hong Kong Liver Cancer staging system with treatment stratification for patients with hepatocellular carcinoma. Gastroenterology. 2014;146(7):1691-700
17. Dong ZR, Zhang PF, Wang CH. et al. Postoperative adjuvant transcatheter arterial chemoembolization for resectable multiple hepatocellular carcinoma beyond the Milan criteria: a retrospective analysis. Am J Cancer Res. 2015;5(1):450-57
18. Li YT, Wu HL, Kao JH. et al. Expression of Metastatic Tumor Antigen 1 Splice Variant Correlates With Early Recurrence and Aggressive Features of Hepatitis B Virus-Associated Hepatocellular Carcinoma. Hepatology. 2019;70(1):184-97
19. Wu H, Yang TY, Li Y. et al. Tumor Necrosis Factor Receptor-Associated Factor 6 Promotes Hepatocarcinogenesis by Interacting With Histone Deacetylase 3 to Enhance c-Myc Gene Expression and Protein Stability. Hepatology. 2020;71(1):148-63
20. Tomar D, Singh R. TRIM family proteins: emerging class of RING E3 ligases asregulator of NF-kappaB pathway. Biol Cell. 2015;107:22-40
21. Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. 2012;770:27-37
22. Huynh H, Lee LY, Goh KY. et al. Infigratinib Mediates Vascular Normalization, Impairs Metastasis, and Improves Chemotherapy in Hepatocellular Carcinoma. Hepatology. 2019;69(3):943-58
23. Feng LH, Dong H, Lau WY. et al. Novel microvascular invasion-based prognostic nomograms to predict survival outcomes in patients after R0 resection for hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(2):293-303
24. Firl DJ, Sasaki K, Agopian VG. et al. Charting the Path Forward for Risk Prediction in Liver Transplant for Hepatocellular Carcinoma: International Validation of HALTHCC Among 4,089 Patients. Hepatology. 2019 Jun 27. doi: 10.1002/hep.30838
25. Thuluvath PJ. Vascular invasion is the most important predictor of survival in HCC, but how do we find it? J Clin Gastroenterol. 2009;43(2):101-2
26. Finn RS, Zhu AX, Farah W. et al. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: A systematic review and meta-analysis. Hepatology. 2018;67(1):422-35
27. Liu PH, Hsu CY, Lee YH. et al. Hong Kong Liver Cancer Staging System Is Associated With Better Performance for Hepatocellular Carcinoma: Special Emphasis on Viral Etiology. Medicine (Baltimore). 2015;94(41):e1772
28. Wang L, Heidt DG, Lee CJ. et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-caten in stabilization. Cancer Cell. 2009;15(3):207-19
29. Wang S, Kollipara RK, Humphries CG. et al. The ubiquitin ligase TRIM25 targets ERG for degradation in prostate cancer. Oncotarget. 2016;7(40):64921-31
30. Zhang P, Elabd S, Hammer S. et al. TRIM25 has adual function in the p53/Mdm2 circuit. Oncogene. 2015;34(46):5729-38
Author contact
![]() Corresponding author: Tao Li, M.D, Department of General Surgery, Qilu Hospital, Shandong University, 107 West Wen Hua Road, Jinan, 250012, China. Tel & Fax: +86-531-82166341. Email: litao7706com.
Corresponding author: Tao Li, M.D, Department of General Surgery, Qilu Hospital, Shandong University, 107 West Wen Hua Road, Jinan, 250012, China. Tel & Fax: +86-531-82166341. Email: litao7706com.

 Global reach, higher impact
Global reach, higher impact