Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(14):4073-4080. doi:10.7150/jca.40964 This issue Cite
Research Paper
Quantitative proteomic analysis of bile in extrahepatic cholangiocarcinoma patients
1. Department of Thoracic and Cardiovascular Surgery, Gachon University Gil Medical Center, Gachon University, Incheon, 21565, Republic of Korea
2. Center for information security technologies, Korea University
3. Department of Internal Medicine, Korea University Guro Hospital
Received 2019-10-7; Accepted 2020-3-29; Published 2020-4-7
Abstract
Background and Aims: Extrahepatic cholangiocarcinoma (CCA) without liver-fluke is increasing. Multifactorial carcinogenesis makes it hard to find biomarkers related to CCA. Although there are a few studies of bile proteomics, these showed different protein profiles because of having heterogeneous groups of patients and different sampling methods. Our aim was to identify the specific bile proteins of extrahepatic CCA patients.
Methods: We collected bile from 23 patients undergoing endoscopic nasobiliary drainage in Korea University Guro Hospital from May 2018 to January 2019. The CCA group included 18 patients diagnosed with extrahepatic CCA, and the control group included 5 patients with benign biliary conditions. We analyzed bile proteome using liquid chromatography mass spectrometry. We compared the relative abundance of various proteins in the CCA and control groups.
Results: In all, we identified a total of 245 proteins in the bile of CCA and control patients. Increased top 14 proteins in CCA patients were immunoglobulin kappa light chain, apolipoprotein B, inter-alpha-trypsin inhibitor heavy chain H4, apolipoprotein E, Mucin 5B, inter-alpha-trypsin inhibitor heavy chain H1, apolipoprotein A-IV, intercellular adhesion molecule 1, complement C7, complement C5, apolipoprotein C-III, albumin, antithrombin-III, and apolipoprotein A-II. However, the significantly increased proteins in bile of CCA patients comparing with control patients were immunoglobulin kappa light chain, apolipoprotein E, albumin, apolipoprotein A-I, antithrombin-III, α1-antitrypsin, serotransferrin, immunoglobulin heavy constant mu, immunoglobulin J chain, complement C4-A, and complement C3 (p<0.05).
Conclusions: In this study, we identified several proteins that were significantly increased in the bile of extrahepatic CCA. Further study is needed to validate them as potential tumor-associated proteins that may be potential biomarkers for CCA.
Keywords: Cholangiocarcinoma, Bile, Proteins
Introduction
Cholangiocarcinoma (CCA) is a lethal malignant tumor with dismal outcomes; diagnosis in its early stage is very important. The diagnosis of CCA, especially when CCA is accompanied with extrahepatic bile-duct stricture, is very difficult, even using blood tests, imaging studies, and endoscopic methods. Although the direct tissue sampling using endoscopic retrograde cholangiopancreatography (ERCP) is a proper diagnostic tool, there are some limitations for achieving adequate sample [1].
Several studies have investigated the possibilities of liquid biopsy using blood, bile juice, sweat, urine, and feces as a novel and noninvasive diagnostic method [2-5]. Cancer-derived protein can be released into bile by necrosis and apoptosis of tumor cells, and bile includes tumor-specific molecules and provides useful information during malignant transformation in real time [5,6]. Use of proteomics to detect biomarkers in bile may hold promise in aiding differentiation of malignant from benign biliary strictures. To date, a few studies have investigated the proteins in bile. Despite improvements in bile analysis, a promising diagnostic biomarker remains unknown, because the results were not consistent. Besides the diversity of bile proteins, there are the diversity of bile sampling, enrolled patients, and the anatomic location of CCA.
Thereafter, we explore the comparative bile analysis of patients with extrahepatic CCA and control patients without bile-duct disease.
Materials and Methods
Patients
Patients who had undergone ERCP with endoscopic nasobiliary drainage (ENBD) were enrolled in Korea University Guro Hospital from May 2018 to December 2019. The inclusion criterion was age > 18 years.
The patient group was 18 patients who had extrahepatic CCA based on histologic diagnosis. Patients with gallbladder or ampullary caner were excluded.
The control group was 5 patients who had undergone ENBD without biliary disease. They needed bile drainage for bile-duct injury after a hepatobiliary operation (2 patients) or benign ampullary stenosis (3 patients).
In both groups, we excluded patients with symptoms of cholangitis or turbid bile on visual exam and patients who could not have a normal diet. All recruited patients had at least six months follow-up to exclude the possibility of misdiagnosis as malignancy of a benign condition in three patients who had benign ampullary stenosis.
Informed consent was obtained from all patients. This study was approved by the Ethics Committee at Korea University Guro Hospital (approval number 2018GR0133).
Bile sampling
We obtained approximately 5 mL of fresh bile through the ENBD tube after drainage of stagnant old bile, about two or three days after the ERCP procedure. On same day, we obtained blood tests and clinical information. The collected bile samples were directly frozen at -80°C until analysis.
Experimental procedures
(1) Sample preparation
A bile sample of 1 mL was centrifuged at 10,000 x g and 4°C for 15 minutes. We mixed the supernatant collected after centrifugation with 9 mL of ice-cold methanol. The mixture was incubated overnight at -20°C. After confirming the precipitate, we centrifuged the sample at 14,000 x g, 4°C for 30 minutes and the supernatant was removed. After we repeated the centrifuging of the sample and removed of the supernatant, the pellet was dried in the air, then dissolved in lysis buffer (20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 10 % Glycerol, 1 % NP-40). We then measured its protein concentration with the Bradford Assay.
An aliquot of 100 ul (100 μg of lysate resuspended with 50 mM ammonium bicarbonate solution) was reduced with 10 mM DL-Dithiothreitol Solution by shaking for 1 hour at 37°C and was alkylated with 50 mM Iodoacetamide at room temperature and was shaken for 1 hour in the dark. To reduce the Dithiothreitol concentration to a final 1 mM with 1.000 μl volume, we added the 50 mM ammonium bicarbonate. The sample was then digested with trypsin at a protein-to-enzyme ratio of 40:1 at 37°C overnight.
To quench the digestion reaction, we added 1% trifluoroacetic acid (TFA). We used Waters Oasis MCX (Waters, Irand, Waters cat No. 186000252) cartridge to clean up the sample (MCX column work). The sorbent was equilibrated by adding 1 mL of methanol, and then washed with 1 mL of 0.1 % TFA. After adding 500 to 1,000 μl of acidified sample (< pH 2) to load, we washed it with 1 mL of 0.1 % TFA and 1 mL of methanol. The sample was eluted with 500 to 1,000 μl of elution buffer (50 % acetonitrile, 5 % Ammonium hydroxide). The eluate was dried to completion in a Speedvac (Thermo Fischer Scientific, San Jose, CA, USA), and then dissolved in 0.1 % formic acid for LC injection or stored at -20°C before the analysis.
(2) Liquid chromatography-mass spectrometry (LC-MS) configuration
We did LC analysis using a Thermo Scientific Eazynano LC II autosampler with a reversed-phase peptide trap EASY-Column (100 μm inner diameter, 2 cm length) and a reversed-phase analytical EASY-Column (75 μm inner diameter, 10 cm length, 3 μm particle size). The extract was injected in 1/10 volumes, and we did LC using 0.1% formic acid in water as buffer (A) and 0.1% formic acid in acetonitrile as buffer (B), at a flow rate of 300 nL/min.
Collision-induced dissociation (CID)-type fragmentations were activated by Automatic data-dependent LTQ-Orbitrap (20 MS2 (LTQ) + 1 Full MS (Orbitrap)) switching modes. The MS source conditions included the capillary voltage optimized to 1.9 kV, at 275°C, range 300 - 2,000 m/z. Resolution was 100,000. The dynamic exclusion with a repeat count of 1, exclusion duration of 180 seconds, list size of 300 activated.
(3) Mass spectrometry data analysis system
Each processed databit was subsequently transformed to the Sorcerer 2 and Scaffold 4 program. The search parameters used were as follows: 25 ppm tolerance for precursor ion mass and 1.0 Da for fragment ion mass. Finally, protein and peptide were displayed with threshold 80 %, minimum peptides 2, and quantified by Top 3 Total Ion Chromatogram (minimum value =0.01, Normalization) method. We also did protein classification based on functional annotations using Gene Ontology for molecular function, biological processes, and cellular component categories.
Results
We analyzed a total of 23 bile samples, from the 18 patients with extrahepatic cholangiocarcinoma and the 5 patients with benign biliary conditions in the control group. The baseline characteristics of the enrolled patients are summarized in Table 1.
Malignant vs. benign condition
Each of the 23 bile samples was processed by LC-MS analysis for protein identification and quantification.
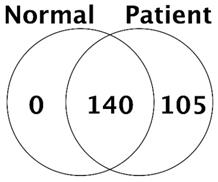
In all, we identified a total of 245 proteins in the bile of CCA patients and control patients. The number of common protein between control and CCA patient was 140, and 105 of protein was confirmed only in bile of CCA patients (Fig. 1).
Number of proteins identified in control (normal) and cancer patients
The increased proteins with spectral counts of more than twofold were immunoglobulin kappa light chain, apolipoprotein B, inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4), apolipoprotein E, Mucin 5B, inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1), apolipoprotein A-IV, intercellular adhesion molecule 1, complement C7, complement C5, apolipoprotein C-III, albumin, antithrombin-III, and apolipoprotein A-II (Table 2).
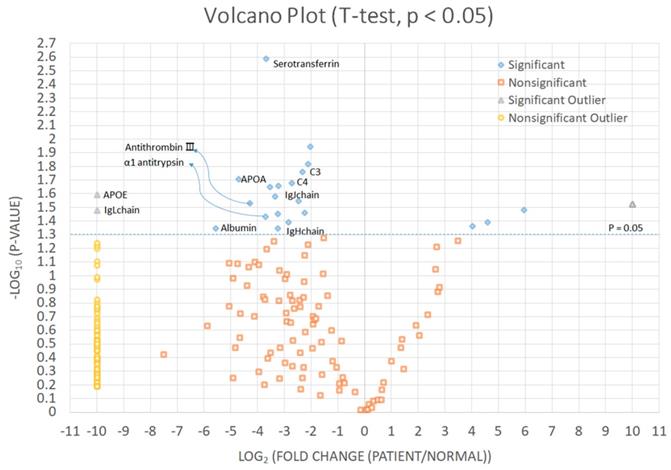
We identified significantly increased proteins in CCA patients than in the control group (Fig. 2 and Table 3). Immunoglobulin kappa light chain, apolipoprotein E, albumin, apolipoprotein A-I, antithrombin-III, α1-antitrypsin, serotransferrin, immunoglobulin heavy constant mu, immunoglobulin J chain, complement C4-A, and complement C3 were significantly increased in bile of CCA patients comparing with control patients.
Patient characteristics
| Number | Pathology | Age (yrs) | Sex | Albumin (g/dL) | Bilirubin (mg/dL) | AST (IU/L) | ALT (IU/L) | CA 19-9 (U/ml) |
|---|---|---|---|---|---|---|---|---|
| 1 | Control | 70 | F | 3.6 | 0.32 | 27 | 30 | |
| 2 | Control | 33 | F | 4.5 | 0.55 | 64 | 55 | |
| 3 | Control | 59 | F | 4.1 | 0.37 | 16 | 10 | |
| 4 | Control | 67 | F | 3.6 | 2.04 | 41 | 55 | |
| 5 | Control | 60 | M | 3.6 | 0.35 | 22 | 22 | |
| 6 | Hilar CC | 67 | M | 3.7 | 13.6 | 70 | 185 | 320 |
| 7 | CBD ca | 47 | F | 4.1 | 1.96 | 135 | 170 | 0.4 |
| 8 | CBD ca | 58 | M | 3.0 | 5.24 | 51 | 69 | 255 |
| 9 | CBD ca | 72 | M | 2.9 | 12.3 | 111 | 112 | 960 |
| 10 | CBD ca | 77 | M | 3.3 | 0.48 | 36 | 23 | 27 |
| 11 | CBD ca | 72 | F | 3.3 | 0.85 | 22 | 18 | 42 |
| 12 | CBD ca | 75 | M | 3.5 | 1.35 | 141 | 127 | 50.6 |
| 13 | Hilar CC | 56 | M | 3.9 | 24.5 | 32 | 46 | 12.8 |
| 14 | CBD ca | 70 | F | 3.3 | 7.34 | 93 | 88 | 476 |
| 15 | CBD ca | 75 | F | 3.3 | 0.41 | 26 | 81 | 20.5 |
| 16 | Hilar CC | 65 | M | 4.2 | 0.89 | 19 | 59 | 36.6 |
| 17 | Hilar CC | 83 | F | 2.7 | 24.3 | 118 | 75 | 950 |
| 18 | CBD ca | 79 | M | 3.9 | 4.19 | 138 | 153 | 13.6 |
| 19 | CBD ca | 62 | M | 3.8 | 4.2 | 55 | 105 | 253 |
| 20 | Hilar CC | 66 | M | 4.4 | 3.5 | 61 | 89 | 497 |
| 21 | CBD ca | 66 | F | 3.2 | 3.9 | 24 | 33 | 70.0 |
| 22 | CBD ca | 77 | M | 3.8 | 1.9 | 62 | 183 | 41.0 |
| 23 | Hilar CC | 90 | F | 3.7 | 4.3 | 84 | 54 | 341 |
CC, Cholangiocarcinoma; ca, cancer; CBD, common bile duct; AST, aspartate aminotransferase; ALT, alanine aminotransferase; CA 19-9, carbohydrate antigen 19-9.
Top 14 Proteins identified as more abundant in cholangiocarcinoma than control
| Protein | Gene | MW (kDa) | Average Spectral Counts | Log (Fold) | P value | |
|---|---|---|---|---|---|---|
| control | cancer | |||||
| Immunoglobulin kappa light chain | 23 | 0.00 | 2.27 | 27.50 | 0.03 | |
| Apolipoprotein B | APOB | 516 | 0.00 | 14.56 | 25.25 | 0.08 |
| Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 101 | 0.00 | 5.31 | 24.98 | 0.06 |
| Apolipoprotein E | APOE | 36 | 0.00 | 3.47 | 24.89 | 0.03 |
| Mucin-5B | MUC5B | 596 | 0.00 | 5.67 | 24.10 | 0.08 |
| Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1 | 101 | 0.00 | 3.54 | 23.84 | 0.06 |
| Apolipoprotein A-IV | APOA4 | 45 | 0.00 | 2.30 | 23.05 | 0.18 |
| Intercellular adhesion molecule 1 | ICAM1 | 58 | 0.00 | 2.36 | 22.78 | 0.06 |
| Complement C7 | C7 | 94 | 0.00 | 1.78 | 21.56 | 0.29 |
| Complement C5 | C5 | 188 | 0.00 | 1.50 | 20.61 | 0.44 |
| Apolipoprotein C-III | APOC3 | 13 | 0.00 | 1.00 | 20.61 | 0.19 |
| Albumin | ALB | 69 | 45.00 | 422.61 | 5.55 | 0.045 |
| Antithrombin-III | SERPINC1 | 53 | 1.00 | 6.25 | 4.25 | 0.03 |
| Apolipoprotein A-II | APOA2 | 11 | 1.00 | 2.45 | 2.20 | 0.26 |
Proteins identified as significantly increased in cholangiocarcinoma than control
| Protein | Gene | MW (kDa) | Average Spectral Counts | Log (Fold) | P value | |
|---|---|---|---|---|---|---|
| control | cancer | |||||
| Immunoglobulin kappa light chain | 23 | 0.00 | 2.27 | 27.50 | 0.033 | |
| Apolipoprotein E | APOE | 36 | 0.00 | 3.47 | 24.89 | 0.026 |
| Albumin | ALB | 69 | 45.00 | 422.61 | 5.55 | 0.045 |
| Apolipoprotein A-I | APOA1 | 31 | 2.50 | 17.28 | 4.70 | 0.02 |
| Antithrombin-III | SERPINC1 | 53 | 1.00 | 6.25 | 4.25 | 0.03 |
| Α1-antitrypsin | SERPINA1 | 47 | 17.75 | 61.33 | 3.70 | 0.037 |
| Serotransferrin | TF | 77 | 7.33 | 45.78 | 3.70 | 0.002 |
| Immunoglobulin heavy constant mu | IGHM | 49 | 3.00 | 6.78 | 3.25 | 0.045 |
| Immunoglobulin J chain | JCHAIN | 18 | 2.00 | 2.82 | 3.22 | 0.022 |
| Complement C4-A | C4A | 193 | 3.00 | 22.06 | 2.72 | 0.021 |
| Complement C3 | C3 | 187 | 11.67 | 39.61 | 2.32 | 0.017 |
Significantly increased proteins in cholangiocarcinoma patients
Bile of patients with CCA
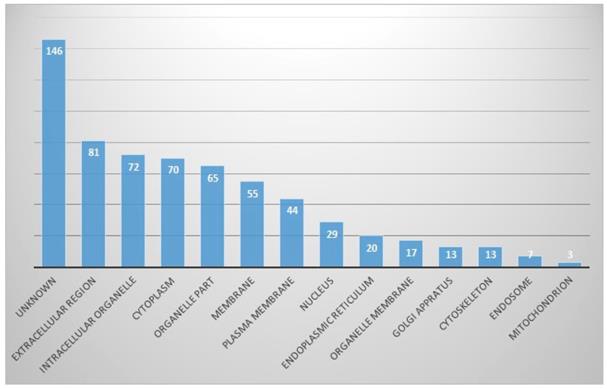
In the cellular component part, proteins originating from the extracellular region were the most common (13%), followed by the intracellular organelles (11%) and cytoplasmic part. Other identified proteins were from organelle parts, membranes, plasma membranes, nuclei, endoplasmic reticula, and so on. However, proteins of unknown origin were considerably high, at 14.2% (Fig. 3).
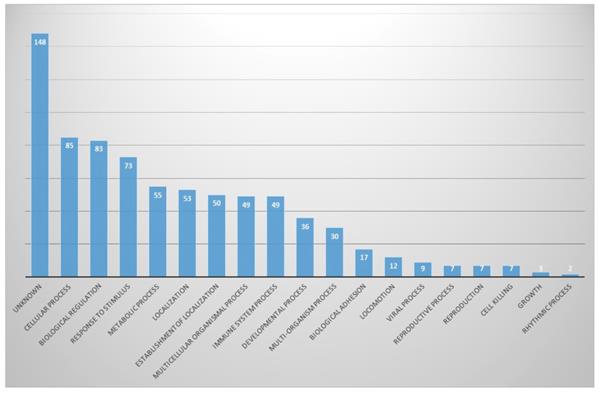
In biological aspects, proteins involved in cellular processes (11%) and biological regulation (10.7%) were the most abundant, and proteins involved in response to stimuli, metabolic process and localization were next. There were also many unknown proteins (19.0%) (Fig. 4).
Proteins with molecular functions were the most abundant (21.9%), and then proteins in categories of binding and catalytic activities (Fig. 5).
Discussion
Tissue sampling such as cytology or biopsy through ERCP is not enough. The sensitivity for malignancy diagnosis by ERCP with radiologic evaluation and biopsy is only 9-57% [7,8].
In cellular component part, proportion of identified proteins in bile
In biological process part, proportion of identified proteins in bile
In Molecular function part, proportion of identified proteins in bile
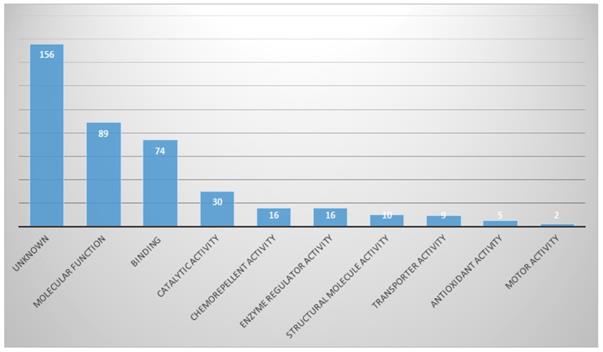
Bile analysis for diagnosis of CCA is a promising method, especially in differentiation of biliary stricture, because CCA which originates from the biliary epithelium might release proteins from cancer cells into bile [9]. Thus, proteomic analyses of bile for discovering CCA biomarkers have been done in many studies [9-15]. However, the bile-protein profiles of CCA were different from the studies. One reason for those differences is the diversity of CCA types enrolled in studies. Previous studies enrolled both of intrahepatic and extrahepatic CCA in the same study. CCA is a heterogeneous entity and is defined by anatomic location as intra or extrahepatic CCA [15]. For intrahepatic CCA, the different characterization depended on whether the liver is cirrhotic or not [15]. Thus, the tumor character differences by anatomical location might affect the bile-protein profile. One study demonstrated that intrahepatic CCA is a heterogeneous group affected by background liver conditions on tumor biology, while extrahepatic CCA is a more homogeneous subgroup [15]. For elimination of heterogeneity of cancer tissue, we enrolled only extrahepatic CCA in our study.
Previous studies of bile proteome analysis for CCA biomarkers were usually based on comparison between CCA and control groups. The control group showed diversity in the studies and contained various disease entities, such as benign biliary conditions, choledocholithiasis, and even primary sclerosing cholangitis (PSC) [9-15]. It has been known that proteins associated with the Wnt signaling pathway were highly expressed in CCA without PSC [16,17]. However, pathogenesis of PSC-associated CCA is more complex, and pro-inflammatory cytokines might have an important role [18]. One study of bile proteomic analysis showed that the most elevated pathways in PSC-associated CCA were inflammation-associated cytokine and chemokine pathway; however, the Wnt signaling pathway was mainly elevated in CCA without PSC [14], which suggested that enrolled patient diversity in the control group might affect the diversity of the bile-protein profile in the studies. Thus, we enrolled only benign biliary conditions that needed bile drainage and were not induced by biliary disease. Those patients seemed to be closer to a normal control.
Bile is produced by the hepatocytes and is secreted into bile ducts. It is stored in the gallbladder, which concentrates hepatic bile, hence the bile profile might be different from the collecting site of the biliary tree. Previous studies used concentrated bile from the gallbladder or common bile duct during cholecystectomy or ERCP, however, we used fresh secreted bile instead of concentrated bile in our study in order to avoid the change of bile profile during concentration or staying in the gallbladder.
Apolipoprotein E is involved in DNA synthesis, cell proliferation, angiogenesis, and metastasis, and changes of these functions might induce tumorigenesis or progression [19].
Apolipoprotein E overexpression has been reported in various cancers, such as gastric, lung, and prostate [20-22]. In our study, apolipoprotein E and apolipoprotein A-I in the bile of CCA were significantly increased. Apolipoprotein A-I is synthesized in the liver and small intestine [23]. Several studies have suggested that the serum level of apolipoprotein A-I could be a potential biomarker for CCA [24]. Apolipoprotein has a key role in lipoprotein metabolism but it's role in bile is not well known. It is presumed that apolipoproteins transport lipid in bile as in the serum, and that apoproteins are transported in the hepatocytes as vesicles and released into the bile across the canalicular membrane [25]. It is suggested that release of apolipoprotein into bile could be increased according to the change of bile lipid in cholangiocrcinoma.
Antithrombin is known as a protein related with hemostasis, but it has been studied as an anti-tumor effect through inhibiting a protease involved in metastasis and generating an anti-angiogenic molecule [26]. It was reported as a modulator of tumor cell migration and invasion in gastric cancer [27]. And the novel function as a diagnostic biomarker was studied in hepato-pancreato-biliary malignancy [28, 29]. In our result, antithrombin-III was significantly increased in the bile of CCA.
α1-antitrypsin is a glycoprotein produced by hepatocytes and mononuclear phagocytes, and is the principal human inhibitor of neutrophil elastase [30]. α1-antitrypsin was expressed in the tumors of CCA patients [31]. One study that analyzed the bile of six CCA patients showed α1-antitrypsin was an overexpressed protein and might be a marker for diagnosis of CCA [9]. In our study, α1-antitrypsin was elevated more than in the control group.
Serotransferrin is serum transferrin synthesized in the liver. Previous studies found that iron metabolism and iron regulatory proteins were altered in cancer and transferrin receptor-1 was increased in cholangiocarcinoma [32,33]. Changes in glycosylation of serotransferrin occur in CCA patients, and these glycoforms could be used as risk biomarkers and prognosis and diagnosis markers of CCA [34]. In our study, the level of serotransferrin was increased in CCA.
Liver has Kupffer cells and several immune cells and plays an important role in immune response [35]. Immunoglobulin A is known as an abundant protein in normal bile, contributes to immunological surveillance within the biliary system [36]. Clinically, increase in serum immunoglobulins are observed in specific hapatobiliary diseases such as autoimmune hepatitis (elevated IgG), primary biliary cirrhosis (elevated IgM) and alcoholic liver disease (elevated IgA), Immunoglobulin G4-related sclerosing cholangitis (elevated IgG) [36]. It has not been known whether immunoglobulins are increased in bile of biliary tract cancer. In our study, immunoglobulin kappa light chain showed the greatest change between the CCA and control groups. In addition to Immunoglobulin, complement proteins were reported to participate in local immune response in the biliary tract [35]. A recent study of plasma proteins analysis found that complement C3 and apolipoprotein C-III were essential proteins in HCC [37]. Our results showed that complement C4 and C3 were significantly elevated in CCA patients. Another study of bile proteomic analysis that distinguished CCA from PCS showed that ITIH4 was elevated in CCA [9], as is similar to our study but it was not significant. ITIH4 is a serum glycoprotein secreted mainly by the liver [38-40], and involved in stabilization of the extracellular matrix [40,41]. A previous study showed that ITIH4 plays a significant role in the pathway involved in TGF-β and fibrosis-independent carcinogenesis [38, 42]. In a study of pancreatic cancer biomarkers using pancreatic juice samples, potential biomarkers complement C5 and ITIH3 were elevated in cancer, but biliary obstruction had a significant effect on the performance of the markers [42].
Albumin is produced solely by hepatocytes and relatively abundant protein in bile. It was founded that the neonatal Fc Receptor is required for delivery of synthesized albumin to the blood stream and absence of this receptor results in increased albumin levels in the bile [43]. Cholangiocyte is differentiated from liver progenitor cells and alterations in bile secretion and abnormal bile composition can result in hepatocellular and/or bile duct injury [44,45].
Cholangiocarcinoma cell arising from cell proliferation after bile duct injury could be a form of undifferenciated cholangiocyte producing albumin. There are several reports on aberrant expression of albumin; it could be used to distinguish bile duct neoplasm from other metastatic adenocarcinoma [46-48].
In conclusion, we identified the bile proteins that were elevated in the extrahepatic CCA more than in the control group, who did not have a biliary disease, and the significantly increased proteins were immunoglobulin kappa light chain, apolipoprotein E, albumin, apolipoprotein A-I, antithrombin-III, α1-antitrypsin, serotransferrin, immunoglobulin heavy constant mu, immunoglobulin J chain, complement C4-A, and complement C3. Even though further study is needed, those proteins in bile have potential as biomarkers of CCA.
Acknowledgements
This research was supported by Korea University Grant (K1723411).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tavan J, Kawin L, Sittiruk R. et al. Novel serum biomarkers to differentiate cholangiocarcinoma from benign biliary tract diseases using a proteomic approach. Dis Markers. 2015;2015:105358
2. Ferhan AR, Jackman JA, Park JH. et al. Nanoplasmonic sensors for detecting circulating cancer biomarkers. Adv Drug Deliv Rev. 2018;125:48-77
3. Laohaviroj M, Potriquet J, Jia X. et al. A comparative proteomic analysis of bile for biomarkers of cholangiocarcinoma. Tumour Biol. 2017;39:1010428317705764
4. Li L, Masica D, Ishida M. et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60:896-907
5. Srinivas PR, Srivastava S, Hanash S. et al. Proteomics in early detection of cancer. Clin Chem. 2001;47:1901-11
6. Voigtländer T, Metzger J, Schönemeier B. et al. A combined bile and urine proteomic test for cholangiocarcinoma diagnosis in patients with biliary strictures of unknown origin. United European Gastroenterol J. 2017;5:668-76
7. Baron TH, Harewood GC, Rumalla A. et al. A prospective comparison of digital image analysis and routine cytology for the identification of malignancy in biliary tract strictures. Clin Gastroenterol Hepatol. 2004;2:214-9
8. Harewood GC, Baron TH, Stadheim LM. et al. Prospective, blinded assessment of factors influencing the accuracy of biliary cytology interpretation. Am J Gastroenterol. 2004;99:1464-9
9. Lankisch TO, Metzger J, Negm AA. et al. Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology. 2011;53:875-84
10. Lukic N, Visentin R, Delhaye M. et al. An integrated approach for comparative proteomic analysis of human bile reveals overexpressed cancer-associated proteins in malignant biliary stenosis. Biochim Biophys Acta. 2014;1844:1026-33
11. Shen J, Wang W, Wu J. et al. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS One. 2012;7:e47476
12. Sriwanitchrak P, Viyanant V, Chaijaroenkul W. et al. Proteomics analysis and evaluation of biomarkers for detection of cholangiocarcinoma. Asian Pac J Cancer Prev. 2011;12:1503-10
13. Navaneethan U, Lourdusamy V, Gk Venkatesh P. et al. Bile proteomics for differentiation of malignant from benign biliary strictures: a pilot study. Gastroenterol Rep (Oxf). 2015;3:136-43
14. Rupp C, Bode KA, Leopold Y. et al. Pathological features of primary sclerosing cholangitis identified by bile proteomic analysis. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1380-9
15. Le Faouder J, Gigante E, Léger T. et al. Proteomic landscape of cholangiocarcinomas reveals three different subgroups according to their localization and the aspect of non-tumor liver. Proteomics Clin Appl. 2019;13(1):e1800128
16. Carotenuto P, Fassan M, Pandolfo R. et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268-77
17. Boulter L, Guest RV, Kendall TJ. et al. WNT signaling drives cholangiocarcinoma growth and can be pharmacologically inhibited. J Clin Invest. 2015;125:1269-85
18. Timmer MR, Beuers U, Fockens P. et al. Genetic and epigenetic abnormalities in primary sclerosing cholangitis-associated cholangiocarcinoma. Inflamm Bowel Dis. 2013;19:1789-97
19. Zhao Z, Zou S, Guan X. et al. Apolipoprotein E overexpression Is associated with tumor progression and poor survival in colorectal cancer. Front Genet. 2018;9:650
20. Oue N, Hamai Y, Mitani Y. et al. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64:2397-405
21. Su WP, Chen YT, Lai WW. et al. Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer. 2011;71:28-33
22. Venanzoni MC, Giunta S, Muraro GB. et al. Apolipoprotein E expression in localized prostate cancers. Int J Oncol. 2003;22:779-86
23. Brewer HB Jr, Fairwell T, LaRue A. et al. The amino acid sequence of human APOA I, an apolipoprotein isolated from high density lipoproteins. Biochem Biophys Res Commun. 1978;80:623-30
24. Wang X, Dai S, Zhang Z. et al. Characterization of apolipoprotein A I as a potential biomarker for cholangiocarcinoma. Eur J Cancer Care (Engl). 2009;18:625-35
25. Reshetnyak VI. Physiological and molecular biochemical mechanisms of bile formation. World J Gastroenterol. 2013;19:7341-60
26. Luengo-Gil G, Calvo MI, Martín-Villar E. et al. Antithrombin controls tumor migration, invasion and angiogenesis by inhibition of enteropeptidase. Sci Rep. 2016;6:27544
27. Repetto O, De Re V. Coagulation and fibrinolysis in gastric cancer. Ann N Y Acad Sci. 2017;1404:27-48
28. Pasanen PA, Eskelinen M, Partanen K. et al. Multivariate analysis of six serum tumor markers (CEA, CA 50, CA 242, TPA, TPS, TATI) and conventional laboratory tests in the diagnosis of hepatopancreatobiliary malignancy. Anticancer Res. 1995;15:2731-7
29. Awan FM, Naz A, Obaid A. et al. Identification of circulating biomarker candidates for hepatocellular carcinoma (HCC): an integrated prioritization approach. PLoS One. 2015;10:e0138913
30. Jie Z, Cai Y, Yang W. et al. Protective effects of α1 antitrypsin on acute lung injury inrabbits induced by endotoxin. Chin Med J (Engl). 2003;116:1678-82
31. Jamnongkan W, Techasen A, Thanan R. et al. Oxidized alpha 1 antitrypsin as a predictive risk marker of opisthorchiasis associated cholangiocarcinoma. Tumour Biol. 2013;34:695-704
32. Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342-55
33. Jamnongkan W, Thanan R, Techasen A. et al. Upregulation of transferrin receptor-1 induces cholangiocarcinoma progression via induction of labile iron pool. Tumour Biol. 2017;39:1010428317717655
34. Jamnongkan W, Lebrilla CB, Barboza M. et al. Discovery of serotransferrin glycoforms: novel markers for diagnosis of liver periductal fibrosis and prediction of cholangiocarcinoma. Biomolecules. 2019;9:E538
35. Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:54-62
36. Lee CW, Ronnekleiv-Kelly S. Autoimmune diseases of the biliary Tract: a review. Surg Clin North Am. 2019;99:185-201
37. Sumiyoshi K, Andoh A, Fujiyama Y. et al. Characterization of complement C3, C4, and factor B molecules in human bile. J Gastroenterol. 1997;32:230-5
38. Nakamura N, Hatano E, Iguchi K. et al. Elevated levels of circulating ITIH4 are associated with hepatocellular carcinoma with nonalcoholic fatty liver disease: from pig model to human study. BMC Cancer. 2019;19(1):621
39. Tobe T, Saguchi K, Hashimoto K. et al. Mapping of human interalpha-trypsin inhibitor family heavy chain-related protein gene (ITIHL1) to human chromosome 3p21→p14. Cytogenet Cell Genet. 1995;71:296-8
40. Salier JP, Rouet P, Raguenez G. et al. The inter-alpha-inhibitor family: from structure to regulation. Biochem J. 1996;315:1-9
41. Bost F, Diarra-Mehrpour M, Martin JP. Inter-alpha-trypsin inhibitor proteoglycan family-a group of proteins binding and stabilizing the extracellular matrix. Eur J Biochem. 1998;252:339-46
42. Tang Y, Kitisin K, Jogunoori W. et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105:2445-50
43. Pyzik M, Sand KMK, Hubbard JJ. et al. The neonatal Fc receptor (FcRn): a misnomer? Front Immunol. 2019;10:1540
44. Pozniak KN, Pearen MA, Pereira TN. et al. Taurocholat induces biliary differentiation of liver progenitor cells causing hepatic stellate cell chemotaxis in the ductular reaction: role in pediatric cystic fibrosis liver disease. Am J Pathol. 2017;187:2744-57
45. Trauner M, Fickert P, Halilbasic E. et al. Lessons from the toxic bile concept for the pathogenesis and treatment of cholestatic liver diseases. Wien Med Wochenschr. 2008;158:542-8
46. Moy AP, Arora K, Deshpande V. Albumin expression distinguishes bile duct adenomas from metastatic adenocarcinoma. Histopathology. 2016;69:423-30
47. Lin F, Shi J, Wang HL. et al. Detection of albumin expression by RNA in situ hybridization is a sensitive and specific method for identification of hepatocellular carcinomas and intrahepatic cholangiocarcinomas. Am J Clin Pathol. 2018;150:58-64
48. Brackett DG, Neyaz A, Arora K. et al. Cholangiolar pattern and albumin in situ hybridisation enable a diagnosis of intrahepatic cholangiocarcinoma. J Clin Pathol. 2020;73:23-9
Author contact
![]() Corresponding author: Hyo Jung Kim, M.D., PhD. Division of Gastroenterology, Department of Internal Medicine, Korea University Guro Hospital, Korea University College of Medicine, 148 Gurodong-ro, Guro-gu, Seoul 08308, Korea. Phone; +82-2-2626-1779, Fax; +82-2-2626-1770; E-mail: hjkimmdac.kr
Corresponding author: Hyo Jung Kim, M.D., PhD. Division of Gastroenterology, Department of Internal Medicine, Korea University Guro Hospital, Korea University College of Medicine, 148 Gurodong-ro, Guro-gu, Seoul 08308, Korea. Phone; +82-2-2626-1779, Fax; +82-2-2626-1770; E-mail: hjkimmdac.kr

 Global reach, higher impact
Global reach, higher impact