Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(14):4106-4114. doi:10.7150/jca.37842 This issue Cite
Research Paper
microRNA Expression Profiling in Young Prostate Cancer Patients
1. Urologic Oncology Branch, National Cancer Institute, National Institutes of Health. Bethesda MD.
2. Translational Surgical Pathology Section, Laboratory of Pathology, National Cancer Institute, National Institutes of Health, Bethesda MD.
3. Faculty of Natural Science and Mathematics, Universidad del Rosario, Bogotá, Colombia
*These authors contributed equally.
Received 2019-6-23; Accepted 2020-3-27; Published 2020-4-7
Abstract
MicroRNAs (miRNAs) are small, non-coding RNA molecules with multiple roles in many biological processes. Few studies have shown the molecular characteristics in younger prostate cancer (PCa) patients. In this study, we performed miRNA profiling in young PCa (EO-PCa) cases compared with PCa arising in older men (LO-PCa).
Experimental Design: Formalin-fixed, paraffin embedded tissue was used. miRNA was extracted for PCR array and NanoString methods. Relative miRNAs expression levels were obtained by comparing young vs older men, and young PCa tumor samples vs normal epithelium.
Results: miRNA profiling showed a different expression pattern in PCa arising in younger men, and young PCa tumoral and its normal counterpart. Nine miRNAs (hsa-miR-140-5p, hsa-miR-146a, hsa-miR-29b, hsa-miR-9, hsa-miR-124-3p, hsa-let-7f-5p, hsa-miR-184, hsa-miR-373, hsa-miR-146b-5p) showed differences in the expression compared to LO-PCa. Fourteen miRNAs were significantly up-regulated (miR-1973, miR-663a, miR-575, miR-93-5p, miR-630, miR-600, miR-494, miR-150-5p, miR-137, miR-25-3p, miR-375, miR-489, miR-888-5p, miR-142-3p), while 9 were found down-regulated (miR-21-5p, miR-363-3p, miR-205-5p, miR-548ai, miR-3195, 145-5p, miR-143-3p, miR-222-3p, miR-221-3p) comparing young PCa tumoral tissue compared to normal counterpart. The higher expression of miR-600 and miR-137 were associated with high Gleason score, extraprostatic extension and lymphatic invasion.
Conclusion: These results suggest that PCa in younger patients has a different expression profile compared to normal tissue and PCa arising in older man. Differentially expressed miRNAs provide insights of molecular mechanisms involve in this PCa subtype.
Keywords: MicroRNAs, prostate, early onset, young
Introduction
Prostate cancer (PCa) is the most common tumor in men and the fifth cause of cancer death. It is estimated that there were almost 1.3 million new cases of PCa and 359.000 associated deaths worldwide in 2018 [1]. The incidence of PCa has been increasing across the time. Between 1986 and 2008 the incidence of PCa in young men, defined as PCa arising in men under 55 years has been increased by 5.7-fold from 5.6 to 32 cases per 100.00 persons years (IC 95% CI 5.0-6.7), making PCa in young men an important emerging issue for public health [2,3]. In 2012, 10% of men (241.740 persons) with newly diagnosed PCa were 55 years old or younger [4]. Different risk factors as ethnicity, familiar history and genetic factors has been associated in this setting [5].
The genetic profile between young PCa and 'classical' or PCa arising in older men are different [3,6-9]. Previous studies also have revealed different genetic alterations in young PCa, such as a greater number of single nucleotide polymorphism in the germinal DNA [6-8], different expressed genes involved in the inflammatory and immune-related pathways (CTL4, IDO1/TDO2) [9], and gene mutations in BRCA 1 and 2, and HOXB13 [5]. The relative risk to develop PCa in patients with BRCA 1 mutations has been reported as 1.8-4.5, while the relative risk reported in patients with mutations in BRCA 2 is 23, and patients with HOXB13 mutation have eightfold higher risk [5]. PCa in young man with mutations in BRCA 1 and 2 has been associated with unfavorable prognosis, in contrast to patients with HOXB13 mutations were the genetic alteration has been associated with a favorable prognosis [5].
The molecular pathway of PCa in younger men is unknown. Weischenfeldt, et al. [7] recognized the important role of androgen regulated transmembrane serine protease 2 (TMPRSS2) gene fusion with the ERG gene. This fusion is a very early event in prostate oncogenesis and might be driven by increased androgen stimulation. Young PCa patients have a significantly increased tumor androgen receptor levels and positive correlation with ERG rearrangements. The frequency of ERG rearrangements in younger PCa patients is approximately 64% [10,11]. TMPRSS2-ERG fusion positive cases are associated with loss of PTEN suppressor gene and TMPRSS2-ERG fusion negative cases with loss of 5q21 and 6q15 [12].
Differences in clinical behavior between young and older PCa patients are controversial [3]. Some studies report aggressive biological behavior and higher Gleason score in the young [3,11,13,14] while other studies report no significant difference in survival across age groups after prostatectomy, brachytherapy and radiation therapy [3,15,16]. Schaefer et al. [10] observed that ERG-positive status was associated with low-serum PSA and lower prostate volume, while Huang et al. [11] further confirmed the ERG-positive status was associated with Gleason score and higher biochemical relapse rate but not with presurgical PSA levels, tumor volume, pathological stage, surgical margin or lymphovascular invasion.
MicroRNAs (miRNAs) are small, non-coding RNA molecules with multiple roles in many biological processes. They can prevent protein expression through cleavage of specific target mRNAs or through inhibition of their translation [17]. Since the discovery of miRNAs, numerous studies have demonstrated their relevance in carcinogenesis of several cancers [18]. Recently, it has been demonstrated the relevance of miRNAs in PCa as biomarkers in diagnosis, treatment, and prognosis.
In this study, we investigated whether PCa arising on younger men has a different microRNA profile compared to PCA in older men in order to further characterize its potential role in tumorigenesis, tumor progression and disease prognosis.
Material and Methods
Patient Samples
Formalin-fixed, paraffin embedded (FFPE) tissue samples from younger (age <55 y.o) patients with PCa and older (>55 y.o) PCa were retrieved from the surgical archives of the Laboratory of Pathology, National Cancer Institute, Bethesda, MD, USA after IRB approval. Samples without sufficient tumor tissue were excluded. Hematoxylin and eosin (H&E) stained slides were reviewed to confirm the diagnosis. Tumoral and normal tissue adjacent to the tumor (epithelial cells from the prostate glands) were selected from the same patients. The clinicopathological features were reviewed, including Gleason score, extraprostatic extension, margins, seminal vesicle, perineural invasion, lymphatic invasion, and pTNM (pathological tumor-node, metastasis) staging.
microRNA Isolation
Isolation of total miRNA from FFPE specimens was performed as described previously [21,22]. In brief, tumoral and normal tissue were manually microdissected under light microscope followed by miRNA isolation using the RecoverAll™ Total Nucleic Acid Isolations Kit (Ambion by Life Technologies, Foster City, CA, USA). The concentration of all RNA samples was quantified using NanoDrop 2000 (Thermo Scientific, USA). RNA concentration of samples used for profiling was normalized at 33 ng/µl following the recommendations from NanoString Technologies.
microRNA expression profiling using PCR arrays
As we described previously [21], the extracted total RNA including miRNAs (10 ng/µl concentration) was first reverse transcribed into first strand cDNA using the RT2- miRNA First Strand Kit following manufacturer's recommendations (SA Biosciences, Rockville, MD). One µl cDNA per well was then mixed with SYBR Green qPCR Master Mix and placed into a 96-well PCR-array plate containing a panel of 88 mature miRNAs sequences. The arrays also contain appropriate small nucleolar RNA sequences that are used as housekeeping assays and quality controls. One µl was used in a 12 µl final volume reaction for Real-time PCR analysis on an Applied Biosystems Step-One Plus Real Time PCR system. Relative amounts were calculated by the ΔΔ CT method. Samples without good RNA quality were excluded in the statistical analysis.
microRNA profiling using NanoString nCounter miRNA assay and data analysis
Total RNA samples were analyzed according to the manufacturer's instructions for the nCounter Human miRNA Expression Assay kit (NanoString Technologies, Seattle, WA). From each sample 100 ng from total RNA sample was used as input into the nCounter Human miRNA sample preparation. Hybridization was conducted for 16 h at 65°C. Subsequently, probes were purified and counted on the nCounter Prep Station. Each sample was scanned for 600 FOV (fields of view) on the nCounter Digital Analyzer. Data was extracted using the nCounter RCC Collector. The analysis was made as we described previously [22]. miRNAs raw data was normalized for lane-to-lane variation with a dilution series of six spike-in positive controls. The sum of the six positive controls for a given lane was divided by the average sum across lanes to yield a normalization factor, which was then multiplied by the raw counts in each lane to give normalized values. For each sample, the mean plus 2 times the standard deviation of the 8 negative controls was subtracted from each miRNA count in that sample. Only miRNAs with non-negative counts across all samples were retained for downstream analysis. The relative miRNA levels were indicated as median fold changes (tumor/normal tissue) and a cutoff of 1.5-fold-change (up or down) was used.
Assessment of prognostic significance of miRNAs associated with clinicopathological features
With PROGmir V2 we compared the overall survival, relapse-free survival, and metastasis-free survival of prostate adenocarcinoma patients with high and low expression of miRNAs associated with clinicopathological features. PROGmiR V2 is an online free tool that combines the prognostic data of miRNAs for different kinds of cancers [23].
Statistical Analysis
Only mature miRNAs that showed at least a 1.5-fold change in expression are reported. p-values were calculated for each miRNA between the normal and tumor samples using the biological replicates and genes. They were considered differentially expressed and statistically significant if their p value was < 0.05. To compare relative miRNA fold changes between Young PCa and older PCa patients we used Mann-Whitney and Kruskal-Wallis non-parametric tests. According to edgeR, t-tests were carried out to compare the two groups (tumor vs. normal), p value was adjusted for multiple comparisons with the calculation of the false discovery rate (FDR) (<0.05). To evaluate differences between miRNA expression and clinicopathological features (Gleason score was categorized in low grade (3+3; 3+4) and high grade (4+3; 4+4;5+5)) we used t test. Supervised and non-supervised hierarchical clustering was conducted based on the Euclidean distance of miRNAs in samples using the Pheatmap package in R3.5.1. Analysis was performed using STATA SE 15 and R3.5.1.
Results
Characterization of studied population
In all, ten cases of young PCa and nineteen cases of older PCa patients were included. The clinicopathologic characteristics of the cases are shown in Table 1. The median age of young PCa was 46 years (range 40-55), 7 had low or intermediate risk Gleason score (3 + 3 or 3 + 4) and 3 had high-grade Gleason score (4 + 5 and 5 + 5). Perineural invasion was observed in seven patients, extraprostatic extension in three patients (high-grade Gleason score). While the average age of older PCa patients was 63 years (range 58-71), nine were low-grade Gleason score and ten were high-grade Gleason score. Perineural invasion was observed in 12 patients, extraprostatic extension and positive margins in six, and lymphatic invasion in four.
microRNA expression profile comparing young PCa to older PCa patients
To determine if young PCa patients have a tumor specific pattern of miRNAs expression, the expression level of 88 mature miRNAs using PCR based assay was compared. Different expression was recognized in nine miRNAs between the groups (hsa-miR-140-5p, hsa-miR-146a, hsa-miR-29b, hsa-miR-9, hsa-miR-124-3p, hsa-let-7f-5p, hsa-miR-184, hsa-miR-373, hsa-miR-146b-5p) (Figure 1). Three were upregulated (Fold change >1.5) (hsa-miR-140-5p (p 0.008), hsa-miR-146a (p 0.01), hsa-miR-29b (p0.01)) in younger PCa patients and one (hsa-let-7f-5p ( p 0.02)) was downregulated (Fold change <1.5) (Table 2).
microRNA expression profile between tumoral tissue to its normal counterpart in young PCa tumors
A panel of 800 miRNAs was analyzed in 6 young PCa patients. A t-test was performed to comparing the tumor versus its corresponding normal prostate epithelium. In total, 14 miRNAs were up-regulated ranging from 1.51-fold to 2.17 while nine miRNAs showed to be down-regulated ranging from -1.52-fold to -12.42-fold change. Only two miRNAs showed FDR <0.05 (hsa-miR-205 (FDR: 0.002) and hsa-miR-21-5p (FDR: 0.02) (Table 3). Among the 14 miRNAs up-regulated (hsa-miR-1973, hsa-miR-663a, hsa-miR-575, hsa-miR-93-5p, hsa-miR-630, hsa-miR-600, hsa-miR-494, hsa-miR-150-5p, hsa-miR-137, hsa-miR-25-3p, hsa-miR-375, hsa-miR-489, hsa-miR-888-5p, hsa-miR-142-3p), two miRNAs (hsa-miR-1973 and hsa-miR-93-5p) were the most prominently up-regulated (p<0.05).
Clinicopathological features of prostate cancer patients included in the study.
| Variable | Young-PCa (N:10) | Older-PCa (N:19) | p value |
|---|---|---|---|
| Age, median (Years) (range) | 46 (40-55) | 63 (58-71) | |
| Gleason score groups | 0.06 | ||
| ≤3+4 | 7 | 9 | |
| ≥4+4 | 3 | 10 | |
| Extraprostatic extension | 0.63 | ||
| Yes | 3 | 6 | |
| No | 7 | 13 | |
| Positive margins | 0.42 | ||
| Yes | 2 | 6 | |
| No | 8 | 13 | |
| Seminal vesicle invasion | 0.34 | ||
| Yes | 1 | 0 | |
| No | 9 | 19 | |
| Perineural invasion | 0.52 | ||
| Yes | 7 | 12 | |
| No | 3 | 7 | |
| Lymphatic invasion | 0.66 | ||
| Yes | 2 | 4 | |
| No | 8 | 15 | |
| T Stage | 0.24 | ||
| pT2a ≤ pT2C | 8 | 13 | |
| pT3a≥ pT3 | 1 | 6 | |
| N Stage | 0.003 | ||
| N0 | 8 | 4 | |
| N1 | 2 | 15 | |
| M Stage | 1.000 | ||
| M0 | 10 | 19 | |
| M1 | 0 | 0 |
MicroRNAs differentially expressed in young PCa patients versus older PCa patients.
| miRNA | Young PCa (mean, range) | Older-PCa (mean, range) | p Value |
|---|---|---|---|
| hsa-miR-140-5p | 1.95 (1.69-2.15) | -2.28 (-5.53- -0.3) | 0.008 |
| hsa-miR-146a | 4.78 (2.07-7.3) | 0.79 (-3.21- 7.3) | 0.01 |
| hsa-miR-29b | 5.77 (3.76-9.54) | 0.8 (-1.66-2.8) | 0.01 |
| hsa-miR-9 | 1.21 (-3.38-4.11) | -4.84 (-9.63 - 1.9) | 0.02 |
| hsa-miR-124-3p | 0.11 (-0.15-0.34) | 2.55 (-7.03-8.61) | 0.01 |
| hsa-let-7f-5p | 1.27 (0.43-2.67) | 5.84 (1.88-9.73) | 0.02 |
| hsa-miR-184 | 0.09 (-1.14-1.27) | 4.13 (-4.31-10.14) | 0.02 |
| hsa-miR-373 | -1.43 (-4.48-0.41) | 2.65 (-5.93-9.24) | 0.03 |
| hsa-miR-146b-5p | -0.69 (-1.32-0.03) | 1.92 (-3.77-9.95) | 0.04 |
ΔΔ CT of miRNAs differentially expressed between young PCa and older PCa tumoral tissue
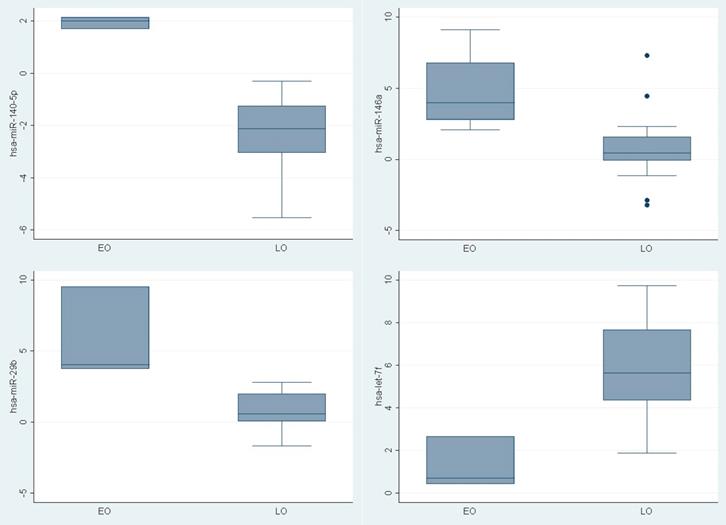
Among the nine miRNAs down-regulated, three miRNAs (hsa-miR-21-5p, hsa-miR-363-3p, hsa-miR-205-5p) were the most prominently down-regulated (p<0.05).
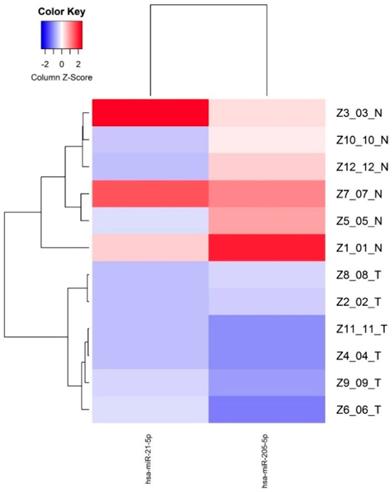
Supervised hierarchical clustering of the miRNAs down-regulated with FDR <0.05 was made based on miRNAs normalized expression, showing two groups separating normal and tumor epithelium mainly based the miR-205 expression, indicating a cancer specific expression pattern (Figure 2).
Differentially expressed miRNAs (FDR<0.05) between normal epithelium and tumor tissue from young PCa patients.
microRNA expression and clinicopathological features
Using the expression profile data, we also evaluated the possible correlation between clinicopathological features and the expression of deregulated miRNAs in tumoral tissue. We analyzed the group of Young PCa patients for expression profiling regarding the Gleason score grade (low or high), extraprostatic extension, margins, seminal vesicle invasion, perineural invasion, lymphatic invasion, and pTNM stage. We found association between high levels of hsa-miR-575, hsa-miR-663a, hsa-miR-600, hsa-miR-137 with high grade Gleason score and presence of extraprostatic extension, as well the low levels of hsa-miR-143 (Table 4). In contrast, low levels of hsa-miR-221 was associated with low grade Gleason score and absence of extraprostatic extension. High levels of hsa-miR-137 and hsa-miR-600 were associated with presence of lymphatic invasion, while high levels hsa-miR-663, and low levels of hsa-miR-221 and hsa-miR-143 were associated with absence of lymphatic invasion. Low levels of hsa-miR-143 and high levels has-miR-1973 were associated with absence of perineural invasion (Table 4).
MicroRNAs differentially expressed in tumoral young PCa tissue versus normal epithelium.
| miRNA | Fold change | p Value |
|---|---|---|
| hsa-miR-93-5p | 1.87 | 0.006 |
| hsa-miR-1973 | 2.17 | 0.03 |
| hsa-miR-25-3p | 1.6 | 0.07 |
| hsa-miR-137 | 1.6 | 0.05 |
| hsa-miR-575 | 2.01 | 0.09 |
| hsa-miR-150-5p | 1.61 | 0.09 |
| hsa-miR-375 | 1.6 | 0.10 |
| hsa-miR-663a | 2.03 | 0.11 |
| hsa-miR-142-3p | 1.51 | 0.12 |
| hsa-miR-630 | 1.83 | 0.16 |
| hsa-miR-600 | 1.74 | 0.16 |
| hsa-miR-888-5p | 1.55 | 0.16 |
| hsa-miR-489 | 1.6 | 0.17 |
| hsa-miR-494 | 1.73 | 0.29 |
| hsa-miR-205-5p | -4.81 | 2.48E-06 |
| hsa-miR-21-5p | -12.42 | 6.43E-05 |
| hsa-miR-363-3p | -5.47 | 0.0089 |
| hsa-miR-145-5p | -1.61 | 0.05 |
| hsa-miR-222-3p | -1.55 | 0.07 |
| hsa-miR-3195 | -1.87 | 0.08 |
| hsa-miR-548ai | -2.83 | 0.09 |
| hsa-miR-143-3p | -1.55 | 0.13 |
| hsa-miR-221-3p | -1.52 | 0.12 |
Correlation between clinicopathological features and miRNA expression levels
| Gleason score | |||
| High | Low | P-value | |
| hsa-miR-575 | 465.43 | 144.34 | 0.01 |
| hsa-miR-663 | 91.95 | 26.56 | 0.04 |
| hsa -miR-600 | 50.41 | 26.89 | 0.02 |
| hsa -miR-137 | 88.25 | 68.24 | 0.03 |
| hsa-miR-143 | 1356.32 | 532 | 0.01 |
| hsa -miR-221 | 44.36 | 112.32 | 0.04 |
| Extraprostatic extension | |||
| Yes | No | ||
| hsa-miR-575 | 465.43 | 144.34 | 0.01 |
| hsa-miR-663 | 91.95 | 26.56 | 0.04 |
| hsa -miR-600 | 50.41 | 26.89 | 0.02 |
| hsa -miR-137 | 88.25 | 68.24 | 0.03 |
| hsa-miR-143 | 1356.32 | 532 | 0.01 |
| hsa -miR-221 | 44.36 | 112.32 | 0.04 |
| Perineural invasion | |||
| Yes | No | ||
| hsa-miR-973 | 29.38 | 80.94 | 0.03 |
| hsa -miR-143 | 824.02 | 1596.64 | 0.04 |
| Lymphatic invasion | |||
| Yes | No | ||
| hsa -miR-137 | 88.82 | 68.24 | 0.03 |
| hsa -miR-600 | 50.41 | 26.89 | 0.02 |
| hsa -miR-663 | 91.95 | 26.56 | 0.04 |
| hsa -miR-221 | 44.36 | 112.32 | 0.04 |
| hsa -miR-143 | 532.05 | 1356.32 | 0.01 |
Relapse-free survival curves based on hsa-miR-137 expression levels in prostate cancer from PROGmiR V2.
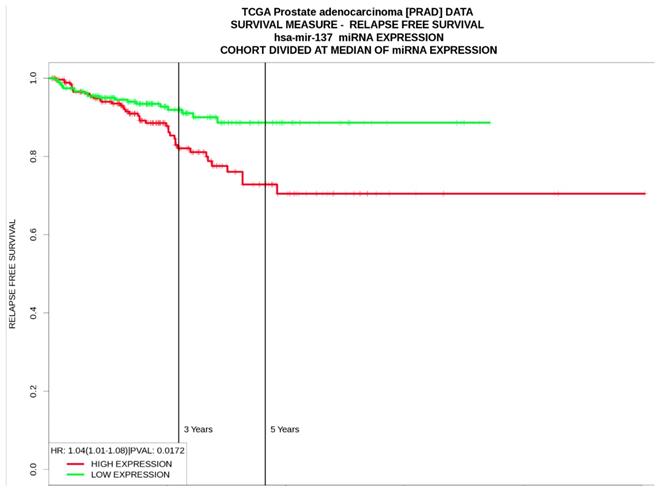
Assessment of prognostic significance of miRNAs associated with clinicopathological features from PROGmir V2
According to the results from PROGmir V2, prostate cancer patients with high expression levels of hsa-miR-137 had significant poor relapse‐free survival (HR: 1.04 (1.01-1.08, p=0.01) (Figure 3). In contrast, high levels of hsa-miR-143 was associated as protective factor for relapse‐free survival (HR= 0.64 (0.48-0.84, p=0.001), same as hsa-miR-221 (HR= 0.68 (0.51-0.89, p=0.005) (Table 5).
Prognostic significance of miRNAs with clinicopathological associations
| miRNA's | Overall survival | Relapse-free survival | Metastasis-free survival |
|---|---|---|---|
| hsa -miR-137 | HR: 0.98 (0.91-1.05) | HR: 1.04 (1.01-1.08) | HR: 1.05 (0.9-1.22) |
| hsa-miR-143 | HR: 0.53 (0.27-1.06) | HR: 0.64 (0.48-0.84) | HR: 1.94(0.27-13.69) |
| hsa -miR-221 | HR: 0.52 (0.27-1.02) | HR: 0.08 (0.51-0.89) | HR: 0.65 (0.19-2.3) |
| hsa-miR-663 | HR: 0.96 (0.88-1.04) | HR: 0.97 (0.94-1.01) | HR: 0.91 (0.78-1.07) |
Discussion
In the present study, we observed a different expression profile of miRNAs in young PCa compared to older PCa patients (Table 2) and compared tumoral to normal tissue (Table 3), suggesting a cancer-specific miRNAs expression profile for Young PCa. Due to lack of studies about the miRNAs in young PCa, the current knowledge about its biology is limited. In the study by Diung et al [9] they found the different miRNAs expression between young PCa with GS 7 (3+4) and PCa in older patients. Like them, we observed differences in the expression of hsa-miR-146a, hsa-miR-9, hsa-miR-124-3p, hsa-miR-146b-5p. Other study tested the miRNA's expression in eleven young patients with PCa. They measured the expression of genomic alterations in younger PCa and found rearranged genes in androgen pathway. They focused on finding miRNAs that had as target to PTEN and identified 13 miRNAs (hsa-miR-17-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-20a-5p, hsa-miR-92a-3p, hsa-miR-106b-5p, hsa-miR-93-5p, hsa-miR-25-3p, hsa-miR-141-3p, hsa-miR-214-3p, hsa-miR-494, hsa-miR-222-3p, hsa-miR-21-5p) differentially expressed(> 1.5-fold change). In our study we also observed upregulation of hsa-miR-93-5p, hsa-miR-25-3p and hsa-miR-494, and downregulation of hsa-miR-222-3p when we compare tumoral vs normal tissue. Unlike of them, we found downregulation of hsa-miR-21-5p.
Among the miRNAs with differences between young and older PCa patients, we observed nine miRNAs with different levels of expression between them (hsa-miR-140-5p, hsa-miR-146a, hsa-miR-29b, hsa-miR-9, hsa-miR-124-3p, hsa-let-7f-5p, hsa-miR-184, hsa-miR-373, hsa-miR-146b-5p). Three were upregulated (Fold change >1.5) (hsa-miR-140-5p (p 0.008), hsa-miR-146a (p 0.01), hsa-miR-29b (p0.01)) in young PCa patients and one was downregulated (Fold change <1.5) (hsa-let-7f-5p ( p 0.02)). This in in agreement with published work that shows that hsa-miR-140-5p and hsa-let-7f-5p in PCa are upregulated [19,20,24] while hsa-miR-29b is downregulated [19,20]. High expression levels of hsa-miR-140-5p has been observed mainly in metastatic PCa [24]. hsa-miR-146a has shown to modulate androgen-Independent prostate cancer cells apoptosis through regulation of ROCK/Caspase 3 pathway [25]. Furthermore, hsa-miR-29b has been observed downregulated in PCa [19,20] and may be involved in the epithelial-mesenchymal transition by the interaction with different targets such as e-cadherin, MMP-2, snail and twist [26].
Several miRNAs were observed dysregulated when we compared tumoral tissue versus tissue in Young PCa. Fourteen miRNAs were upregulated (Table 3). hsa-miR-93-5p has been reported overexpressed in PCa [27] and may are involved in cell proliferation, migration, invasion, block cell cycle, and promote the early apoptosis [27]. In a previous study, it was found that hsa-miR-93 with hsa-miR-106b and miR-375 may downregulate CIC-CIC-CRABP1 and promote the progression of PCa [28]. In young PCa, Weischenfeldt et al. found hypomethylation in the promotor region (R: -0.682, p<0.001) [7]. On the other hand, hsa-miR-25-3p is part of a cluster with hsa-miR-93, hsa-miR-25 and hsa-miR-106b [29]. This miR-106b-25 cluster promotes cell-cycle progression and hyperproliferation due to relationship with several actors in different oncological pathways such as PTEN, E2F1, and p21/WAF1 [30-32]. In PCa, the expression of this cluster is high and has been associated with tumor progression and metastasis [21,33]. In a previous study, we had already observed upregulation of miR-25 in tumor cells versus normal epithelium [21]. In young PCa the hypomethylation of the hsa-miR-25-3p promotor region (R: -0.625, p<0.001) has also been observed [7].
Among the miRNAs without previously reported association and whose biological functions have not been characterized in PCa are hsa-miR-1973, hsa-miR-575, hsa-miR-630 and hsa-miR-600. hsa-miR-575 and the hsa-miR-630 are proposed oncomiRNAs in other tumors such as gastric cancer, lung cancer, renal cell carcinoma, hepatocellular carcinoma, and breast cancer [34-40]. hsa-miR-630 has different gene targets such as BCL-2, MTDH, YAP-1, SNAI2 [37-39] involved in PCa oncogenesis [41-44]. hsa-miR-575 has as a target BLID (BH3-like motif containing, BRCC2). BLID is a tumor-suppressor gene involved in DNA repair and gene integrity [40]. In breast cancer, BLID inhibited cancer cell growth and metastasis via downregulating AKT pathway [40]. In prostate cancer cell lines, it has been demonstrated that BLID induce a caspase-dependent mitochondrial pathway of cell death [45]. Expression of hsa-miR-1973 has noted in other tumors as breast cancer, Hodgkin lymphoma and ovarian cancer [46-48]. Other miRNA without previously reported in PCa is hsa-miR-600, this has been involved in breast cancer, lung and colorectal cancer [49-51].
Among the differentially expressed miRNAs, some were associated with Gleason score, extraprostatic extension and lymphatic invasion. high expression levels for hsa-miR-137 and hsa-miR-600 were associated with high Gleason score, presence of extraprostatic extension, and lymphatic invasion. hsa-miR-137 also was associated with poor relapse‐free survival (HR= 1.04). In the present study, we found that hsa-miR-137 is upregulated in young PCa, while in the literature has been reported downregulated and associated with recurrence following prostatectomy [52]. hsa-miR-137 has a function as an androgen regulated suppressor of androgen signaling by modulating expression of an extended network of transcriptional coregulators [53].
Conclusion
The present study supports the hypothesis that young PCa have a different miRNAs signature compared with normal tissue and older PCa patients. We present new miRNAs that may be involved in PCa pathogenesis (e.g. hsa -miR-1973, hsa-miR-575, hsa-miR-630, hsa-miR-3195), and present miRNAs with different expression (high or low) compared with previous studies (e.g hsa -miR-494, 150-5p, hsa -miR-137, hsa -miR-548ai, hsa -miR-21). In addition, we show that low expression of miR-600 and miR-137 were associated with clinicopathological features with poor prognosis and with poor relapse‐free survival. The identification of these miRNAs may provide insights into understanding this subtype of PCa and these miRNAs may prove to be useful as a biomarker in the diagnosis, prognostic as well in the development of new therapeutic approaches. More studies with larger sample size are needed to confirm the results presented in this study and to correlate them with clinicopathological outcomes in young patients with PCa.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A J Clin. 2018;68:394-424
2. Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975-2014, National Cancer Institute. Bethesda, MD, n.d.
3. Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: An important clinical entity. Nat Rev Urol. 2014;11:317-23
4. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
5. Hussein S, Satturwar S, Van der Kwast T. Young-age prostate cancer. J Clin Pathol. 2015;68:511-5
6. Lange EM, Salinas CA, Zuhlke KA, Ray AM, Wang Y, Lu Y. et al. Early onset prostate cancer has a significant genetic component. Prostate. 2012;72:147-56
7. Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S. et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159-70
8. Lange EM, Johnson AM, Wang Y, Zuhlke KA, Lu Y, Ribado J V. et al. Genome-wide association scan for variants associated with early-onset prostate cancer. PLoS One. 2014;9:e93436
9. Ding Y, Wu H, Warden C, Steele L, Liu X, Iterson M van. et al. Gene Expression Differences in Prostate Cancers between Young and Old Men. PLOS Genet. 2016;12:e1006477
10. Schaefer G, Mosquera J-M, Ramoner R, Park K, Romanel A, Steiner E. et al. Distinct ERG rearrangement prevalence in prostate cancer: higher frequency in young age and in low PSA prostate cancer. Prostate Cancer Prostatic Dis. 2013;16:132-8
11. Huang KC, Dolph M, Donnelly B, Bismar TA. ERG expression is associated with increased risk of biochemical relapse following radical prostatectomy in early onset prostate cancer. Clin Transl Oncol. 2014;16:973-9
12. Steurer S, Mayer PS, Adam M, Krohn A, Koop C, Ospina-Klinck D. et al. TMPRSS2-ERG fusions are strongly linked to young patient age in low-grade prostate cancer. Eur Urol. 2014;66:978-81
13. Wilson JM, Kemp IW, Stein GJ. Cancer of the prostate. Do younger men have a poorer survival rate? Br J Urol. 1984;56:391-6
14. Lin DW, Porter M, Montgomery B. Treatment and survival outcomes in young men diagnosed with prostate cancer: a Population-based Cohort Study. Cancer. 2009;115:2863-71
15. Huang T-H, Kuo J-Y, Huang Y-H, Chung H-J, Huang WJS, Wu HHH. et al. Prostate cancer in young adults-Seventeen-year clinical experience of a single center. J Chin Med Assoc. 2017;80:39-43
16. Grönberg H, Damber JE, Jonsson H, Lenner P. Patient age as a prognostic factor in prostate cancer. J Urol. 1994;152:892-5
17. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
18. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004
19. Vanacore D, Boccellino M, Rossetti S, Cavaliere C, D'Aniello C, Di Franco R. et al. Micrornas in prostate cancer: an overview. Oncotarget. 2017;8:50240-51
20. Cannistraci A, Di Pace AL, De Maria R, Bonci D. MicroRNA as new tools for prostate cancer risk assessment and therapeutic intervention: results from clinical data set and patients' samples. Biomed Res Int. 2014;2014:146170
21. Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA Profiling of Prostate Cancer. J Cancer. 2013;4:350-7
22. Merino MJ, Gil S, Macias CG, Lara K. The Unknown microRNA Expression of Male Breast Cancer. Similarities and Differences with Female Ductal Carcinoma. Their Role as Tumor Biomarker. J Cancer. 2018;9:450-9
23. Goswami CP, Nakshatri H. PROGmiR: a tool for identifying prognostic miRNA biomarkers in multiple cancers using publicly available data. J Clin Bioinforma. 2012;2:23
24. Watahiki A, Wang Y, Morris J, Dennis K, O'Dwyer HM, Gleave M. et al. MicroRNAs Associated with Metastatic Prostate Cancer. PLoS One. 2011;6:e24950
25. Xu B, Huang Y, Niu X, Tao T, Jiang L, Tong N. et al. Hsa-miR-146a-5p modulates androgen-independent prostate cancer cells apoptosis by targeting ROCK1. Prostate. 2015;75:1896-903
26. Ru P, Steele R, Newhall P, Phillips NJ, Toth K, Ray RB. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166-73
27. Yang Y, Jia B, Zhao X, Wang Y, Ye W. miR-93-5p may be an important oncogene in prostate cancer by bioinformatics analysis. J Cell Biochem. 2019;120:10463-83
28. Choi N, Park J, Lee J-S, Yoe J, Park GY, Kim E. et al. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015;6:23533-47
29. Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191-4
30. Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM. et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167-74
31. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I. et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-86
32. Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM. et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29
33. Hudson RS, Yi M, Esposito D, Glynn SA, Starks AM, Yang Y. et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene. 2013;32:4139-47
34. Chu D, Zhao Z, Li Y, Li J, Zheng J, Wang W. et al. Increased microRNA-630 expression in gastric cancer is associated with poor overall survival. PLoS One. 2014;9:e90526
35. Zhao J-J, Chen P-J, Duan R-Q, Li K-J, Wang Y-Z, Li Y. miR-630 functions as a tumor oncogene in renal cell carcinoma. Arch Med Sci. 2016;12:473-8
36. Chen M-J, Wu D-W, Wang G-C, Wang Y-C, Chen C-Y, Lee H. MicroRNA-630 may confer favorable cisplatin-based chemotherapy and clinical outcomes in non-small cell lung cancer by targeting Bcl-2. Oncotarget. 2018;9:13758-67
37. Zhou C-X, Wang C-L, Yu A-L, Wang Q-Y, Zhan M-N, Tang J. et al. MiR-630 suppresses breast cancer progression by targeting metadherin. Oncotarget. 2016;7:1288-99
38. Wu D-W, Wang Y-C, Wang L, Chen CY, Lee H. A low microRNA-630 expression confers resistance to tyrosine kinase inhibitors in EGFR-mutated lung adenocarcinomas via miR-630/YAP1/ERK feedback loop. Theranostics. 2018;8:1256-69
39. Chen W-X, Zhang Z-G, Ding Z-Y, Liang H-F, Song J, Tan X-L. et al. MicroRNA-630 suppresses tumor metastasis through the TGF-β- miR-630-Slug signaling pathway and correlates inversely with poor prognosis in hepatocellular carcinoma. Oncotarget. 2016;7:22674-86
40. Li X, Kong X, Wang Y, Yang Q. BRCC2 inhibits breast cancer cell growth and metastasis in vitro and in vivo via downregulating AKT pathway. Cell Death Dis. 2013;4:e757
41. Wang Z, Wei Y-B, Gao Y-L, Yan B, Yang J-R, Guo Q. Metadherin in prostate, bladder, and kidney cancer: A systematic review. Mol Clin Oncol. 2014;2:1139-44
42. Emadi Baygi M, Soheili Z-S, Essmann F, Deezagi A, Engers R, Goering W. et al. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol. 2010;31:297-307
43. Collak FK, Demir U, Ozkanli S, Kurum E, Zerk PE. Increased expression of YAP1 in prostate cancer correlates with extraprostatic extension. Cancer Biol Med. 2017;14:405-13
44. Renner W, Langsenlehner U, Krenn-Pilko S, Eder P, Langsenlehner T. BCL2 genotypes and prostate cancer survival. Strahlenther Onkol. 2017;193:466-71
45. Broustas CG, Gokhale PC, Rahman A, Dritschilo A, Ahmad I, Kasid U. BRCC2, a novel BH3-like domain-containing protein, induces apoptosis in a caspase-dependent manner. J Biol Chem. 2004;279:26780-8
46. Suryawanshi S, Vlad AM, Lin H-M, Mantia-Smaldone G, Laskey R, Lee M. et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19:1213-24
47. Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clin Cancer Res. 2014;20:253-64
48. Fomicheva KA, Knyazev EN, Mal'tseva D V. hsa-miR-1973 MicroRNA is Significantly and Differentially Expressed in MDA-MB-231 Cells of Breast Adenocarcinoma and Xenografts Derived from the Tumor. Bull Exp Biol Med. 2017;163:660-2
49. Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177-87
50. El Helou R, Pinna G, Cabaud O, Wicinski J, Bhajun R, Guyon L. et al. miR-600 Acts as a Bimodal Switch that Regulates Breast Cancer Stem Cell Fate through WNT Signaling. Cell Rep. 2017;18:2256-68
51. Zhang P, Zuo Z, Wu A, Shang W, Bi R, Jin Q. et al. miR-600 inhibits cell proliferation, migration and invasion by targeting p53 in mutant p53-expressing human colorectal cancer cell lines. Oncol Lett. 2017;13:1789-96
52. Pashaei E, Pashaei E, Ahmady M, Ozen M, Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS One. 2017;12:e0179543
53. Nilsson EM, Laursen KB, Whitchurch J, McWilliam A, Ødum N, Persson JL. et al. MiR137 is an androgen regulated repressor of an extended network of transcriptional coregulators. Oncotarget. 2015;6:35710-25
Author contact
![]() Corresponding author: Maria J. Merino M.D. Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441 Fax: (301) 480-1458. E-mail: mjmerinonih.gov.
Corresponding author: Maria J. Merino M.D. Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441 Fax: (301) 480-1458. E-mail: mjmerinonih.gov.

 Global reach, higher impact
Global reach, higher impact