Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(14):4284-4296. doi:10.7150/jca.42093 This issue Cite
Review
Aberrant LncRNA Expression in Leukemia
Department of Hematology, West China Hospital, Sichuan University, Chengdu 610041, Sichuan, China
Received 2019-11-14; Accepted 2020-3-31; Published 2020-4-27
Abstract
Leukemia is a common malignant cancer of the hematopoietic system, whose pathogenesis has not been fully elucidated. Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without protein-coding function. Recent studies report their role in cellular processes such as the regulation of gene expression, as well as in the carcinogenesis, occurrence, development, and prognosis of various tumors. Evidence indicating relationships between a variety of lncRNAs and leukemia pathophysiology has increased dramatically in the previous decade, with specific lncRNAs expected to serve as diagnostic biomarkers, novel therapeutic targets, and predictors of clinical outcomes. Furthermore, these lncRNAs might offer insight into disease pathogenesis and novel treatment options. This review summarizes progress in studies on the role(s) of lncRNAs in leukemia.
Keywords: long non-coding RNAs (lncRNAs), leukemia, transcription regulation, expression profiling, therapeutic targets
Introduction
Leukemia is a malignant clonal disease of hematopoietic stem and progenitor cells, in which abnormally cloned leukemia cells accumulate in the bone marrow and other non-hematopoietic tissues owing to uncontrolled cell proliferation, blocked differentiation, and apoptosis obstruction, thus inhibiting normal hematopoiesis and immune function. Rapid advances in cell and molecular biology have enabled the discovery of dysregulated molecules associated with leukemia, suggesting that the disease might be related to the heterogeneity of cellular and molecular genetics [1-4]. Chromosomal abnormalities, including the appearance of hyperdiploid and hypodiploid characteristics, amplification, translocation, changes in DNA copy number, as well as insertion, deletion, and point mutations, are commonly observed in leukemia [5]. Moreover, modifications of transcription factors, tumor-suppressor genes, oncogenic mutations, and epigenetic changes have been reported [6]; however, the specific pathogenesis of leukemia remains poorly understood.
Less than 2% of the human genome encodes proteins, with the remaining 98% considered as genetic byproducts. However, studies demonstrate that areas of the genome are transcribed as noncoding RNAs [7, 8]; these include various types of small-noncoding RNAs such as microRNAs (miRNAs), small-interfering (si)RNAs, small-nucleolar RNAs, and piwi-associated RNAs. Some dysregulated noncoding RNAs, particularly miRNAs, are well-known gene silencers [9]. Among noncoding RNAs, long noncoding (lnc)RNAs are a class of transcripts with a length >200 nucleotides and no protein-coding function. It was thought that lncRNAs lack biological functions; however, recent application of high-throughput sequencing and the rapid development of biological techniques have resulted in lncRNAs becoming a subject of tremendous research interest. Increasing evidence indicates that lncRNAs can regulate gene expression at multiple levels, including the epigenetic, transcriptional, and post-transcriptional stages. Additionally, lncRNAs are considered to be involved in the induction of chromatin remodeling and nucleosome modification, transcriptional activation and inhibition, regulation of variable splicing modes and protein activity, generation of endogenous siRNAs, and changing the protein localization (Figure 1) [10-12]. Moreover, lncRNAs are involved in the pathogenesis of cancer-related diseases, and therefore represent potential biomarkers and therapeutic targets [11]. Additionally, studies on the relationship between lncRNAs and hematological malignancies, such as leukemia, multiple myeloma, and lymphoma, are increasing. A variety of lncRNAs with potential as oncogenes or tumor-suppressors have been identified as significantly associated with the development and progression of these diseases [13-15].
Mechanisms of LncRNA action: lncRNAs (indicated in red) regulate gene expression at multiple levels: (a). LncRNAs can interact with the nuclear chromatin remodeling complex to achieve epigenetic regulation of target loci. (b). LncRNAs can regulate transcription by acting as a decoy or guide for transcription factors (indicated in yellow), thereby inhibiting or promoting their binding to target promoter sequences, respectively. (c). LncRNAs can interact with Staufen homolog proteins, thereby regulating mRNA stability. (d). LncRNAs can modulate mRNA levels by competing for microRNA (indicated in grey) binding. (e). Translation of mRNA can be modulated by lncRNAs. (f). LncRNAs can directly alter protein (indicated in blue) functions.
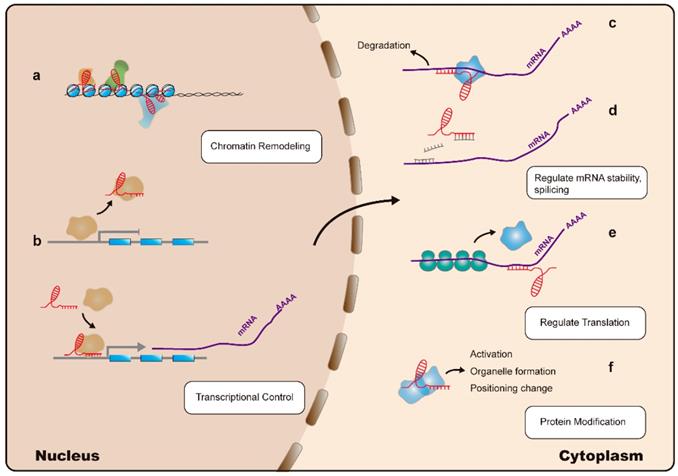
This review summarizes the current knowledge regarding lncRNA involvement in leukemia. The data indicate that several lncRNAs might have clinically promising applications in the diagnosis, prognosis, and treatment of leukemia.
Overview of LncRNAs
LncRNAs exhibit high functional heterogeneity, are generally located in the cytoplasm or nucleus, and possess diverse biological functions and complex regulatory mechanisms. According to the relative positional relationship between protein-coding genes and lncRNAs, lncRNAs can be roughly classified into five types: intron lncRNAs, antisense lncRNAs, intergenic lncRNAs, UTR (untranslated region)-associated lncRNAs, and promoter-associated lncRNAs [16]. Functionally speaking, lncRNAs can regulate gene expression at the chromatin-modification, gene-transcription, and post-transcriptional levels. Furthermore, mechanistically, lncRNA activity can be classified into four modes: signal, decoy, guide, and scaffold [17]. In the signal mode of action, lncRNAs can be associated with the genome-imprinting process: for example, when two X chromosomes are present, one is in a suppressed state, with this phenotype capable of being stably transmitted to a subsequent generation, where the X-inactive-specific transcript (Xist) plays an important regulatory role [18, 19]. The mode in which lncRNA functions as a decoy can be described as the binding of lncRNA to a protein with transcription-regulatory functions, such as transcription factors or nuclear receptors in the nucleus, in order to regulate transcriptional inhibition of target genes downstream. For example, when DNA damage occurs, p53 binds to the cyclin-dependent kinase inhibitor (CDKN)1 and activates expression of the lncRNA PANDAR, which blocks the expression of pro-apoptotic genes by binding to the nuclear transcription factor Y subunit α (NF-YA), thus prolonging cell survival [20]. As a guide, lncRNA binds to a protein molecule (usually a transcription factor) and promotes its localization to a specific DNA sequence to regulate downstream signaling pathways and gene expression. An example of this is the lncRNA Xist acting as a guide to target gene-silencing activity in an allele-specific manner. As a scaffold, lncRNA binds to multiple effector molecules simultaneously to provide a platform for interaction. For example, the 3′ domain of the lncRNA HOX antisense intergenic RNA (HOTAIR) binds the histone demethylase complex lysine-specific demethylase (LSD)1/CoRE1-silencing transcription factor (REST)/REST, whereas the 5′ domain of HOTAIR binds Polycomb repressive complex 2 (PRC2). The resulting interaction promotes assembly of selected histone-modification enzymes by providing a binding surface, thereby resulting in the chromosome being in a closed state; this results in gene silencing [21, 22]. In this review, we focused on the functions and mechanisms of lncRNAs involved in leukemia pathogenesis (Table 1).
Aberrant LncRNA expression in different subsets of leukemia
| Disease | LncRNA | Expression level in patients/cell lines | Mechanism | Clinical parameters and treatment responses | Ref. |
|---|---|---|---|---|---|
| AML | MEG3 | Downregulated | Inhibits tumorigenesis in a p53-dependent and -independent manner | Abnormal methylation of MEG3 confers worse OS | [23, 24] |
| H19 | Upregulated | Possibly correlated with ID2 expression | Highest in M2 AML, correlated with sex, older age, higher WBC counts, intermediate karyotype, FLT3-ITD DNMT3A mutations, lower CR rate, and shorter OS | [25]. | |
| UCA1 | Upregulated | Sponges for miR-126, miR-125a, miR-16; activates PI3K/AKT and JAK/STAT signaling | Elevated in patients carrying CEBPA mutations; elevated in ADR-resistant pediatric AML cases | [26-28] | |
| HOTAIR | Upregulated | Sponges for miR-193a and modulates c-KIT expression; regulates LSC self-renewal | Higher peripheral leukocyte and BM blast counts, lower platelet and hemoglobin counts, poor DFS and OS | [29-33] | |
| CRNDE | Upregulated | Promotes cell proliferation and cell cycle progression, inhibits apoptosis | Higher in M4 and M5 than in M1, M2, and M3 patients; negatively correlated with total survival time. | [34] | |
| PANDAR | Upregulated | Interacts with NF-YA and inhibits pro-apoptotic gene expression | Older age, higher BM blasts, poor karyotypes, lower CR rate, and shorter OS. | [35] | |
| PVT1 | Upregulated | Sponge for miR-1204 and regulates MYC activation | Elevated in t (8;21) AML and APL. Corelated with high-risk clinical criteria; shorter LFS and OS | [36-39] | |
| CASC15 | Downregulated | Regulates SOX4 expression | Elevated in t (8;21) AML, relatively better prognosis | [40] | |
| IRAIN | Downregulated | Interacts with the IGF1R promoter | Higher WBC counts, blast counts and shorter RFS, OS; refractory response to chemotherapy | [41, 42] | |
| RUNXOR | Upregulated | Interacts with the H3K27 methylase EZH2 and RUNX1 | Elevated in t (8;21) AML | [43] | |
| CCAT1 | Upregulated | Represses monocyte differentiation and promotes AML cell growth by sequestering miR-155 | Significantly elevated in M4 and M5 subtypes | [36, 44] | |
| CCDC26 | Upregulated | Regulates AML cell proliferation via c-KIT expression | Older age, anemia, poor/intermediate risk, partial/no remission, shorter OS | [45, 46] | |
| TUG1 | Upregulated | Targets AURKA and induces AML cell proliferation; reduces miR-34a expression and contributes to ADR resistance | Higher WBC counts, FLT3-ITD mutation, monosomal karyotype, poor-risk stratification, and worse event-free survival and OS | [47, 48] | |
| MALAT | Upregulated | Influences proliferation, apoptosis and Ara-C sensitivity by upregulating miR-96 | Markedly upregulated in M5 subtype, correlated with higher WBC and platelet counts, shorter OS | [49, 50] | |
| HOXA-AS2 | Upregulated | Suppresses ATRA-induced apoptosis via TRAIL; increases ADR resistance via the miR-520c-3p /S100A4 pathway | [51, 52] | ||
| MONC | Upregulated | Enhances proliferation of immature erythroid progenitor cells | [53] | ||
| NEAT1 | Downregulated | Impairs myeloid differentiation, regulates miR‐23a‐3p/SMC1A | [54] | ||
| ALL | BALR-2 | Upregulated | Inhibits downstream glucocorticoid receptor genes FOS, JUN, and BIM | Shorter OS and poor response to prednisone | [55] |
| BALR-6 | Upregulated | Regulation of the transcriptome downstream of SP1 | Highest expression in patients carrying MLL rearrangement | [56] | |
| CASC15 | Downregulated | Regulates SOX4 expression | Elevated in pediatric B-ALL with t (12; 21); associated with relatively better survival | [40] | |
| GAS5 | Downregulated | Sponge for miR-222; modulates B lymphocytic leukemia cell tumorigenesis and metastasis; essential for mTOR-related inhibition of T cell proliferation | Elevated on day 15, but decreased on day 33 after glucocorticoid therapy | [57-59] | |
| HOXA-AS2 | Upregulated | Enhances glucocorticoid resistance, upregulates HOXA3 to activate EGFR/Ras/Raf/MEK/ERK signaling | Higher in pediatric prednisone-poor response ALL cases | [60] | |
| ZEB1-AS1 | Upregulated | Promotes activation of IL-11/STAT3 signaling | Correlated with poor prognosis | [61] | |
| NEAT1 | Upregulated | Related to dysregulation of miR-335-3p and indirectly regulates multidrug-resistance genes | [62] | ||
| PVT1 | Upregulated | Participates in cell cycle progression and proliferation regulation | [63] | ||
| SNHG16 | Upregulated | Host of miR-124-3p and promotes ALL cell proliferation | [64] | ||
| NALT | Upregulated | Regulates NOTCH1 signaling | [65] | ||
| T-ALL-R-LncR1 | Upregulated | Inhibits formation of the Par-THAP1 complex and caspase-3 activation | [66] | ||
| LUNAR1 | Upregulated | Enhances IGF1R expression to sustain IGF1 signaling | [67] | ||
| XLOC_001561 | Downregulated | Involved in T cell differentiation and possible tumorigenesis | [68] | ||
| ANRIL | Upregulated | Alters CDKN2A/B expression; targets EZH2, and activates the NF-κB pathway | [69, 70] | ||
| Linc-PINT | Downregulated | Induces HMOX1 transcription and reduces ALL cell viability | [71] | ||
| Lnc-INSR | Upregulated | Promotes immune suppression by enhancing Treg cell differentiation | [72] | ||
| ARIEL | Upregulated | Activates ARID5B expression, thereby upregulating TAL1-induced transcriptional programs and MYC oncogene expression | [73] | ||
| RP11-137H2.4 | Upregulated | Involved in proliferation, apoptosis, cell migration, and glucocorticoid resistance | [74] | ||
| CML | HOTAIR | Upregulated | Contributes to IM resistance by activating the PI3K/AKT pathway | Upregulated in MRP1-high patients | [75] |
| MEG3 | Downregulated | Regulates miRNA-21, miRNA-147, and the JAK/STAT pathway | Lower in AP and BP than in CP patients. Lower in imatinib-resistant compared to imatinib-sensitive patients | [76-78] | |
| H19 | Upregulated | Required for BCR-ABL-mediated tumorigenesis | A tendency toward higher WBC counts and BCR-ABL transcript | [79, 80] | |
| HAND2-AS1 | Upregulated | Host of miR-1275 and promotes CML cell proliferation | Expression level in AP/BP stages was much lower than that in CP | [81] | |
| HULC | Upregulated | Sponge for miR-200a and modulates c-Myc and Bcl-2 levels | Positively correlated with clinical stages | [82] | |
| MALAT1 | Upregulated | MALAT1/miR-328 axis promotes CML cell proliferation and imatinib resistance | [83] | ||
| NEAT1 | Upregulated | Essential mediator of apoptosis induced by imatinib | [84] | ||
| SNHG5 | Upregulated | Affects DR4 methylation; promotes IM resistance by attenuating miR-205-5p expression | [85, 86] | ||
| UCA1 | Upregulated | Sponge for miRNA-16 and contributes to IM resistance | [87] | ||
| PLIN2 | Upregulated | Promotes tumor growth via Wnt/β-catenin signaling | [88] | ||
| FENDRR | Downregulated | HuR/FENDRR/miR-184 interaction contributes to MDR1 activity | [89] | ||
| BGL3 | Downregulated | Host of miR-17, miR-20a, miR-20b, miR-93, miR-106a, and miR-106b; regulates PTEN expression | [90] | ||
| CLL | DLEU1/2 | Downregulated | Host of miR-12a and miR-16-1; regulates NF-κB signaling | Corelated with poor prognosis | [91-94] |
| MALAT1 | Upregulated | Involved in tumorigenic processes | No statistically significance difference between the prognosis categories | [95] | |
| MIAT | Upregulated | Initiates a regulatory loop with OCT4 in malignant mature B cells | Correlated with rapid death cases | [96] | |
| GATA6-AS1 | Downregulated | Inhibits cell proliferation and enhances apoptosis in the caspase-9-dependent intrinsic apoptosis pathway | Methylation of GATA6-AS1 corelated with advanced Rai stage | [97] | |
| TRERNA1 | Upregulated | Enhances protection against cytotoxicity mediated DNA damage | Associated with aggressive disease markers, shorter time to treatment, shorter PFS and OS | [98] | |
| lncRNA-p21 | Downregulated | Activated by p53 and binds hnRNP-K to induce apoptosis | [99, 100] |
Aberrant LncRNA Expression in Leukemia
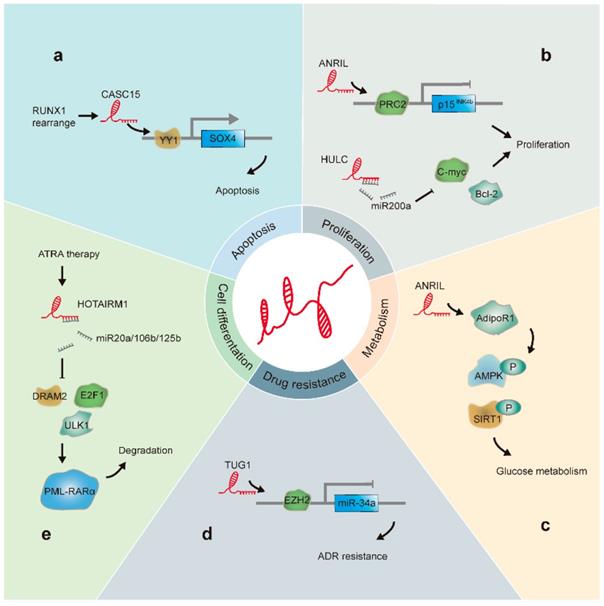
The positive or negative role of lncRNAs in leukemia progression is determined by their activity in terms of their specific roles in differentiation, energy metabolism, malignant proliferation, apoptosis, and drug resistance of leukemia cells (Figure 2). Here, we focused on lncRNAs that have been well studied in association with leukemia and describe the progress in this field and mechanisms elucidated.
Maternal Expression Gene 3 (MEG3)
MEG3, a putative tumor-suppressor gene located on chromosome 14q32, suppresses the proliferation of various tumor cells by directly regulating retinoblastoma protein phosphorylation and indirectly activating the p16INK4a pathway[23, 101]. Benetatos et al. observed that, in a sample of 42 acute myelogenous leukemia (AML) patients, 47.6% of patients displayed hypermethylation of MEG3, with MEG3 methylation associated with significantly reduced overall survival (OS); these findings suggest that this methylation status represents a useful biomarker for leukemia [24]. Previous studies have found that MEG3 plays a regulatory role in carcinogenesis and metastasis in chronic myelogenous leukemia (CML) by interacting with miRNA-21 [76]. Li et al. found that patients in accelerated and blast phases showed lower expression of miR-147 and MEG3. Furthermore, it was shown that MEG3 was capable of binding to several members of the Janus kinase (JAK)-signal transducer and activators of the transcription (STAT) pathway, resulting in reduced signaling. This activity regulated leukemia progression, suggesting a role for MEG3 and its target miR-147 as new therapeutic targets [77]. Furthermore, Zhou et al. showed that MEG3 might be involved in imatinib resistance by regulating miR-21, thereby affecting cell proliferation, multidrug-resistance transporter expression, and cell apoptosis [78].
LncRNAs involved in leukemia progression: (a). CASC15 is upregulated in RUNX1-rearranged AML. Additionally, CASC15 enhances Yin and Yang-1(YY1)-mediated regulation of the SOX4 promoter, thus increasing apoptosis. (b). HULC acts as a sponge for miR-200a and modulates c-Myc and Bcl-2 levels, promoting CML cell proliferation. ANRIL recruits PRC2 to the p15INK4b locus and silences the p15INK4b tumor suppressor gene, resulting in cell proliferation. (c) ANRIL regulates the expression of the adiponectin receptor (AdipoR1), a key regulator of glucose metabolism; this results in the regulation of AMPK and SIRT1 phosphorylation levels. (d). TUG1 epigenetically suppresses miR-34a expression by increasing EZH2 recruitment and H3K27me3 levels at the miR-34a promoter in AML cells, thus contributing to ADR resistance. (e). HOTAIRM1 acts as a sponge for miRNA20a/106b/125b, regulates the expression of autophagy-associated genes, and enhances PML-RARα degradation.
Nuclear Paraspeckle Assembly Transcript 1 (NEAT1)
NEAT1 is located on chromosome 11 and encodes transcripts that localize specifically to nuclear paraspeckles [102]. Studies show that NEAT1 is overexpressed in several types of solid tumors including childhood ALL samples[103]; in addition, NEAT1 is associated with aberrant expression of miR-335-3p, resulting in indirect modulation of the expression of multidrug-resistant genes, including ATP-binding cassette subfamily A member 3 [62]. However, in certain types of cancers, such as AML, NEAT1 might act as a tumor suppressor. Zhao et al. showed that the expression of NEAT1 and structural maintenance of chromosome 1α (SMC1A) were decreased in primary AML patients and THP-1 cells. Additionally, they found elevated levels of miR-23a-3p, which indicate that NEAT1 binds miR-23a-3p to regulate SMC1A expression, thereby inhibiting leukemia cell proliferation and enhancing apoptosis [54]. In de novo acute promyelocytic leukemia (APL), NEAT1 levels are significantly reduced: Zeng et al. reported that the promyelocytic leukemia-retinoic acid receptor α (PML-RARα) fusion protein inhibits NEAT1 expression, while the latter is significantly upregulated in APL cells treated with all-trans retinoic acid (ATRA) [104]. This group subsequently found downregulated NEAT1 expression in primary CML cells, which was restored by inhibition of BCR-ABL kinase activity [84]. Moreover, their results indicated that NEAT1 was transcriptionally regulated by c-Myc via binding to the NEAT1 promoter, and that splicing factor and proline- and glutamine-rich protein were required for NEAT1-mediated K562 cell apoptosis [84]. These results suggest that targeting NEAT1 represents a new treatment strategy for leukemia, and contribute to a more comprehensive understanding of the pathogenesis of this cancer.
HOX Antisense Intergenic RNA Myeloid 1 (HOTAIRM1)
HOTAIRM1 is a lncRNA located in the HOXA gene cluster; this lncRNA is generated by RNA polymerase II antisense transcription and expressed in the myeloid lineage. Retinoic acid induces HOTAIRM1 expression and thereby regulates the expression of genes involved in myeloid differentiation. Studies show that HOTAIRM1 levels are significantly elevated during ATRA-induced NB4 cell lineage differentiation; in contrast, HOTAIRMI knockdown inhibits ATRA-induced granulocyte differentiation and releases cell cycle arrest in the G1/S phase, revealing that HOTAIRM1 can regulate the maturation of myeloid cells by affecting integrin gene expression [105]. Additionally, Chen et al. revealed that HOTAIRM1 acts as a miRNA sponge for miR-20a/106b and miR-125b. Further, downregulation of HOTAIRM1 levels inhibits ATRA-induced PML-RARα degradation via miRNA-mediated pathways to suppress the expression of autophagy-related genes and granulocyte differentiation of APL cells [106]. These results suggest that HOTAIRM1 plays an essential role in myeloid differentiation in leukemia. However, another study showed that in 241 AML patient specimens, elevated HOTAIRM1 levels were associated with shorter leukemia-free survival, shorter OS, and higher cumulative incidence of relapse [106].
Leukemia-induced Noncoding Activator RNA-1 (LUNAR1)
LUNAR1 is a NOTCH-regulated oncogenic lncRNA, located on chromosome 15q26.3, and specifically expressed in T cell acute lymphoblastic leukemia (T-ALL), thereby playing a crucial role in its progression. The gene encoding LUNAR1 is located in close proximity to the insulin-like growth factor 1 receptor (IGF1R) gene. Upon activation, LUNAR1 recruits the mediator complex on the IGF1R promoter to regulate its transcription, thereby promoting T-ALL cell proliferation [67]. In vivo experiments involving the transplantation of tumor cells into mice revealed that tumor proliferation was blocked only in mice in which LUNAR1 was inactivated. This suggests that LUNAR1 inhibitors can potentially be used for targeted therapy.
Antisense Noncoding RNA in the INK4 Locus (ANRIL)
ANRIL, which is located on chromosome 9p21, exerts an oncogenic function through modulation of p15INK4b and p16INK4a expression[107]. In the development of B-cell precursor (BP)-ALL and AML, ANRIL is overexpressed, thereby aggravating inhibition of the p15INK4b locus. Iacobucci et al. compared ALL blood samples with nonmalignant controls and showed an apparent correlation between ANRIL and BCR-ABL-associated ALL nucleotide polymorphisms. They speculated that this association reflects the ability of certain ANRIL polymorphisms to contribute to their own transcriptional alteration and increased susceptibility to ALL[69]. In AML, ANRIL is upregulated in patients and downregulated after complete remission (CR) [106]. Additionally, this study showed that ANRIL regulated the expression of the adiponectin receptor and its downstream factors adenosine monophosphate-activated protein kinase (AMPK)/sirtuin 1 (SIRT1), thereby modulating disease progression by regulating glucose metabolism [108]; these findings suggest that ANRIL represents both a potential candidate for AML diagnosis and a therapeutic target. Furthermore, Song et al. showed that ANRIL could promote the proliferation of T-ALL cells by targeting enhancer of zeste homolog 2 (EZH2) and activating the nuclear factor kappaB (NF-κB) pathway, indicating that aberrant ANRIL expression was involved in T-ALL leukemogenesis [70].
Deleted in Leukemia (DLEU)1/2
Over 50% of patients with chronic B-cell lymphocytic leukemia have a 13q14.3 deficiency. This critical deleted region comprises two adjacent subregions: DLEU1 and DLEU2 [109]. DLEU2 negatively regulates cyclins D1 and E1 via miR-15a and miR-16-1, which play important roles in chronic lymphocytic leukemia (CLL) pathogenesis by regulating B-cell lymphoma 2 expression [92, 110]. Additionally, Garding et al. found that DLEU1 and DLEU2 were significantly demethylated at their respective 5′ end in almost all CLL patients, resulting in attenuated transcription of a series of adjacent sequences encoding tumor-suppressor genes [93]. Moreover, DLEU1 is reportedly poorly expressed in other solid tumors and negatively correlated with prognosis [111]. Therefore, its clinical application in leukemia treatment requires further investigation.
β Globin Locus Transcript 3 (BGL3)
BGL3 is located on chromosome 11p15.4 and negatively regulated by c-Myc-dependent DNA methylation. BGL3 is a host of miR-17, miR-20a, miR-20b, miR-93, miR-106a, and miR-106b, and acts as a competing endogenous (ce)RNA to alter the expression of the tumor suppressor PTEN. Additionally, BGL3 overexpression significantly reduces the survival of K562 cells and promotes imatinib-induced apoptosis [90]. These results indicate that BCR-ABL-mediated cell transformation requires silencing of the tumor suppressor BGL3, thereby offering a potential strategy for treatment of BCR-ABL-positive leukemia.
HOX Transcript Antisense RNA (HOTAIR)
HOTAIR is located on chromosome 12 and is transcribed from the antisense strand of the homeobox C gene locus. HOTAIR plays a repressive role by interacting with and guiding various chromatin-modifying complexes, including LSD1 and PRC2, to target-gene promoter regions, resulting in gene silencing [112]. In hematological malignancies, HOTAIR regulates self-renewal of leukemia stem cells (LSCs) to promote uncontrolled self-renewal and proliferation [33]. Previous studies report HOTAIR overexpression in AML patients, and show that these elevated levels are associated with higher peripheral leukocyte and bone marrow blast counts and lower platelet and hemoglobin counts, as well as poor clinical prognosis [29-31]. Furthermore, HOTAIR acts as a ceRNA by binding to miR-193a, which targets c-KIT, thus modulating c-KIT expression [32]. In CML, HOTAIR is upregulated in patients with elevated levels of multidrug-resistance protein 1. In K562 imatinib-resistant cells, HOTAIR knockdown leads to higher sensitivity to imatinib via the inactivation of phosphoinositide 3-kinase (PI3K)/AKT signaling [75]. These results suggest HOTAIR is involved in the development of imatinib resistance.
Urothelial Carcinoma-associated 1 (UCA1)
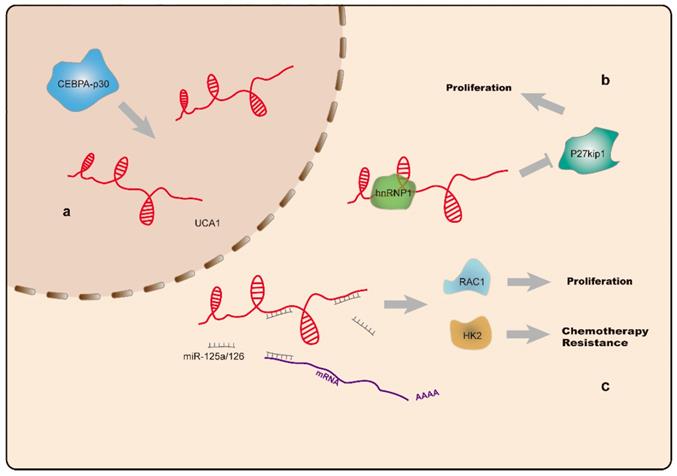
UCA1 is located on chromosome 19p13 [113] and highly expressed as a proto-oncogene in a variety of tumors [114-118]. Additionally, UCA1 levels are elevated in AML and CML cell lines. The oncogenic effect of UCA1 is achieved by sponging miR-126, which precludes degradation of Ras-related C3 botulinum toxin substrate-1(RAC1) and activates JAK/STAT and PI3K/AKT signaling [26]. Hughes et al. found elevated UCA1 levels in AML patients carrying CCAAT enhancer binding protein α (CEBPA) mutations, and that UCA1 sustained the proliferation of AML cells by inhibiting expression of the cell cycle regulator p27kip1 [27]. Additionally, they found that abnormally expressed UCA1 acted as a ceRNA targeting miR-125a, which resulted in upregulated hexokinase-2 (HK2) expression, a key enzyme involved in glycolysis. Moreover, UCA1 is associated with resistance to chemotherapy, with elevation of UCA1 expression following doxorubicin (ADR)-based chemotherapy. UCA1 knockdown in ADR-resistant HL60 cells partially reversed AML chemoresistance via the miR-125a/HK2 axis(Figure 3) [28]. Another study reported that UCA1 is capable of binding miR-16 to regulate MDR1 expression and promote imatinib resistance in CML cells [87]. These findings support UCA1 as a potential diagnostic biomarker and therapeutic target for leukemia treatment and reversal of drug resistance.
a. CEBPA-p30 protein promotes UCA1 (indicated in red) expression in AML cells with CEBPA mutations. b. UCA1 sustains proliferation of AML cells by repressing the expression of the cell cycle regulator p27kip1. c. The UCA1 transcript functions as a sponge for miR-125a and miR-126, thus modulating RAC1 and HK2 expression, and promotes AML cell proliferation and resistance to chemotherapy.
H19
H19 is an endogenous gene located on chromosome 11p15 [119]. H19 maintains hematopoietic stem cell (HSC) quiescence at the transcriptional and post-transcriptional levels by regulating IGF2-IGFR1 activity [120]. Other studies have identified H19 as either an oncogene or tumor suppressor, depending on tumor type [121-125]. Guo et al. reported that H19 expression is positively regulated by c-Myc and required for tumorigenesis, as H19 knockdown enhanced the sensitivity of CML cells to imatinib, inhibited BCR-ABL-induced tumor proliferation, and promoted apoptosis [79]. A subsequent study indicated that hypomethylation of the H19 differentially methylated region/imprinting control region might mediate its overexpression in CML [80]. Zhang et al. identified upregulated H19 levels in AML patients that were correlated with lower CR rates and shorter OS [25]. These findings suggest that H19 plays different roles in different malignancies. Therefore, further research is needed to comprehensively elucidate the H19-specific mechanisms of action in leukemia.
LncRNA Expression Profiles in Leukemia
Although the study of global lncRNA expression in leukemia remains limited, the expression patterns of various lncRNAs related to leukemia tumorigenesis and specific subtypes have been examined through expression profile analysis.
Lei et al. performed transcriptome analysis of lncRNA-expression profiles of AML patient samples and healthy controls, and identified differentially expressed lncRNAs, revealing that upregulated lncRNAs in AML were related to higher levels of binding to transcription factors such as STAT4, SP1 and ELK1, and lower levels of DNA methylation. Additionally, they found that LOC285758 stimulates the proliferation of AML cell lines by increasing levels of histone deacetylase 2, with elevated LOC285758 levels in patients associated with worse prognosis [126]. By comparing in-depth sequencing data for various RNA-seq libraries and integrating RNA-seq data from 179 AML patients, Zhang et al. showed that lncRNAs are associated with specific AML subtypes [127]. They observed that a subset of lncRNAs were abundantly expressed in patients with M3 subtypes, which are initiated following expression of the PML-RARα fusion gene. Schwarzer et al. revealed noncoding RNA stem cell characteristics as prognostic features shared by healthy HSCs and AML blasts cells, and identified lncRNA signatures specific for acute megakaryoblastic leukemia (AMKL), Down syndrome-associated AMKL, inv(16), t(8; 21), and mixed-lineage leukemia-rearranged samples [128]. Ghavazi et al. performed a comprehensive analysis of the lncRNA transcriptome in ETS variant 6 (ETV6)/Runt-related transcription factor 1 (RUNX1)-positive BP-ALL, and found a specific lncRNA signature comprising 596 lncRNA transcripts [129] . Following data integration with RNA-seq results from other BP-ALL cell lines, they identified 16 unique lncRNA-expression profiles associated with the ETV6/RUNX1 fusion protein, including a potential carcinogenic lncRNA (DBH-AS1) [130]. Another study revealed differential lncRNA expression between AML and ALL patients (1168 mRNAs and 2101 lncRNAs differed between leukemia subsets), with subsequent analysis of co-expression networks revealing single mRNAs potentially associated with more than one lncRNA, and vice versa. These results indicate that lncRNAs play important roles in regulating AML and ALL development [131].
LncRNA expression represents a potential prognostic marker for leukemia, and may enable risk stratification in leukemia patients. Feng et al. identified three lncRNAs (RP11-305O.6, AC092580.4, and RP11-222K16.2) related to the OS of AML patients, with further experiments suggesting that RP11-222K16.2 might affect the differentiation of natural killer cells to promote immune escape of AML [132]. Tsai et al. recruited 275 newly diagnosed non-M3 AML patients and established a prognostic lncRNA score system comprising 5 lncRNAs, demonstrating lncRNA score as an independent prognostic factor for AML [133]. Garzon et al. used a custom microarray platform to study lncRNA-expression profiles in elderly patients with normal cytogenetic (CN)-AML, with an emphasis on evaluating associations with conventional mutations and phenotypes. This led to the identification of dysregulated lncRNAs associated with select gene mutations and clinical outcomes [134]. Additionally, they obtained a lncRNA score from 48 lncRNAs, with patients who had unfavorable lncRNA scores also displaying lower CR rates and shorter disease-free survival and OS. Because the clinical features, molecular abnormalities, and outcomes of older patients with CN-AML differ from those in younger adults, they also studied the prognostic value and biological significance of lncRNA expression in 377 adult patients (<60-years old) with CN-AML. Their results revealed no overlap between the 48 prognostic lncRNAs found in elderly CN-AML patients and the 24 transcripts reported in younger patients [135]. This might be attributable to additional biological differences between the two cohorts, such as age-dependent differences in the frequency of mutations in recurrent prognostic genes. Mer et al. used RNA-seq analysis to detect lncRNA expression in 274 AML patients, finding that 33 lncRNAs were associated with OS[136]. A study identified 24 lncRNA signatures showing differential expression in CLL relative to normal B cell controls, with an independent risk model based on the expression of lnc-KIAA1755-4 and lnc-IRF2-3 capable of distinguishing between three different prognostic groups [137].
Conclusions and Future Directions
LncRNAs are a large class of transcripts that play important roles in biological processes in malignant cells. The number of identified human lncRNAs has increased in the previous 15 years, with many of these also identified in leukemia. However, an understanding of the roles played by lncRNAs in leukemia occurrence and development remains insufficient. In this review, we briefly describe lncRNAs involved in leukemia progression and the underlying mechanisms.
LncRNA functions include regulation of cell differentiation, energy metabolism, malignant proliferation, and drug resistance. Future work should explore these functions more extensively, including subcellular localization to promote function prediction. Most lncRNAs localized to the nucleus modulate transcription and epigenetic modification, whereas those localized to the cytoplasm are likely to be involved in regulation at the post-transcriptional level. Further in-depth investigation of abnormally expressed lncRNAs in leukemia will enable elucidation of leukemia pathogenesis, and potentially provide feasible approaches for its treatment. Additionally, lncRNAs represent potential biomarkers for leukemia diagnosis and prognosis, with reports that abnormal expression of specific lncRNAs is related to leukemia-specific clinicopathological parameters. For example, HOTAIR and H19 correlate with poor OS in AML cases. Furthermore, model scores constructed based on differentially expressed lncRNAs obtained from sequencing or microarray data represent good prognostic predictors. Accordingly, expression of specific lncRNAs could represent a novel diagnostic biomarker and provide guidance for the prediction of clinical outcomes of leukemia; however, unified evaluation criteria based on a large sample are required. Prior to their use as leukemia biomarkers, large-scale prospective trials should be conducted in order to confirm their clinical usefulness and verify their accuracy and sensitivity as diagnostic and prognostic tools.
Although numerous lncRNAs have been identified, their regulatory functions remain largely unclear. Because lncRNAs exhibit low levels of expression and show poor species conservation relative to protein-coding genes, rapidly evolving RNA-seq technology can be used as a faster and more accurate detection system. Laboratory studies show that siRNA-targeting technology and CRISPR-Cas9 can effectively knockdown lncRNAs. The potential clinical utility of lncRNA-interference therapy should be explored in animal models and clinical trials in order to develop novel therapeutic strategies based on lncRNA targets in leukemia.
Abbreviations
ADR: doxorubicin; ALL: acute lymphoblastic leukemia; AMKL: acute megakaryoblastic leukemia; AML: acute myeloblastic leukemia; AMPK: Adenosine monophosphate-activated protein kinase; ANRIL: Antisense noncoding RNA in the INK4 locus; APL: acute promyelocytic leukemia; ARIEL: ARID5B-inducing enhancer associated long noncoding RNA; ATL: acute T cell leukemia; ATRA: all-trans retinoic acid; BALR: B-ALL associated long RNA; BGL3: β globin locus transcript 3; BP-ALL: B-cell precursor ALL; CASC15: cancer susceptibility candidate 15; CCAT1: colon cancer associated transcript-1; CDKN: cyclin-dependent kinase inhibitor; CEBPA: CCAAT enhancer binding protein α; CLL: chronic lymphocytic leukemia; CML: chronic lymphocytic leukemia; CN-AML: normal cytogenetic-AML; CRNDE: colorectal neoplasia differentially expressed; DLEU: deleted in leukemia; EZH2: enhancer of zeste homolog 2; FENDRR: FOXF1 adjacent non-coding developmental regulatory RNA; GAS5: growth arrest-specific 5; GATA6-AS1: homo sapiens GATA6 antisense RNA 1; H19: H19 imprinted maternally expressed transcript; HANDS-AS1: HAND2 antisense RNA 1; HK2: hexokinase 2; HSC: hematopoietic stem cell; HOTAIR: HOX antisense intergenic RNA; HOXA-AS2: HOXA cluster antisense RNA 2; HULC: highly upregulated in liver cancer RNA; LFS: leukemia-free survival; IGFR1: insulin-like growth factor receptor 1; IRAIN: IGF1R antisense imprinted non-protein coding RNA; JAK: Janus kinase; LincRNA-p21: long intergenic non-coding RNA p21; Linc-PINT: long intergenic non-protein coding RNA, p53 induced transcript; Lnc-INSR: lnc-insulin receptor precursor; LncRNA: long noncoding RNA; LSC: leukemia stem cells; LSD: leukemia stem cells; LUNAR1: leukemia-induced noncoding activator RNA-1; MALAT1: metastasis-associated lung adenocarcinoma transcript 1; MEG3: maternally expressed 3; MIAT: myocardial infarction associated transcript; mRNA: messenger RNA; miRNA: microRNA; MONC: megakaryocytic oncogenic non-coding RNA; ncRNA: non-coding RNA; NEAT1: nuclear paraspeckle assembly protein 1; NF-YA: nuclear paraspeckle assembly protein 1; PANDAR: promoter of CDKN1A antisense DNA damage activated RNA; PML-RARα: promyelocytic leukemia-retinoic acid receptor α; PRC2: Polycomb repressive complex 2; PVT1: plasmacytoma variant translocation 1; REST: RE1-silencing transcription factor; siRNA: small-interfering RNA; SIRT1: sirtuin-1; SMC1A: structural maintenance of chromosome 1α; SNHG: small nucleolar RNA host gene; SP1: Specificity Protein 1; STAT: signal transducer and activators of transcription; TRERNA1: translation regulatory long non-coding RNA 1; TUG1: taurine up-regulated 1; UCA1: urothelial carcinoma-associated 1; Xist: X-inactive-specific transcript; ZEB1-AS1: ZEB1 antisense RNA 1.
Acknowledgements
Author Contributions
J.G. drafted the work. F.W., P.W., and Y.C. substantively revised the manuscript and contributed intellectually. Y.J. conceived the review. All authors read and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070-82
2. Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood cancer journal. 2017;7:e577
3. Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563-72
4. Stengel A, Schnittger S, Weissmann S, Kuznia S, Kern W, Kohlmann A. et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood. 2014;124:251-8
5. Mullighan CG. Genomic profiling of B-progenitor acute lymphoblastic leukemia. Best practice & research Clinical haematology. 2011;24:489-503
6. Nordlund J, Kiialainen A, Karlberg O, Berglund EC, Goransson-Kultima H, Sonderkaer M. et al. Digital gene expression profiling of primary acute lymphoblastic leukemia cells. Leukemia. 2012;26:1218-27
7. Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Advances in experimental medicine and biology. 2017;1008:1-46
8. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-8
9. St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends in genetics: TIG. 2015;31:239-51
10. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cellular and molecular life sciences: CMLS. 2016;73:2491-509
11. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. The Journal of clinical investigation. 2017;127:761-71
12. Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Advances in experimental medicine and biology. 2017;1008:47-74
13. Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792-804
14. Han BW, Chen YQ. Potential pathological and functional links between long noncoding RNAs and hematopoiesis. Science signaling. 2013;6:re5
15. Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965-75
16. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends in biochemical sciences. 2016;41:761-72
17. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904-14
18. da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nature structural & molecular biology. 2017;24:197-204
19. Lu Z, Carter AC, Chang HY. Mechanistic insights in X-chromosome inactivation. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2017;372:20160356
20. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621-9
21. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (New York, NY). 2010;329:689-93
22. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-23
23. Lyu Y, Lou J, Yang Y, Feng J, Hao Y, Huang S. et al. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017;31:2543-51
24. Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M. et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leukemia research. 2010;34:148-53
25. Zhang TJ, Zhou JD, Zhang W, Lin J, Ma JC, Wen XM. et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clinical epigenetics. 2018;10:47
26. Sun MD, Zheng YQ, Wang LP, Zhao HT, Yang S. Long noncoding RNA UCA1 promotes cell proliferation, migration and invasion of human leukemia cells via sponging miR-126. European review for medical and pharmacological sciences. 2018;22:2233-45
27. Hughes JM, Legnini I, Salvatori B, Masciarelli S, Marchioni M, Fazi F. et al. C/EBPalpha-p30 protein induces expression of the oncogenic long non-coding RNA UCA1 in acute myeloid leukemia. Oncotarget. 2015;6:18534-44
28. Zhang Y, Liu Y, Xu X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. Journal of cellular biochemistry. 2018;119:6296-308
29. Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. International journal of clinical and experimental pathology. 2015;8:7223-8
30. Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B. et al. Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncology letters. 2015;10:2410-4
31. Zhang YY, Huang SH, Zhou HR, Chen CJ, Tian LH, Shen JZ. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncology reports. 2016;36:3113-22
32. Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Bin Z. et al. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS letters. 2015;589:1981-7
33. Gao S, Zhou B, Li H, Huang X, Wu Y, Xing C. et al. Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Experimental hematology. 2018;67:32-40
34. Wang Y, Zhou Q, Ma JJ. High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. European review for medical and pharmacological sciences. 2018;22:763-70
35. Yang L, Zhou JD, Zhang TJ, Ma JC, Xiao GF, Chen Q. et al. Overexpression of lncRNA PANDAR predicts adverse prognosis in acute myeloid leukemia. Cancer management and research. 2018;10:4999-5007
36. El-Khazragy N, Elayat W, Matbouly S, Seliman S, Sami A, Safwat G. et al. The prognostic significance of the long non-coding RNAs "CCAT1, PVT1" in t(8;21) associated Acute Myeloid Leukemia. Gene. 2019;707:172-7
37. Salehi M, Sharifi M, Bagheri M. Knockdown of Long Noncoding RNA Plasmacytoma Variant Translocation 1 with Antisense Locked Nucleic Acid GapmeRs Exerts Tumor-Suppressive Functions in Human Acute Erythroleukemia Cells Through Downregulation of C-MYC Expression. Cancer biotherapy & radiopharmaceuticals. 2019;34:371-9
38. Salehi M, Sharifi M. Induction of apoptosis and necrosis in human acute erythroleukemia cells by inhibition of long non-coding RNA PVT1. Molecular biology research communications. 2018;7:89-96
39. Zeng C, Yu X, Lai J, Yang L, Chen S, Li Y. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. Journal of hematology & oncology. 2015;8:126
40. Fernando TR, Contreras JR, Zampini M, Rodriguez-Malave NI, Alberti MO, Anguiano J. et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Molecular cancer. 2017;16:126
41. Sun J, Li W, Sun Y, Yu D, Wen X, Wang H. et al. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic acids research. 2014;42:9588-601
42. Pashaiefar H, Izadifard M, Yaghmaie M, Montazeri M, Gheisari E, Ahmadvand M. et al. Low Expression of Long Noncoding RNA IRAIN Is Associated with Poor Prognosis in Non-M3 Acute Myeloid Leukemia Patients. Genetic testing and molecular biomarkers. 2018;22:288-94
43. Wang H, Li W, Guo R, Sun J, Cui J, Wang G. et al. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. International journal of cancer. 2014;135:2783-94
44. Chen L, Wang W, Cao L, Li Z, Wang X. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to Regulate Cell Growth and Differentiation in Acute Myeloid Leukemia. Molecules and cells. 2016;39:330-6
45. Chen C, Wang P, Mo W, Zhang Y, Zhou W, Deng T. et al. lncRNA-CCDC26, as a novel biomarker, predicts prognosis in acute myeloid leukemia. Oncology letters. 2019;18:2203-11
46. Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, Yamazaki T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Molecular cancer. 2015;14:90
47. Li Q, Song W, Wang J. TUG1 confers Adriamycin resistance in acute myeloid leukemia by epigenetically suppressing miR-34a expression via EZH2. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;109:1793-801
48. Wang X, Zhang L, Zhao F, Xu R, Jiang J, Zhang C. et al. Long non-coding RNA taurine-upregulated gene 1 correlates with poor prognosis, induces cell proliferation, and represses cell apoptosis via targeting aurora kinase A in adult acute myeloid leukemia. Annals of hematology. 2018;97:1375-89
49. Hu N, Chen L, Wang C, Zhao H. MALAT1 knockdown inhibits proliferation and enhances cytarabine chemosensitivity by upregulating miR-96 in acute myeloid leukemia cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;112:108720
50. Huang JL, Liu W, Tian LH, Chai TT, Liu Y, Zhang F. et al. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncology reports. 2017;38:1353-62
51. Zhao H, Zhang X, Frazao JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. Journal of cellular biochemistry. 2013;114:2375-83
52. Dong X, Fang Z, Yu M, Zhang L, Xiao R, Li X. et al. Knockdown of Long Noncoding RNA HOXA-AS2 Suppresses Chemoresistance of Acute Myeloid Leukemia via the miR-520c-3p/S100A4 Axis. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;51:886-96
53. Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Molecular cancer. 2014;13:171
54. Zhao C, Wang S, Zhao Y, Du F, Wang W, Lv P. et al. Long noncoding RNA NEAT1 modulates cell proliferation and apoptosis by regulating miR-23a-3p/SMC1A in acute myeloid leukemia. Journal of cellular physiology. 2019;234:6161-72
55. Fernando TR, Rodriguez-Malave NI, Waters EV, Yan W, Casero D, Basso G. et al. LncRNA Expression Discriminates Karyotype and Predicts Survival in B-Lymphoblastic Leukemia. Molecular cancer research: MCR. 2015;13:839-51
56. Rodriguez-Malave NI, Fernando TR, Patel PC, Contreras JR, Palanichamy JK, Tran TM. et al. BALR-6 regulates cell growth and cell survival in B-lymphoblastic leukemia. Molecular cancer. 2015;14:214
57. Jing Z, Gao L, Wang H, Chen J, Nie B, Hong Q. Long non-coding RNA GAS5 regulates human B lymphocytic leukaemia tumourigenesis and metastasis by sponging miR-222. Cancer biomarkers: section A of Disease markers. 2019;26:385-92
58. Gasic V, Stankovic B, Zukic B, Janic D, Dokmanovic L, Krstovski N. et al. Expression Pattern of Long Non-coding RNA Growth Arrest-specific 5 in the Remission Induction Therapy in Childhood Acute Lymphoblastic Leukemia. Journal of medical biochemistry. 2019;38:292-8
59. Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Molecular pharmacology. 2010;78:19-28
60. Zhao Q, Zhao S, Li J, Zhang H, Qian C, Wang H. et al. TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;109:1640-9
61. Wang Q, Du X, Yang M, Xiao S, Cao J, Song J. et al. LncRNA ZEB1-AS1 contributes to STAT3 activation by associating with IL-11 in B-lymphoblastic leukemia. Biotechnology letters. 2017;39:1801-10
62. Pouyanrad S, Rahgozar S, Ghodousi ES. Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long non-coding RNAs, is associated with poor prognosis in childhood acute lymphoblastic leukemia. Gene. 2019;692:35-43
63. Yazdi N, Houshmand M, Atashi A, Kazemi A, Najmedini AA, Zarif MN. Long noncoding RNA PVT1: potential oncogene in the development of acute lymphoblastic leukemia. Turkish journal of biology = Turk biyoloji dergisi. 2018;42:405-13
64. Yang T, Jin X, Lan J, Wang W. Long non-coding RNA SNHG16 has Tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB life. 2019;71:134-42
65. Wang Y, Wu P, Lin R, Rong L, Xue Y, Fang Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Scientific reports. 2015;5:13749
66. Zhang L, Xu HG, Lu C. A novel long non-coding RNA T-ALL-R-LncR1 knockdown and Par-4 cooperate to induce cellular apoptosis in T-cell acute lymphoblastic leukemia cells. Leukemia & lymphoma. 2014;55:1373-82
67. Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A. et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593-606
68. Ngoc PCT, Tan SH, Tan TK, Chan MM, Li Z, Yeoh AEJ. et al. Identification of novel lncRNAs regulated by the TAL1 complex in T-cell acute lymphoblastic leukemia. Leukemia. 2018;32:2138-51
69. Iacobucci I, Sazzini M, Garagnani P, Ferrari A, Boattini A, Lonetti A. et al. A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leukemia research. 2011;35:1052-9
70. Song Z, Wu W, Chen M, Cheng W, Yu J, Fang J. et al. Long Noncoding RNA ANRIL Supports Proliferation of Adult T-Cell Leukemia Cells through Cooperation with EZH2. Journal of virology. 2018;92:e00909-18
71. Garitano-Trojaola A, Jose-Eneriz ES, Ezponda T, Unfried JP, Carrasco-Leon A, Razquin N. et al. Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget. 2018;9:12842-52
72. Wang Y, Yang X, Sun X, Rong L, Kang M, Wu P. et al. Bone marrow infiltrated Lnc-INSR induced suppressive immune microenvironment in pediatric acute lymphoblastic leukemia. Cell death & disease. 2018;9:1043
73. Tan SH, Leong WZ, Ngoc PCT, Tan TK, Bertulfo FC, Lim MC. et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood. 2019;134:239-51
74. Ouimet M, Drouin S, Lajoie M, Caron M, St-Onge P, Gioia R. et al. A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget. 2017;8:7477-88
75. Wang H, Li Q, Tang S, Li M, Feng A, Qin L. et al. The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology (Amsterdam, Netherlands). 2017;22:208-16
76. Li Z, Yang L, Liu X, Nie Z, Luo J. Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;104:181-92
77. Li ZY, Yang L, Liu XJ, Wang XZ, Pan YX, Luo JM. The Long Noncoding RNA MEG3 and its Target miR-147 Regulate JAK/STAT Pathway in Advanced Chronic Myeloid Leukemia. EBioMedicine. 2018;34:61-75
78. Zhou X, Yuan P, Liu Q, Liu Z. LncRNA MEG3 Regulates Imatinib Resistance in Chronic Myeloid Leukemia via Suppressing MicroRNA-21. Biomolecules & therapeutics. 2017;25:490-6
79. Guo G, Kang Q, Chen Q, Chen Z, Wang J, Tan L. et al. High expression of long non-coding RNA H19 is required for efficient tumorigenesis induced by Bcr-Abl oncogene. FEBS letters. 2014;588:1780-6
80. Zhou JD, Lin J, Zhang TJ, Ma JC, Li XX, Wen XM. et al. Hypomethylation-mediated H19 overexpression increases the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukemia. Journal of cellular physiology. 2018;233:2444-50
81. Yang JR, Shi MX, Zeng Y. LncRNA HAND2-AS1 inhibits proliferation and promotes apoptosis of chronic myeloid leukemia cells by sponging with micRNA-1275. European review for medical and pharmacological sciences. 2019;23:2103-11
82. Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J. et al. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene. 2017;607:41-6
83. Wen F, Cao YX, Luo ZY, Liao P, Lu ZW. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochemical and biophysical research communications. 2018;507:1-8
84. Zeng C, Liu S, Lu S, Yu X, Lai J, Wu Y. et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Molecular cancer. 2018;17:130
85. He B, Bai Y, Kang W, Zhang X, Jiang X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. American journal of cancer research. 2017;7:1704-13
86. Gao B, Li S, Li G. Long Noncoding RNA (lncRNA) Small Nucleolar RNA Host Gene 5 (SNHG5) Regulates Proliferation, Differentiation, and Apoptosis of K562 Cells in Chronic Myeliod Leukemia. Medical science monitor: international medical journal of experimental and clinical research. 2019;25:6812-9
87. Xiao Y, Jiao C, Lin Y, Chen M, Zhang J, Wang J. et al. lncRNA UCA1 Contributes to Imatinib Resistance by Acting as a ceRNA Against miR-16 in Chronic Myeloid Leukemia Cells. DNA and cell biology. 2017;36:18-25
88. Sun C, Luan S, Zhang G, Wang N, Shao H, Luan C. CEBPA-mediated upregulation of the lncRNA PLIN2 promotes the development of chronic myelogenous leukemia via the GSK3 and Wnt/beta-catenin signaling pathways. American journal of cancer research. 2017;7:1054-67
89. Zhang F, Ni H, Li X, Liu H, Xi T, Zheng L. LncRNA FENDRR attenuates adriamycin resistance via suppressing MDR1 expression through sponging HuR and miR-184 in chronic myelogenous leukaemia cells. FEBS letters. 2019;593:1993-2007
90. Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y. et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34:1768-79
91. Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE. et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England journal of medicine. 2005;353:1793-801
92. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13944-9
93. Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K. et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS genetics. 2013;9:e1003373
94. Dal Bo M, Rossi FM, Rossi D, Deambrogi C, Bertoni F, Del Giudice I. et al. 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes, chromosomes & cancer. 2011;50:633-43
95. Ahmadi A, Kaviani S, Yaghmaie M, Pashaiefar H, Ahmadvand M, Jalili M. et al. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood research. 2018;53:320-4
96. Sattari A, Siddiqui H, Moshiri F, Ngankeu A, Nakamura T, Kipps TJ. et al. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget. 2016;7:54174-82
97. Wang LQ, Wong KY, Li ZH, Chim CS. Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget. 2016;7:82400-10
98. Miller CR, Ruppert AS, Fobare S, Chen TL, Liu C, Lehman A. et al. The long noncoding RNA, treRNA, decreases DNA damage and is associated with poor response to chemotherapy in chronic lymphocytic leukemia. Oncotarget. 2017;8:25942-54
99. Blume CJ, Hotz-Wagenblatt A, Hullein J, Sellner L, Jethwa A, Stolz T. et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29:2015-23
100. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-19
101. Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. International journal of cancer. 2011;129:773-9
102. Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell proliferation. 2017;50:e12329
103. Li S, Li J, Chen C, Zhang R, Wang K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes & diseases. 2018;5:27-35
104. Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L. et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC cancer. 2014;14:693
105. Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA biology. 2014;11:777-87
106. Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR. et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell death and differentiation. 2017;24:212-24
107. Y K, T N, K K, S S, N L, M K. et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956-62
108. Sun LY, Li XJ, Sun YM, Huang W, Fang K, Han C. et al. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Molecular cancer. 2018;17:127
109. Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D. et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15:2463-73
110. Lerner M, Harada M, Loven J, Castro J, Davis Z, Oscier D. et al. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Experimental cell research. 2009;315:2941-52
111. Li X, Li Z, Liu Z, Xiao J, Yu S, Song Y. Long non-coding RNA DLEU1 predicts poor prognosis of gastric cancer and contributes to cell proliferation by epigenetically suppressing KLF2. Cancer gene therapy. 2018;25:58-67
112. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
113. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ. et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:4851-8
114. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10:103-21
115. Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer medicine. 2017;6:2897-908
116. Wang X, Peng F, Cheng L, Yang G, Zhang D, Liu J. et al. Prognostic and clinicopathological role of long non-coding RNA UCA1 in various carcinomas. Oncotarget. 2017;8:28373-84
117. Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu QL. et al. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer letters. 2017;408:10-21
118. Zhang X, Gao F, Zhou L, Wang H, Shi G, Tan X. UCA1 Regulates the Growth and Metastasis of Pancreatic Cancer by Sponging miR-135a. Oncology research. 2017;25:1529-41
119. Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Molecular cancer. 2015;14:184
120. Xue-Ming Z, Jing-Jing R, Wei-Hua D, Hai-Sheng H, Dong W, Yan L. et al. Effect of 5-aza-2'-deoxycytidine on methylation of the putative imprinted control region of H19 during the in vitro development of vitrified bovine two-cell embryos. Fertility & Sterility. 2012;98:222-7
121. Iizuka N, Oka M, Tamesa T, Hamamoto Y, Yamada-Okabe H. Imbalance in expression levels of insulin-like growth factor 2 and H19 transcripts linked to progression of hepatocellular carcinoma. Anticancer research. 2004;24:4085-9
122. Li H, Yu B, Li J, Su L, Yan M, Zhu Z. et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318-29
123. Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J. et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159-73
124. Zhang T, Wang YR, Zeng F, Cao HY, Zhou HD, Wang YJ. LncRNA H19 is overexpressed in glioma tissue, is negatively associated with patient survival, and promotes tumor growth through its derivative miR-675. European review for medical and pharmacological sciences. 2016;20:4891-7
125. Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY. et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Science signaling. 2017;10:483
126. Lei L, Xia S, Liu D, Li X, Feng J, Zhu Y. et al. Genome-wide characterization of lncRNAs in acute myeloid leukemia. Briefings in bioinformatics. 2018;19:627-35
127. Zhang J, Griffith M, Miller CA, Griffith OL, Spencer DH, Walker JR. et al. Comprehensive discovery of noncoding RNAs in acute myeloid leukemia cell transcriptomes. Experimental hematology. 2017;55:19-33
128. Schwarzer A, Emmrich S, Schmidt F, Beck D, Ng M, Reimer C. et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nature communications. 2017;8:218
129. Ghazavi F, De Moerloose B, Van Loocke W, Wallaert A, Helsmoortel HH, Ferster A. et al. Unique long non-coding RNA expression signature in ETV6/RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Oncotarget. 2016;7:73769-80
130. Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM, Hu YW. et al. HBx-related long non-coding RNA DBH-AS1 promotes cell proliferation and survival by activating MAPK signaling in hepatocellular carcinoma. Oncotarget. 2015;6:33791-804
131. Cheng H, Huang CM, Wang Y, Hu XX, Xu XQ, Song XM. et al. Microarray profiling and co-expression network analysis of the lncRNAs and mRNAs associated with acute leukemia in adults. Molecular bioSystems. 2017;13:1102-8
132. Feng Y, Shen Y, Chen H, Wang X, Zhang R, Peng Y. et al. Expression profile analysis of long non-coding RNA in acute myeloid leukemia by microarray and bioinformatics. Cancer science. 2018;109:340-53
133. Tsai CH, Yao CY, Tien FM, Tang JL, Kuo YY, Chiu YC. et al. Incorporation of long non-coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients. EBioMedicine. 2019;40:240-50
134. Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS. et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18679-84
135. Papaioannou D, Nicolet D, Volinia S, Mrozek K, Yan P, Bundschuh R. et al. Prognostic and biologic significance of long non-coding RNA profiling in younger adults with cytogenetically normal acute myeloid leukemia. Haematologica. 2017;102:1391-400
136. Mer AS, Lindberg J, Nilsson C, Klevebring D, Wang M, Gronberg H. et al. Expression levels of long non-coding RNAs are prognostic for AML outcome. Journal of hematology & oncology. 2018;11:52
137. Ronchetti D, Manzoni M, Agnelli L, Vinci C, Fabris S, Cutrona G. et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood cancer journal. 2016;6:e468
Author contact
![]() Corresponding author: Yongqian Jia, Department of Hematology, West China Hospital, Sichuan University, Chengdu 610041, Sichuan, China. E-mail: jia_yqedu.cn
Corresponding author: Yongqian Jia, Department of Hematology, West China Hospital, Sichuan University, Chengdu 610041, Sichuan, China. E-mail: jia_yqedu.cn

 Global reach, higher impact
Global reach, higher impact