3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(17):5069-5077. doi:10.7150/jca.44697 This issue Cite
Review
CK19-positive Hepatocellular Carcinoma is a Characteristic Subtype
1. Department of Hepatobiliary and Pancreatic Surgery, the First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, 310003, Zhejiang Province, China.
2. NHC Key Laboratory of Combined Multi-organ Transplantation, Key Laboratory of the Diagnosis and Treatment of Organ Transplantation, CAMS, Hangzhou, 310003, Zhejiang Province, China.
3. Department of Hepatobiliary and Pancreatic Surgery, Shulan (Hangzhou) Hospital, Hangzhou, 310003, Zhejiang Province, China.
4. Center for Bionanoengineering and Key Laboratory of Biomass Chemical Engineering of Ministry of Education, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou, 310003, Zhejiang Province, China.
#These authors contributed equally to this work.
Received 2020-2-8; Accepted 2020-6-13; Published 2020-6-28
Abstract
The heterogeneity of hepatocellular carcinoma (HCC) commonly leads to therapeutic failure of HCC. Cytokeratin 19 (CK19) is well acknowledged as a biliary/progenitor cell marker and a marker of tumor stem cell. CK19-positive HCCs demonstrate aggressive behaviors and poor outcomes which including worse overall survival and early tumor recurrence after hepatectomy and liver transplantation. CK19-positive HCCs are resistant to chemotherapies as well as local treatment. This subset of HCC is thought to derive from liver progenitor cells and can be induced by extracellular stimulation such as hypoxia. Besides being a stemness marker, CK19 plays an important role in promoting malignant property of HCC. The regulatory network associated with CK19 expression has been summarized that extracellular stimulations which transmit into cytoplasm through signal transduction pathways (TGF-β, MAKP/JNK and MEK-ERK1/2), further induce important nuclear transcriptional factors (SALL4, AP1, SP1) to activate CK19 promoter. Novel noncoding RNAs are also involved in the regulation of CK19 expression. TGFβR1 becomes a therapeutic target for CK19-positive HCC. In conclusion, CK19 can be a potential biomarker for predicting poor prognosis after surgical and adjuvant therapies. CK19-pisitive HCCs exhibit distinctive molecular profiling, should be diagnosed and treated as a separate subtype of HCCs.
Keywords: cytokeratin 19, hepatocellular carcinoma, subtype
Introduction
Hepatocellular carcinoma (HCC) emerges as a major health care problem globally. It is the fifth most common malignancy and the third cause of cancer-related death worldwide [1, 2]. To date, surgical resection, liver transplantation, transcatheter arterial chemoembolization (TACE), radiofrequency ablation (RFA) and systemic therapies are the main treatment options to prolong life expectancy of patients with HCC [3]. Unfortunately, 5-year overall survival rate is only 30-40% even after surgical resection and 70% of patients suffer from tumor recurrence within 5 years [3, 4]. Besides that, the effectiveness of chemotherapy, molecular-targeted therapy or even immunotherapy to HCC is insubstantial [2, 5].
HCC is heterogeneous not only not only in terms of morphological characteristics and clinical behaviors, but also in genetic and molecular patterns [6]. The inter-tumor and intra-tumor heterogeneity of HCC results in high resistance to therapeutic interventions [7, 8]. Several molecular subtypes based on transcriptomic analysis have shown significant improvement in describing inter-tumor heterogeneity of HCC [9-12]. As shown in previous studies, HCCs are divided into two groups according to their molecular features, namely, the proliferative and non-proliferative subtypes [11, 13, 14]. Notably, cytokeratin 19 (CK19) is the vital marker of the proliferative subtype which indicates a poor prognosis in HCC patients [15].
Approximately 10-30% of HCC patients present CK19 expression [16-19]. CK19-positive HCC is also known as biphenotypic HCC; that is, having the pathological features of both HCC and cholangiocarcinoma (CC). These group of patients often show worse outcomes as compared to the CK19-negative HCC patients. Therefore, inhibiting the incidence of CK19-positive HCC is of great significance.
As of now, the origin of CK19-positive HCCs, as well as the regulation of expression of CK19 in HCC, remains uncertain. In this review, we highlight recent advances in the overall perspective of CK19 in HCC and its relevant therapeutic implications.
Pathophysiological change of CK19 in Liver
CK19 is an intermediate filament with a molecular weight of around 40 kDa. During the embryonic development, CK19 was detected in the primitive hepatic progenitor cells at the 4-10 weeks' gestation. Along with the development of fetal liver, these bipotential progenitor cells differentiate either into hepatocytes or biliary epithelial cells [20]. However, the expression of CK19 is vanished in mature liver hepatocytes while it is constantly present in biliary epithelial cells. [20-22]. Therefore, this explained the reason why CK19 became a significant marker of biliary epithelial cells in pathological diagnosis [23-25].
When hepatocytes suffer from damage in various chronic liver diseases such as chronic hepatitis, (non-) alcoholic steatohepatitis and haemochromatosis, the normal liver cells which are previously quiescent will be activated and transformed into progenitor cells [26-28]. That is, CK19 re-emerges on hepatocytes anew when the resting cells are activated in the presence of inflammation or other stimuli. Meanwhile, a large number of studies showed that CK19 also appears aberrantly in HCC in which the incidence rate varied between 10 to 30% [16-19] (Table 1).
CK19-positive HCC is associated with more aggressive behavior and poorer prognosis
CK19-positive HCC cells showed strong association with invasion, epithelial-mesenchymal transition (EMT) and angiogenesis. It became apparent when knockdown of CK19 successfully inhibited the invasive capacity and EMT in human HCC cells [19, 29, 30]. CK19-positive cells are thought to be positively associated with angiogenesis as well [29]. Kawai et al. [30] demonstrated that FACS-isolated single CK19-positive cells displayed high proliferation capacity and these cells are more resistant to chemotherapy such as doxorubicin and 5-fluorouracil [19, 30]. A considerable number of researches have reported that CK19 is closely related to cancer stem cells which thus explained the resistance of CK19-positive cells to chemotherapy [30-32].
Patients with CK19-positive HCC have poorer prognosis compared with CK19-negative ones. As shown in Table 1, a substantial number of studies demonstrated that CK19 was associated with early tumor recurrence and worse overall survival after surgical resection or liver transplantation [18, 19, 21, 31]. A meta-analysis enrolling 2943 HCC patients also demonstrated that CK19 overexpression was significantly associated with declined OS rate and 1-year DFS rate [33]. After treated with TACE or RFA, CK19-positive HCC patients were also more inclined to early intrahepatic tumor recurrence [34, 35]. Furthermore, vascular invasion and lower degree of differentiation were more commonly seen in CK19-positive HCC patients when compared with the negative counterpart [19, 21, 29]. In order to comprehensively compare the difference of clinical parameters and physio-pathological features between CK19-negative and CK19-positive HCC CK19, we obtained accessible data from published literatures for analysis as shown in Figure 2 [16, 17, 21, 29, 36-39].
CK19-positive HCC and CK19-negative HCC were compared with intrahepatic cholangiocarcinoma (ICC) and combined HCC and cholangiocarcinoma (cHCC-CC). The overall survival rate of CK19-positive HCC was similar with that of cHCC-CC where they were both significantly lower than the overall survival rate of CK19-negative HCC but higher than that in ICC [17]. This indicated that the biological behavior of CK19-positive HCC was close to that of cHCC-CC.
Hoshida (S1-S3), Boyault (G1-G6) etc. have depicted several subtypes of HCC based on transcriptomics. As a matter of fact, CK19 is commonly described as a progenitor feature as it is closely associated with several stemness-related markers (EPCAM, NOTCH) [15, 40]. Furthermore, CK19-positive subtype has been shown to have correlation with Hoshida S2, G1, proliferation subtype, Cluster A, iClust1 [40, 41], and all these subtypes were related to poor differentiation, higher serum AFP level, higher frequency of vascular invasion and worse outcome [41]. Taken together, CK19-positive HCC showed higher malignancy and worse outcome that it should be diagnosed and treated as a separate entity of HCCs.
The origination of CK19-positive HCC
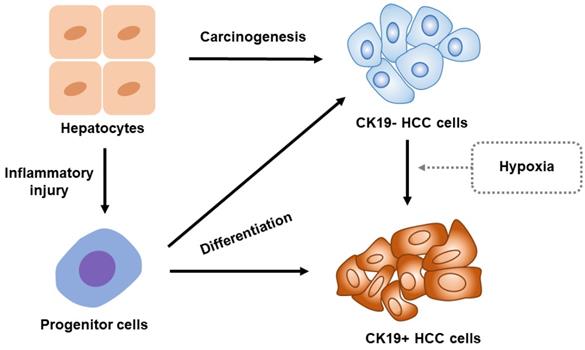
The molecular expressions of the HCCs are highly heterogeneous as they differ even within a single nodule of HCC. The heterogeneity of HCC is related to the origination of tumor cells which include hepatocyte or adult stem/progenitor cells. However, the origination of CK19-positive HCC remains controversial. Therefore, we illustrate an overview of CK19-positive HCC, as shown in Figure 1.
CK19-positive HCCs originate from hepatocytes/liver progenitor cells
In normal liver, progenitor cell, one primordial component, is both positive for hepatocyte-differentiated (HEP-PAR) and biliary-differentiated (CK19) markers [18]. At the time of liver tissue damage, either quiescent progenitor/stem cells are activated [42], or hepatocytes are dedifferentiated into immature progenitors or biliary-type cells [43, 44]. Plentiful evidence suggested that these activated progenitor cells can be potential target cell during the development of HCC [45-47]. Thus, the origination of CK19-positive HCC cells was presumed to be a cluster of progenitor cells.
Overview for the origin of CK19-positive HCC. CK19 expression vanishes in adults' hepatocytes. Once the liver is damaged by inflammation, a portion of hepatocyte will reverse into progenitor cells which express CK19. These progenitor cells will potentially differentiate into CK19-positive HCC cells. Notably, the hepatocytes develop into CK19-negative HCC cells preferentially under normal condition. The hypoxia stimulations including the oxygen-deficient environment in tumor and hypoxic status caused by local treatments such as TACE will, however, induce CK19-negative HCC cells to transform into CK19-positive HCC cells. Abbreviation: CK19-, CK19-negative; CK19+, CK19-positive.
Clinical prognosis comparison between CK19-negative and CK9-positive HCC
| Author | Year | Source of specimen | Cases of CK19+ | Prognosis-related indexes | Note | |
|---|---|---|---|---|---|---|
| CK19- | Ck19+ | |||||
| Wu et al. [18] | 1996 | Needle biopsy/ surgical resection/ autopsy | 79 (27.24%) | 30.7% | 17.6% | 1.5y OS |
| Lee et al. [17] | 2012 | Surgical resection | 21 (30.00%) | 80.4% | 28.9% | 5y OS |
| Fatourou et al. [38] | 2015 | Surgical resection/ liver transplantation | 9 (10.11%) | 56% | 15% | 5y OS $ |
| Miltiadous et al. [40] | 2015 | Liver transplantation | 58 (43.94%) # | 67% | 44.9% | 5y OS |
| Takano et al. [29] | 2016 | Surgical resection | 12 (8.82%) | 90% | 61% | 5y OS $ |
| Lee et al. [37] | 2017 | Liver transplantation | 8 (36.36%) | 82.3% | 55.4% | 5y OS |
| Yang et al. [36] | 2008 | Surgical resection | 45 (18.75%) | 57.2% | 37.8% | 7y OS |
| Yoneda et al [21] | 2011 | Surgical resection | 9 (11.54%) | 64% | 28% | 2y DFS $ |
| Uenishi et al [39] | 2003 | Surgical resection | 15 (9.55%) | 44% | 20% | 3y DFS |
| Durnez et al. [16] | 2006 | Surgical resection/ needle biopsy/ liver transplantation | 18 (16.51%) | 95% | 50% | 3y RFS $ |
| Lee et al. [17] | 2012 | Surgical resection | 21 (30.00%) | 54.5% | 34.3% | 3y DFS |
Abbreviation: HCC, hepatocellular carcinoma; CK19, cytokeratin 19; OS, overall survival; DFS, disease-free survival; RFS, recurrence free survival; CK19-, CK19-negative; CK19+, CK19-positive.
#, CK19/S2 + gene expression signature.
$, the data of overall survival (OS) and disease-free survival (DFS) were measured by computer image-scale comparison.
The clinical parameters and physio-pathological features compared between CK-negative and CK19-positive HCC
| CK19- | CK19+ | P value | ||
|---|---|---|---|---|
| Gender | 0.007 | |||
| Male | 650 (81.4) | 100 (71.4) | ||
| Female | 149 (18.6) | 40 (28.6) | ||
| Cirrhosis | 0.651 | |||
| Yes | 414 (58.6) | 76 (60.8) | ||
| No | 292 (41.4) | 49 (39.2) | ||
| HBV | 0.010 | |||
| Yes | 331 (43.2) | 76 (55.1) | ||
| No | 435 (56.8) | 62 (44.9) | ||
| HCV | 0.003 | |||
| Yes | 269 (35.0) | 30 (21.9) | ||
| No | 499 (65.0) | 107 (78.1) | ||
| Preoperative AFP (ng/ml) | 0.047 | |||
| ≤20 | 221 (49.6) | 30 (37.5) | ||
| >20 | 225 (50.4) | 50 (62.5) | ||
| Tumor number | 0.796 | |||
| Single | 509 (78.9) | 95 (77.9) | ||
| Multiple | 136 (21.1) | 27 (22.1) | ||
| Tumor size (cm) | 0.545 | |||
| >5 | 200 (43.1) | 41 (46.6) | ||
| ≤5 | 264 (56.9) | 47 (53.4) | ||
| Differentiation | <0.001 | |||
| Well/moderate | 338 (71.2) | 34 (45.3) | ||
| Poor | 137 (28.8) | 41 (54.7) | ||
| TNM stage | 0.575 | |||
| I-II | 399 (67.9) | 67 (65.0) | ||
| III-IV | 189 (32.1) | 36 (35.0) | ||
| Microvascular invasion | 0.099 | |||
| Yes | 128 (54.5) | 28 (68.3) | ||
| No | 107 (45.5) | 13 (31.7) | ||
| Portal vein invasion | 0.033 | |||
| Yes | 89 (35.0) | 16 (55.2) | ||
| No | 165 (65.0) | 13 (44.8) | ||
| Metastasis | 0.585 | |||
| Yes | 52 (11.1) | 12 (13.0) | ||
| No | 418 (88.9) | 80 (87.0) | ||
Lee et al. [14] integrated the gene expression data from rat fetal hepatoblasts and adult hepatocytes with HCC from human and mouse models followed by subsequent classification of HCCs into two subtypes (HB and HC subtype). They demonstrated that HCC of the HB subtype arose from bipotential hepatic progenitor cells, and found that expression of CK19 was significantly higher in the HB subtype of HCC. This reflected that CK19-positive HCCs might be derived directly from liver progenitor cells.
Researchers from Columbia University [48] demonstrated that hepatocytes functioned as a cellular source for HCC and these liver progenitor cells found in HCC were derived from hepatocytes. This suggested that hepatocyte-derived HCC may dedifferentiate into a progenitor-like immature phenotype. As CK19 is considered as one of the progenitor cell markers, this may suggest that hepatocyte-derived HCC can dedifferentiate into CK19 positive HCC during the development of tumorigenesis.
If all HCC cells were originated from hepatocytes, these HCCs would only express hepatocyte-related markers (HEP-PAR, CK8 and CK18). As CK19 is increasingly regarded as a marker of bipotential hepatic progenitor cells, it is reasonable to hypothesize that CK19-positive HCCs were developed from hepatic progenitor cells.
CK19-positive HCCs originate from environmental stimulation
The transformation of CK19-negative tumor cells into CK19-positive tumor cells is assumed to be an adaption to the specific challenges in the environment such as hypoxia and physical/chemical stimulation. Several studies have illustrated that carbonic anhydrase IX (CAIX), a hypoxia marker, was positively associated with CK19 after the TACE treatment [49, 50]. Also, the incidence of CK19 positivity was significantly increased as the sessions of TACE increased [50]. As shown in Table 3, the CK19 positive rate in the patients with TACE was higher than that in the non-TACE patients. These studies indicated that HCC with CK19-positive phenotype might originate from the transformations due to the anti-cancer and/or ischemic effects of TACE. Yoshida et al. [51] reported that the residual tumor due to incomplete RFA could be reactivated in the presence of thermal and hypoxic stimulation. These reactivated tumor cells usually portray higher malignancies and more aggressive invasive capabilities. In this study, HCC cell was exposed to high temperature which simulated the marginal zone of RFA treatment. In vitro cells survived from mimic-RFA showed an increase in CK19 expression.
The comparison of CK19 positive rate in HCC patients receiving TACE treatment before operation
| Author | Year | Treatment | Total cases | Treat group | Non-treat group | ||
|---|---|---|---|---|---|---|---|
| Ck19- | Ck19+ | Ck19- | Ck19+ | ||||
| Lai et al. [49] | 2015 | TACE | 57 | 34 | 6 | 17 | 0 |
| Nishihara et al. [35] | 2008 | TACE* | 226 | 61 | 19 | 128 | 18 |
| Zen et al [79] | 2011 | TACE* | 80 | 32 | 8 | 40 | 0 |
| Rhee et al. [50] | 2016 | TACE* | 85 | 38 | 13 | 32 | 2 |
Abbreviations: HCC, hepatocellular carcinoma; CK19, cytokeratin 19; TACE: transcatheter arterial chemoembolization; CK19-, CK19-negative; CK19+, CK19-positive.
*, p<0.05, Fisher's Exact Test.
Taken these evidences together, a new and inspiring hypothesis has been put forward that CK19 negative HCC may transform into CK19 positivity after patients receive several adjuvant therapies (e.g. TACE, RFA) as tumor environmental is stimulated. In addition, it provides a potential possibility for tumor progression and recurrence after treatment, and indicates a potential prevention strategy, that is, the CK19 regulatory network should be controlled in advance.
Molecular characteristics of CK19-positive HCC
Despite the clinical significance of CK19 in HCC, the role of CK19 in tumor is still ambiguous in which whether it functions as a phenotype marker or an oncogenic factor has yet to be speculated. Over the past two decades, CK19-positive cells have gradually been regarded as a kind of stem cell [52] as well as an important prognostic marker of HCC as described above. Recent studies have also showed that CK19 enhanced the tumorous properties in breast cancer, colon cancer and hepatocellular carcinoma [19, 53, 54]. These data implied that CK19 plays an important part in carcinogenesis. However, the molecular network of this phenotype is not unraveled completely. Therefore, we summarized and highlighted the vital pathways involved in regulating the CK19 expression which include the carcinogenic growth factors and corresponding receptors, MAKP/JNK and MEK-ERK1/2 pathways, transcription factors and noncoding RNAs (Figure 2).
Extracellular stimulation: Carcinogenic growth factors
Belgian researchers [19] previously revealed that CK19 was associated with platelet-derived growth factor receptor α (PDGFRα). Furthermore, they demonstrated that PDGF could elevate CK19 expression via PDGFRα-La/SSB-LAMB1 axis [55]. A study from Kanazawa University [21] showed that epidermal growth factor (EGF) had potent effects on promoting CK19 expression in vitro. Another Korean study showed that hepatocyte growth factor (HGF) from cancer-associated fibroblasts (CAF) could upregulate CK19 expression based on cross-talk between CAF and HCC cells [56]. Apparently, the transforming growth factor-β (TGF-β) is another important extracellular factor involved in the progressive features of CK19-positive HCC cells as the inhibition of transforming growth factor-β receptor 1 (TGFβR1) could significantly attenuate the proliferation capability of CK19-positive HCC [30]. Albeit no obvious evidence supporting the direct regulation between TGFβ and CK19, TGFβ/Smad signaling exhibits enormous association with CK19-positive HCC as described above. This collectively indicates that these carcinogenic growth factors with corresponding receptors engage in regulating the CK19 expression.
Signal transduction: JNK, MEK-ERK1/2 and Smads signaling pathway
Yoneda et al. [21] demonstrated that c-Jun-N-terminal kinase (JNK)/stress-activated protein kinase (SAPK) is a downstream signaling pathway involved in regulating CK19 expression via EGF-EGFR. In another study, HCC tumor specimens with matched distal noncancerous liver tissue were divided into two subgroupes according to JNK1 activation status. As a result, CK19 was over expressed in high JNK1 HCC [57]. American investigators [58] evaluated the transcriptomic differences between CK19-positive and CK19-negative foci through the resistant hepatocyte (RH) rat models to select unique genes in each group. The connectivity of the top regulatory networks showed a dominant enrichment of AP-1/JUN in CK19-positive areas. These data suggested a hypothesis that JNK pathway is involved in regulating CK19 expression.
MEK-ERK1/2 pathway is another vital intracellular signaling pathway participated in the modulation of CK19 expression. Rhee et al. proposed that, MET, the receptor of extracellular signal HGF, upregulated CK19 expression via activating MEK-ERK1/2 pathway [56]. MET is also known to be related to poor prognosis and HGF/MET signaling axis currently became an emerging therapeutic target of HCC [59]. Therefore, CK19-positive HCCs are deemed to be accompanied with the activation of HGF/MET signaling. In Kawai's study, TGFβ/Smad signaling is activated in CK19-positive cells [30] and it is always reasonable to assume that Smad pathway is part of the regulatory network of CK19 expression.
Endonuclear activation: Nuclear transcription factors
Regulatory pathways relay the signal to transcription factors and further activate the CK19 promoter. Two important transcription factors, namely activator protein 1 (AP1) and specificity protein 1 (SP1), have been identified as regulators that bind directly to the CK19 promoter site [56]. Besides that, AP1 and SP1 are also known as the downstream transcriptional activators of ERK1/2. JUN and FOS proteins which belong to the AP1 family can be dimerized to form JUN homodimers or JUN/FOS heterodimers in order to become valid transcriptional factors [60]. As mentioned above, JNK pathway is also involved in regulating CK19 expression via EGF-EGFR while FOSL1, a protein of FOS family, can be activated by ERK1/2 pathway [56]. In short, we conclude that AP1 serves as the downstream transcriptional factor of both JNK and ERK pathway for CK19 regulation.
Diagrammatic sketch of the regulatory network of CK19. Abbreviation: CK19, cytokeratin 19; TGFβ, transforming growth factor beta; TGFβR, transforming growth factor beta receptor; HGF, hepatocyte growth factor; MET, MET proto-oncogene receptor tyrosine kinase or hepatocyte growth factor receptor; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; PDGF, platelet derived growth factor; PDGFRα, platelet derived growth factor receptor alpha; Smad, drosophila mothers against decapentaplegic protein; MEK, mitogen-activated protein kinase kinase 7; ERK, mitogen-activated protein kinase 1; JNK, mitogen-activated protein kinase 8; JUN, Jun proto-oncogene AP-1 transcription factor subunit; FOS, Fos proto-oncogene AP-1 transcription factor subunit; AP1, activator protein 1; SP1, specificity protein 1; KLF4, Krüppel like factor 4; KLF10, Krüppel like factor 10; SALL4, spalt-like transcription factor 4; CDH17, cadherin 17; LamB1, laminin subunit beta 1; Src, SRC proto-oncogene non-receptor tyrosine kinase; miR-642, microRNA 642a; miR-141, microRNA 141; miR-200c, microRNA 200c; Lin00974, long intergenic non-protein coding RNA 974.
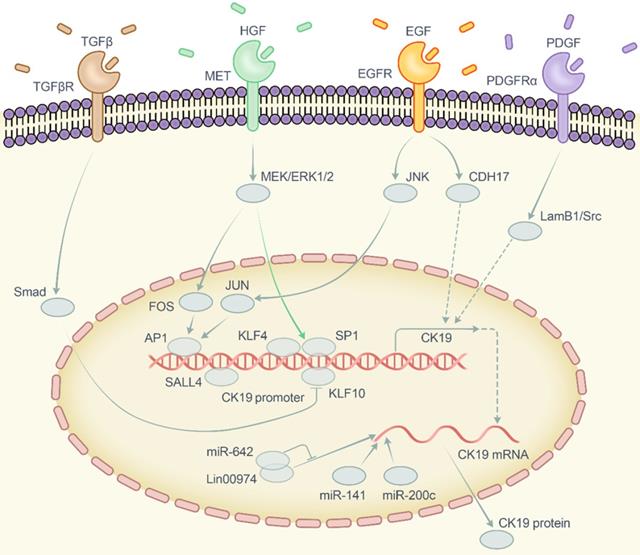
Recently, overexpression of Sal-like 4 (Drosophila) (SALL4) was reported to upregulate CK19 expression at both the mRNA and the protein level [61]. SALL4 is an important transcription factor in HCC which correlates to stemness, 5-FU resistance and differentiation [62]. We suggest that SALL4 might bind to the promoter site of CK19.
Two members from Krüppel-like Factors (KLF) family, which are also DNA-binding transcriptional regulators, have shown an association with CK19. Brembeck from University of Pennsylvania verified that the CK19 gene is regulated by the interplay of Krüppel-like factor 4 (KLF4) and SP-1 through a critical cis-regulatory element in the proximal promoter [63]. On the other hand, Andersen et al. [58] examined the functional connectivity among the significant genes and found that the most predominant feature in the CK19 negative focal lesions was the overexpression of Krüppel-like factor 10 (KLF10) in rat model. As murine CK19 gene exhibits high homology to human counterpart [64], we can translate these findings to human being. Besides, KLF10 has been validated as a tumor suppressor gene and shown association with TGFβ/Smad signaling pathway [65]. Therefore, we hypothesize that KLF10 may inhibit CK19 promoter activity by suppressing TGFβ/Smad signaling. Above all, transcription factors such as AP-1, SP-1, SALL4 and KLF family (KLF4, KLF10) might regulate the expression of CK19 either directly or indirectly by interacting with its promoter binding site.
Non-coding RNA and others
Tang et al. [66] showed that long noncoding RNA (lncRNA), Linc00974, presented positive regulation of CK19 via posttranscriptional modification. It functions as a sponge to endogenously compete with the suppressive effect of miR-642 to CK19. Moreover, miR-200 family (miR-141/miR-200c) especially miR-141 was identified to have strong linkage with CK19 expression [19]. That is, overexpression of miR-141 and miR-200c significantly upregulated the expression of CK19 in CK19-negative or CK19-low HCC cells. Lee et al. [67] detected 20 patients with hepatitis B virus (HBV)-HCC through Affymetrix U133A oligonucleotide microarray. It was shown that cadherin 17 (CDH17) was positively correlated with CK19 expression and CDH17 could enhance the expression of CK19 through EGF/EGFR-CDH17-CK19 pathway.
Make a brief summary, as CK19 has been regarded as a marker of progenitor- and stem-cells, previous publications have done a body of studies to clarify how to regulate the expression of CK19 as shown in Figure 2. The internal regulatory network, however, is more complex and intricate than what we have seen so far. Govaere et al. also reported that CK19 showed a robust positive correlation with other 'aggressive' phenotypes and signatures including 'poor survival HCC subtype', 'proliferation HCC subtype', 'S1 signature with aberrant Wnt activation subtype' and so on [19]. Therefore, attention should not be focused solely on CK19 as a target that function as a biomarker and oncogenic factor. CK19-positive HCC should be regarded as a completely different phenotype and an independent entity with its own characteristics.
The detective and therapeutic strategy for CK19 positive HCC
Non-invasive detection method for CK19 expression
The current detection accesses of CK19 are mostly dependent on the post-operative immunochemistry. Advanced acquisition of CK19 expression level can guide clinical physicians to choose optimal therapeutic methods before surgery or locoregional treatment. CYFRA 21-1 is a soluble fragment of CK19 in peripheral circulation. In fact, serum CYFRA 21‐1 has been detected in various malignancies including non‐small cell lung cancer, esophageal cancer, breast cancer and pancreatic cancer [68-71]. Therefore, serum CYFRA 21‐1 is considered as a useful biomarker to indicate CK19 expression in HCC [72].
More recently, radiological examination has become a novel non-invasive diagnostic method for CK19-positive HCC. Choi et al. [73] determined the preoperative magnetic resonance (MR) imaging characteristics of HCC potentially related to CK19 expression. Another study illustrated that MR features combining with elevated AFP were able to distinguish CK19-positive HCCs from CK19-negative HCC [74]. Therefore, application of these non-invasive detection methods will be conducive to monitor the CK19 expression status.
The therapeutic strategy for CK19-positive HCC
Considering the different molecular features and the distinct invasive properties of CK19-positive HCCs, this subtype of HCCs should be regarded as a separate entity which is different from the CK19-negative HCCs [19]. We would like to regard the presence of CK19 as a therapeutic-phenotype to guide individualized treatment in clinical context as it is of utmost important to deliberate the strategies thoroughly to overcome the highly tortuous CK19-positive HCCs.
As aforementioned, Japanese researchers discovered that TGFb/Smad signaling was activated in CK19-positive HCC cells. In other words, TGFβR1 inhibitor (LY2157299) could effectively inhibit the proliferation of CK19-positive HCC [30]. Therefore, TGFbR1 inhibitor should be considered as a new targeted-therapy against CK19-positive HCC.
A Chinese study enrolling 280 HCC patients who were either treated with or without sorafenib after surgical resection showed no difference between two groups in terms of overall survival. Intriguingly, patients who have received sorafenib treatment after surgery exhibited superior overall survival compared to those who have not received sorafenib treatment after surgery in CK19+OV6+ subgroup [75]. This implied that patients with CK19-positive HCC could benefit from sorafenib administration after surgery. As Govaere et al. [55] demonstrated that CK19 could be progressed through PDGFRα-LAMB1-CK19 axis, inhibitors of PDGFRα such as imatinib [76], regorafenib [77] and lenvatinib [78] might exert specific effect on CK19-positive HCC. Therefore, the above-named first-line or second-line multi-targeted tyrosine kinase inhibitors (TKI) approved by FDA for HCC are supposed to have therapeutic priority to CK19-positive HCC. There is no doubt that more clinical trials are needed to validate the most efficacious drug in dealing with CK19-positive HCC.
Conclusions
CK19-positive HCC demonstrates more aggressive behaviors and poorer prognosis. The complex internal regulatory network makes CK19-positive HCC an independent entity with its own characteristics, which should be diagnosed and treated as an independent subtype. Clarification of the internal molecular mechanisms is urgently needed to prevail over CK19-positive HCCs in the clinical management.
Abbreviations
HCC: hepatocellular carcinoma; CK19: cytokeratin 19; TACE: transcatheter arterial chemoembolization; RFA: radiofrequency ablation; TGFβ: transforming growth factor beta; TGFβR: transforming growth factor beta receptor; HGF: hepatocyte growth factor; MET: MET proto-oncogene receptor tyrosine kinase or hepatocyte growth factor receptor; EGF: epidermal growth factor; EGFR: epidermal growth factor receptor; PDGF: platelet derived growth factor; PDGFRα: platelet derived growth factor receptor alpha; Smad: drosophila mothers against decapentaplegic protein; MEK: mitogen-activated protein kinase kinase 7; ERK: mitogen-activated protein kinase 1; JNK: mitogen-activated protein kinase 8; JUN: Jun proto-oncogene AP-1 transcription factor subunit; FOS: Fos proto-oncogene AP-1 transcription factor subunit; AP1: activator protein 1; SP1: specificity protein 1; KLF4: Krüppel like factor 4; KLF10: Krüppel like factor 10; SALL4: spalt-like transcription factor 4; CDH17: cadherin 17; LamB1: laminin subunit beta 1; Src: SRC proto-oncogene non-receptor tyrosine kinase; miR-642: microRNA 642a; miR-141: microRNA 141; miR-200c: microRNA 200c; Lin00974: long intergenic non-protein coding RNA 974.
Acknowledgements
This work was funded by grants from the National Science and Technology Major Project of China (2017ZX10203205), the National Natural Science Funds for Distinguished Young Scholar of China (81625003).
Authors' Contributions
JYZ, LD and WYT contributed to writing the manuscript. SSZ, YQS and XX contributed to revising it critically for important intellectual content. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mak LY, Cruz-Ramon V, Chinchilla-Lopez P, Torres HA, LoConte NK, Rice JP. et al. Global Epidemiology, Prevention, and Management of Hepatocellular Carcinoma. Am Soc Clin Oncol Educ Book. 2018 p: 262-79
2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-14
3. European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. Journal of hepatology. 2018;69:182-236
4. Nakagawa S, Wei L, Song WM, Higashi T, Ghoshal S, Kim RS. et al. Molecular Liver Cancer Prevention in Cirrhosis by Organ Transcriptome Analysis and Lysophosphatidic Acid Pathway Inhibition. Cancer Cell. 2016;30:879-90
5. Worns MA, Galle PR. HCC therapies-lessons learned. Nat Rev Gastroenterol Hepatol. 2014;11:447-52
6. Llovet JM, Di Bisceglie AM, Bruix J, Kramer BS, Lencioni R, Zhu AX. et al. Design and endpoints of clinical trials in hepatocellular carcinoma. Journal of the National Cancer Institute. 2008;100:698-711
7. Nault JC, Galle PR, Marquardt JU. The role of molecular enrichment on future therapies in hepatocellular carcinoma. Journal of hepatology. 2018;69:237-47
8. Wang XW, Thorgeirsson SS. The biological and clinical challenge of liver cancer heterogeneity. Hepatic oncology. 2014;1:349-53
9. Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35-51
10. Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T. et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667-76
11. Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E. et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42-52
12. Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer research. 2009;69:7385-92
13. Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B. et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer research. 2008;68:6779-88
14. Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF. et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nature medicine. 2006;12:410-6
15. Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nature reviews Clinical oncology. 2015;12:436
16. Durnez A, Verslype C, Nevens F, Fevery J, Aerts R, Pirenne J. et al. The clinicopathological and prognostic relevance of cytokeratin 7 and 19 expression in hepatocellular carcinoma. A possible progenitor cell origin. Histopathology. 2006;49:138-51
17. Lee JI, Lee JW, Kim JM, Kim JK, Chung HJ, Kim YS. Prognosis of hepatocellular carcinoma expressing cytokeratin 19: comparison with other liver cancers. World journal of gastroenterology. 2012;18:4751-7
18. Wu PC, Fang JW, Lau VK, Lai CL, Lo CK, Lau JY. Classification of hepatocellular carcinoma according to hepatocellular and biliary differentiation markers. Clinical and biological implications. Am J Pathol. 1996;149:1167-75
19. Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F. et al. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut. 2014;63:674-85
20. Haruna Y, Saito K, Spaulding S, Nalesnik MA, Gerber MA. Identification of bipotential progenitor cells in human liver development. Hepatology. 1996;23:476-81
21. Yoneda N, Sato Y, Kitao A, Ikeda H, Sawada-Kitamura S, Miyakoshi M. et al. Epidermal growth factor induces cytokeratin 19 expression accompanied by increased growth abilities in human hepatocellular carcinoma. Lab Invest. 2011;91:262-72
22. Schwartz RE, Reyes M, Koodie L, Jiang Y, Blackstad M, Lund T. et al. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. The Journal of clinical investigation. 2002;109:1291-302
23. Terada T, Nakanuma Y. Development of human intrahepatic peribiliary glands. Histological, keratin immunohistochemical, and mucus histochemical analyses. Lab Invest. 1993;68:261-9
24. Shah KD, Gerber MA. Development of intrahepatic bile ducts in humans. Immunohistochemical study using monoclonal cytokeratin antibodies. Arch Pathol Lab Med. 1989;113:1135-8
25. Bartek J, Bartkova J, Taylor-Papadimitriou J, Rejthar A, Kovarik J, Lukas Z. et al. Differential expression of keratin 19 in normal human epithelial tissues revealed by monospecific monoclonal antibodies. Histochem J. 1986;18:565-75
26. Roskams TA, Libbrecht L, Desmet VJ. Progenitor cells in diseased human liver. Semin Liver Dis. 2003;23:385-96
27. Roskams T, Yang SQ, Koteish A, Durnez A, DeVos R, Huang X. et al. Oxidative stress and oval cell accumulation in mice and humans with alcoholic and nonalcoholic fatty liver disease. Am J Pathol. 2003;163:1301-11
28. Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818-22
29. Takano M, Shimada K, Fujii T, Morita K, Takeda M, Nakajima Y. et al. Keratin 19 as a key molecule in progression of human hepatocellular carcinomas through invasion and angiogenesis. BMC cancer. 2016;16:903
30. Kawai T, Yasuchika K, Ishii T, Katayama H, Yoshitoshi EY, Ogiso S. et al. Keratin 19, a Cancer Stem Cell Marker in Human Hepatocellular Carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:3081-91
31. Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE. et al. Human hepatocellular carcinomas with "Stemness"-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54:1707-17
32. Kim H, Park YN. Hepatocellular carcinomas expressing 'stemness'-related markers: clinicopathological characteristics. Dig Dis. 2014;32:778-85
33. Sun DW, Zhang YY, Sun XD, Chen YG, Qiu W, Ji M. et al. Prognostic value of cytokeratin 19 in hepatocellular carcinoma: A meta-analysis. Clin Chim Acta. 2015;448:161-9
34. Tsuchiya K, Komuta M, Yasui Y, Tamaki N, Hosokawa T, Ueda K. et al. Expression of keratin 19 is related to high recurrence of hepatocellular carcinoma after radiofrequency ablation. Oncology. 2011;80:278-88
35. Nishihara Y, Aishima S, Kuroda Y, Iguchi T, Taguchi K, Asayama Y. et al. Biliary phenotype of hepatocellular carcinoma after preoperative transcatheter arterial chemoembolization. J Gastroenterol Hepatol. 2008;23:1860-8
36. Yang XR, Xu Y, Shi GM, Fan J, Zhou J, Ji Y. et al. Cytokeratin 10 and cytokeratin 19: predictive markers for poor prognosis in hepatocellular carcinoma patients after curative resection. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:3850-9
37. Lee SH, Lee JS, Na GH, You YK, Kim DG. Immunohistochemical markers for hepatocellular carcinoma prognosis after liver resection and liver transplantation. Clin Transplant. 2017 p; 31
38. Fatourou E, Koskinas J, Karandrea D, Palaiologou M, Syminelaki T, Karanikolas M. et al. Keratin 19 protein expression is an independent predictor of survival in human hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2015;27:1094-102
39. Uenishi T, Kubo S, Yamamoto T, Shuto T, Ogawa M, Tanaka H. et al. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003;94:851-7
40. Miltiadous O, Sia D, Hoshida Y, Fiel MI, Harrington AN, Thung SN. et al. Progenitor cell markers predict outcome of patients with hepatocellular carcinoma beyond Milan criteria undergoing liver transplantation. Journal of hepatology. 2015;63:1368-77
41. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nature reviews Clinical oncology. 2018;15:599-616
42. Hsia CC, Evarts RP, Nakatsukasa H, Marsden ER, Thorgeirsson SS. Occurrence of oval-type cells in hepatitis B virus-associated human hepatocarcinogenesis. Hepatology. 1992;16:1327-33
43. Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM. et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719-24
44. Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B. et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324-38
45. Komuta M, Spee B, Vander Borght S, De Vos R, Verslype C, Aerts R. et al. Clinicopathological study on cholangiolocellular carcinoma suggesting hepatic progenitor cell origin. Hepatology. 2008;47:1544-56
46. Libbrecht L, Bielen D, Verslype C, Vanbeckevoort D, Pirenne J, Nevens F. et al. Focal lesions in cirrhotic explant livers: pathological evaluation and accuracy of pretransplantation imaging examinations. Liver Transpl. 2002;8:749-61
47. Theise ND, Saxena R, Portmann BC, Thung SN, Yee H, Chiriboga L. et al. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30:1425-33
48. Mu X, Espanol-Suner R, Mederacke I, Affo S, Manco R, Sempoux C. et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. The Journal of clinical investigation. 2015;125:3891-903
49. Lai JP, Conley A, Knudsen BS, Guindi M. Hypoxia after transarterial chemoembolization may trigger a progenitor cell phenotype in hepatocellular carcinoma. Histopathology. 2015;67:442-50
50. Rhee H, Nahm JH, Kim H, Choi GH, Yoo JE, Lee HS. et al. Poor outcome of hepatocellular carcinoma with stemness marker under hypoxia: resistance to transarterial chemoembolization. Mod Pathol. 2016;29:1038-49
51. Yoshida S, Kornek M, Ikenaga N, Schmelzle M, Masuzaki R, Csizmadia E. et al. Sublethal heat treatment promotes epithelial-mesenchymal transition and enhances the malignant potential of hepatocellular carcinoma. Hepatology. 2013;58:1667-80
52. Michel M, Torok N, Godbout MJ, Lussier M, Gaudreau P, Royal A. et al. Keratin 19 as a biochemical marker of skin stem cells in vivo and in vitro: keratin 19 expressing cells are differentially localized in function of anatomic sites, and their number varies with donor age and culture stage. J Cell Sci. 1996;109( Pt 5):1017-28
53. Saha SK, Choi HY, Kim BW, Dayem AA, Yang GM, Kim KS. et al. KRT19 directly interacts with beta-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene. 2017;36:332-49
54. Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE. et al. Krt19(+)/Lgr5(-) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell stem cell. 2015;16:627-38
55. Govaere O, Petz M, Wouters J, Vandewynckel YP, Scott EJ, Topal B. et al. The PDGFRalpha-laminin B1-keratin 19 cascade drives tumor progression at the invasive front of human hepatocellular carcinoma. Oncogene. 2017;36:6605-16
56. Rhee H, Kim HY, Choi JH, Woo HG, Yoo JE, Nahm JH. et al. Keratin 19 Expression in Hepatocellular Carcinoma Is Regulated by Fibroblast-Derived HGF via a MET-ERK1/2-AP1 and SP1 Axis. Cancer research. 2018;78:1619-31
57. Chang Q, Chen J, Beezhold KJ, Castranova V, Shi X, Chen F. JNK1 activation predicts the prognostic outcome of the human hepatocellular carcinoma. Mol Cancer. 2009;8:64
58. Andersen JB, Loi R, Perra A, Factor VM, Ledda-Columbano GM, Columbano A. et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2010;51:1401-9
59. Qi XS, Guo XZ, Han GH, Li HY, Chen J. MET inhibitors for treatment of advanced hepatocellular carcinoma: A review. World journal of gastroenterology. 2015;21:5445-53
60. Hesari A, Ghasemi F, Salarinia R, Biglari H, Tabar Molla Hassan A, Abdoli V. et al. Effects of curcumin on NF-kappaB, AP-1, and Wnt/beta-catenin signaling pathway in hepatitis B virus infection. J Cell Biochem. 2018;119:7898-904
61. Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y. et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. Journal of hepatology. 2014;60:127-34
62. Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y. et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469-83
63. Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000;275:28230-9
64. Lussier M, Filion M, Compton JG, Nadeau JH, Lapointe L, Royal A. The mouse keratin 19-encoding gene: sequence, structure and chromosomal assignment. Gene. 1990;95:203-13
65. Memon A, Lee WK. KLF10 as a Tumor Suppressor Gene and Its TGF-beta Signaling. Cancers (Basel). 2018 p:10
66. Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L. et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549
67. Lee CW, Lin SE, Tsai HI, Su PJ, Hsieh CH, Kuo YC. et al. Cadherin 17 is related to recurrence and poor prognosis of cytokeratin 19-positive hepatocellular carcinoma. Oncol Lett. 2018;15:559-67
68. Shimada H, Nabeya Y, Okazumi S, Matsubara H, Miyazawa Y, Shiratori T. et al. Prognostic significance of CYFRA 21-1 in patients with esophageal squamous cell carcinoma. J Am Coll Surg. 2003;196:573-8
69. Pujol JL, Grenier J, Daures JP, Daver A, Pujol H, Michel FB. Serum fragment of cytokeratin subunit 19 measured by CYFRA 21-1 immunoradiometric assay as a marker of lung cancer. Cancer research. 1993;53:61-6
70. Boeck S, Wittwer C, Heinemann V, Haas M, Kern C, Stieber P. et al. Cytokeratin 19-fragments (CYFRA 21-1) as a novel serum biomarker for response and survival in patients with advanced pancreatic cancer. Br J Cancer. 2013;108:1684-94
71. Nakata B, Takashima T, Ogawa Y, Ishikawa T, Hirakawa K. Serum CYFRA 21-1 (cytokeratin-19 fragments) is a useful tumour marker for detecting disease relapse and assessing treatment efficacy in breast cancer. Br J Cancer. 2004;91:873-8
72. Kawai T, Yasuchika K, Ishii T, Katayama H, Yoshitoshi EY, Ogiso S. et al. Identification of keratin 19-positive cancer stem cells associating human hepatocellular carcinoma using CYFRA 21-1. Cancer Med. 2017;6:2531-40
73. Choi SY, Kim SH, Park CK, Min JH, Lee JE, Choi YH. et al. Imaging Features of Gadoxetic Acid-enhanced and Diffusion-weighted MR Imaging for Identifying Cytokeratin 19-positive Hepatocellular Carcinoma: A Retrospective Observational Study. Radiology. 2018;286:897-908
74. Hu XX, Wang WT, Yang L, Yang ZX, Liang HY, Ding Y. et al. MR features based on LI-RADS identify cytokeratin 19 status of hepatocellular carcinomas. Eur J Radiol. 2019;113:7-14
75. Li XF, Chen C, Xiang DM, Qu L, Sun W, Lu XY. et al. Chronic inflammation-elicited liver progenitor cell conversion to liver cancer stem cell with clinical significance. Hepatology. 2017;66:1934-51
76. Oseini AM, Roberts LR. PDGFRalpha: a new therapeutic target in the treatment of hepatocellular carcinoma? Expert Opin Ther Targets. 2009;13:443-54
77. Belum VR, Wu S, Lacouture ME. Risk of hand-foot skin reaction with the novel multikinase inhibitor regorafenib: a meta-analysis. Invest New Drugs. 2013;31:1078-86
78. Spallanzani A, Orsi G, Andrikou K, Gelsomino F, Rimini M, Riggi L. et al. Lenvatinib as a therapy for unresectable hepatocellular carcinoma. Expert Rev Anticancer Ther. 2018;18:1069-76
79. Zen C, Zen Y, Mitry RR, Corbeil D, Karbanova J, O'Grady J. et al. Mixed phenotype hepatocellular carcinoma after transarterial chemoembolization and liver transplantation. Liver Transpl. 2011;17:943-54
Author contact
![]() Corresponding authors: Xiao Xu, E-mail: zjxuedu.cn; Tel: +86-571-87236567; Fax: +86-571-87236567. You-Qing Shen, E-mail: shenyqedu.cn.
Corresponding authors: Xiao Xu, E-mail: zjxuedu.cn; Tel: +86-571-87236567; Fax: +86-571-87236567. You-Qing Shen, E-mail: shenyqedu.cn.

 Global reach, higher impact
Global reach, higher impact