Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(17):5088-5098. doi:10.7150/jca.44777 This issue Cite
Research Paper
Knockdown of Kif20a inhibits growth of tumors in soft tissue sarcoma in vitro and in vivo
1. Department of Orthopaedic Trauma, The First Hospital of Jilin University, Xinmin Street 71#, Changchun City, Jilin Province, China.
2. Department of Immunology, College of Basic Medical Sciences, Jilin University, Xinmin Street 126#, Changchun City, Jilin Province, China.
3. College of Chemistry, Jilin University, Changchun, Jilin Province, China.
*Equal contributions to this work.
Received 2020-2-10; Accepted 2020-5-29; Published 2020-6-28
Abstract
Kif20a (Kinesin Family Member 20A), plays a role in cell mitosis, cell migration and intracellular transport. Numerous studies have demonstrated that Kif20a is abnormally highly expressed in a variety of tumors and shows poor prognosis. Soft tissue sarcoma (STS) represents a group of malignant tumors with poor prognosis. The role of Kif20a in STSs has not been systematically studied. In the present study, bioinformatics analysis, in vitro and in vivo experiments were conducted to investigate the function of Kif20a in STSs. In bioinformatics analysis higher KIf20a expression indicated a poor prognosis. Functional enrichment analysis indicated that Kif20a may be related to cell cycle, p53 and other signaling pathways. In vitro experiments showed that after the down-regulation of Kif20a, cell proliferation, migration and invasion were decreased, while apoptosis was increased. In vivo experiments revealed that Kif20a affected the proliferation of tumors in tumor-bearing mice. In summary, our findings revealed that Kif20a performs an important role in STS, indicating that it is a potential prognostic biomarker and potentially representing a therapeutic target for the disease.
Keywords: Kif20a, bioinformatics, survival, biomarkers, soft tissue sarcoma
Introduction
Soft tissue sarcomas are a rare group of malignancies of mesenchymal origin and can occur in various parts of the body, especially the extremities [1]. In addition, STSs are highly malignant and prone to lead to lung metastasis [2]. At present, surgery, chemotherapy and radiotherapy remain the primary treatment options for STSs [3]. However, a number of complications exist, such as postoperative recurrence and metastasis, poor prognosis and poor chemotherapeutic efficacy. As molecular pathology has developed, it has been confirmed that multiple genes are associated with the progression of soft tissue sarcomas. However, no clinical biomarkers have been identified that guide prognosis.
Kinesin-like proteins, such as Kif20a, are microtubule-associated motors that mainly play a role in cell mitosis, migration and intracellular transport [4-6]. Kif20a is abnormally highly expressed in various tumors, such as gastric cancer [7], glioma, pancreatic cancer, breast cancer and bladder cancer [8]. Kif20a expression has been identified as a prognostic indicator for ovarian clear cell carcinoma [9], nasopharyngeal cancer [10] and pancreatic cancer [11]. In breast cancer, Kif20a plays a role in the resistance to Taxol [12].
Despite the key role KIF20a plays in numerous tumors, no study has so far explored the relationship between Kif20a and STSs. In this study we intended to investigate the expression of Kif20a in STSs and explore its role in carcinogenesis and metastasis. In this study, the expression and function of Kif20a in STSs were firstly analyzed by bioinformatics analysis. The function of Kif20a was then verified both in vitro and in vivo by constructing Kif20a knockdown cell lines of STS.
Materials and Methods
Data sources
The normalized level 3 RNAseq data and relevant clinical characteristics of TCGA-SARC project were downloaded from the UCSC Xena platform (https://xenabrowser.net) [13].
Survival analysis
Samples from different subtypes were divided into two groups (high vs. low) representing a comparison with the median expression level of Kif20a, then survival analysis was performed that compared the two groups.
Co-expression analysis
A protein-protein interaction (PPI) network of Kif20a was constructed using the STRING database (https://string-db.org/) [14], based on both experimentally validated and predicted connections.
GSEA analysis
Gene set enrichment analysis (GSEA) [15, 16] is also known as functional enrichment analysis, which can determine whether there is statistical difference in the predefined set of genes between two groups of samples. TCGA-SARC data were used to conduct GSEA on samples where Kif20a expression level was within either the bottom or top quartile.
Cell lines and cell culture reagents
Mouse STS cell line WEHI164 was purchased from CCTCC (China Center for Type Culture Collection) (Wuhan, China). Mouse STS cell lines MCA101 and MCA207 and mouse fibrocyte cell line L929 were obtained from Zhu's laboratory (Changchun, China). All the cell lines were maintained in RPIM 1640 (Gibco, USA) supplemented with 10% FBS (Hyclone, USA) and 1% antibiotics (penicillin-streptomycin, Gibco, USA). The culture medium was exchanged every 2-3 days. After culture to 80% confluence, the cells were trypsinized with trypsin-EDTA (0.25%) (Hyclone, USA) for passaging.
Transfection
A plasmid containing Kif20a shRNA was constructed by GIEL (Shanghai, China) using GV102 as the carrier. The sequences of the shRNA and control strands were: 5'- GCCACTCACAAATTTACCTTT-3' and 5'- TTCTCCGAACGTGTCACGT-3', respectively. All the three sarcoma cell lines were seeded into six well plate (Corning, USA) at 5×105 cell/well and cultured overnight. PEI Transfection reagent (Polysciences, UK) was used in accordance with the manufacturer's instructions. After 48h of continuous culture, Geneticin was used to screen the transfected sarcoma cells at concentrations of 400 μg/ml, 600 μg/ml and 600 μg/ml for WEHI164, MCA101 and MCA207, respectively. After expansion culture, RT-qPCR and Western blots were used to confirm that Kif20a expression had been silenced.
Antibodies and Western blotting
The three cell lines (WEHI164, MCA101, MCA207) were collected into EP tubes using a cell scraper, then lysed using RIPA buffer (Beyotime, China) containing protease inhibitor (Complete, Germany). After lysis for 30min on ice, each lysate was centrifuged for 10min at 10,000 g at 4 °C in a microfuge (Eppendorf, Germany). Loading buffer was added and the protein mixtures were heated to 100 °C for 5 minutes for denaturation. Gel electrophoresis was conducted using 12% Bis-Tris prefabricated gel (Genscript, China), an equal volume of protein sample added to each pore. Following separation by electrophoresis, proteins were transferred to PVDF membranes (Sigma, USA) using semi-dry transfer (Bio-Rad, USA) then blocked with blocking buffer (Beyotime, China) for 15min. Membranes were incubated with primary antibodies overnight. After washing with PBST (Genescript, China) 3 times, the corresponding secondary antibody was added and incubated for 1 hour at room temperature. The Western blots were developed using enhanced chemiluminescent reagent (New Cell & Molecular Biotech, China) prior to observation. The primary antibodies used in this study were directed against GAPDH (1:2000, Cell Signaling Technology, USA) and Kif20a (1:100, Santa Cruz, USA). The secondary antibodies were goat-anti mouse (1:2000, Proteintech, USA) and goat-anti rabbit (1:2000, Proteintech, USA).
CCK-8 cell proliferation assay
Cells were counted and seeded into 96-well plates at a density of 5×103 cells/well then cultured in incubator at 37 °C in an atmosphere containing 5% CO2 for 12h, 24h, 36h and 48h. Ten μl CCK-8 reagent (Beyotime, China) were then added to each well, then incubated for a further 1h in the incubator (Thermo, USA). Absorbance at a wavelength of 450nm was measured a microplate reader (Bio-Rad, USA).
Cell cycle analysis
According to the quantity of DNA in each cell in different phases of the cell cycle, the distribution of cells in each phase can be ascertained. The Kif20a knockout cells and control cells were seeded in 6-well plates at 3×105 cells/well then 1 ml serum-free culture medium was added to each well to starve the cells for 12h, to ensure that they were at the same phase in the cell cycle. The culture medium was removed and the cells were washed with PBS 3 times and complete culture medium added. After 48 hours, the cells were trypsinized off and washed with PBS, then placed in 70% ice-cold ethanol in a 4℃ refrigerator overnight. After washing with PBS three times, propidium iodide (PI) dye (Beyotime, China) was added and incubated at room temperature for 30 minutes. The cells were placed into flow cytometry tubes after passing through a 200-mesh filter and analyzed by flow cytometry (BD-FACSCalibur, USA), detecting red fluorescence at an excitation wavelength of 488nm. Output data were analyzed using ModFit LT 5.0 (USA, Verity Software House).
Colony forming assay
The cells were seeded in 6-well plates at a density of 500 cells/well and 1 ml complete medium containing 20% fetal bovine serum was added to each well. When the cell dividing the fastest had formed a colony of at least 50, 1 ml 4% paraformaldehyde was added to each well, fixed at 4℃ for 1h and washed once with PBS. One ml crystal violet was added to each well then stained for 10min. The cells were then washed with double distilled water several times, dried and imaged, then counted using Image J (v 1.52).
Apoptosis analysis
The cells were seeded in 6-well plates at a density of 1×105/ well. After 48h, cells were digest using trypsin (EDTA free) and collected in EP tubes. After washing with PBS and binding buffer, 100μl binding buffer was added to each tube to resuspend the cells. Then 5 μl Annexin V-PE and 5 μl 7-ADD solution (Sungene biotech, China) was added to each tube. After incubated in dark for 15min, cells were detected in flow cytometry (BD-FACSCalibur, USA).
Wound healing and transwell assays
Fifty thousand cells were seeded into each well of 6-well plates, and complete medium containing 10% fetal bovine serum added to each well. After 12 hours, a 200 μl pipette tip was then used to generate a scratch in the cell culture. The medium was replaced with complete medium containing 5% fetal bovine serum and the cells were cultured in an incubator at 37 °C. The wound was then photographed with an inverted microscope (Olympus, Japan) at the point of wound creation and after 24h.
To conduct a migration assay, 100 μl Matrigel (Corning, USA) were added to the chamber (8.0 μm pore) membrane, then 1×105 cells in serum-free medium were seeded into the upper well and 500μl RPMI 1640 medium supplemented with 20% FBS added to the lower well. After incubation at 37℃ for 24 hours, the cells and Matrigel in the upper chamber were gently removed with a cotton tip. The membrane was then soaked in 4% paraformaldehyde (Solarbio, China) and fixed at 4 °C for 1h. After soaking then washing in PBS for 1 min, the membrane was soaked in 1% crystal violet solution for 10 min. The membrane was then washed in double distilled water 3 times then dried. The membrane was viewed using an inverted microscope (Olympus, Japan) and multiple fields imaged at random.
Tumor-bearing mouse model
All animal experiments were performed following the approval of the First Hospital of Jilin University Animal Ethics Committee and in accordance with the Guidelines for the Care and Use of Laboratory Animals of Jilin province. A total of 10 Balb/c or C57 mice per group (6-8 weeks old; weighing 20±2 g) were provided by the Vital River Laboratory (Beijing, China). WEHI164 cells were originally derived from Balb/c mice and MCA101 and MCA207 cells from C57 mice, so both Balb/c and C57 mice were used for the tumor-bearing animal models. Transfected cells were collected and resuspended in 200 μl PBS (Gibco, USA). WEHI164 cells were suspended at a concentration of 1×105 cells/200 μl, and MCA101 and MCA207 cells at 0.5×105 cell/s200 μl. The cells were injected subcutaneously into the lower flank of the mice. After 3 weeks of culture, the animals were sacrificed. Each tumor and main organs were separated, and the length and width of the tumor tissue were measured and weighed. Tumors and organ tissues were fixed with 4% paraformaldehyde, then stained with HE and Ki67 (Proteintech, USA) using immunohistochemical staining.
Immunohistochemical staining
Fixed tumor tissues were embedded in paraffin and cut into 3 μm slices. Sections were processed for antigen retrieval and stained. Anti-Ki67 (Proteintech, China) was used for immunohistochemical staining (IHC).
Histological analysis
Paraffin embedded tumor tissue was sectioned to a thickness of 5 μm then stained with hematoxylin and eosin. All samples were examined immediately and imaged using an Olympus (BX53, Japan, ×200).
Statistical analysis
Bioinformatics analysis was conducted using R programming language (v 3.6.0) and additional software packages. CCK-8 results were analyzed and plotted using GraphPad Prism 8.0.2. The cell cycle results were analyzed by ModFit LT 5.0, and the nucleic acid content in G2 phase was set to be 1.9 times of that in G1 phase. After that, the positions of G1 and G2 peaks were adjusted to make the fitting shape of G1 and G2 phase most consistent with the frequency distribution of nucleic acid content. FlowJ v10.0 software was used for apoptosis analysis to calculate the proportion of quadrants. The clone formation experiment, scratch experiment and Transwell experiment used ImageJ 1.52k for counting statistics. Immunohistochemical staining was assessed using ImageJ 1.52k. After threshold adjustment, the proportion of positive areas was calculated. The difference between groups was calculated using the unpaired t test.
Clinical information of TCGA-SARC project
| Characteristic | Detail |
|---|---|
| Age | 61(20-90) |
| Gender | |
| Female | 141 |
| Male | 118 |
| Race | |
| White | 226 |
| American | 18 |
| Black or African American | 18 |
| Asian | 6 |
| Not Evaluated | 6 |
| Unknown | 3 |
| Histological type | |
| Leiomyosarcoma (LMS) | 105 |
| Dedifferentiated liposarcoma | 59 |
| Undifferentiated Pleomorphic Sarcoma (UPS) | 50 |
| Myxofibrosarcoma | 25 |
| Others | 22 |
| Necrosis | |
| 0% (no necrosis or no mention of necrosis) | 70 |
| <10% (focal necrosis) | 38 |
| Moderate Necrosis (≥10, <50%) | 61 |
| Extensive Necrosis (>50%) | 12 |
| Not Available | 78 |
| Radiation therapy | |
| NO | 140 |
| YES | 73 |
| Unknown | 10 |
| Not Available | 36 |
| Vital status | |
| Alive | 184 |
| Dead | 75 |
Results
High transcription levels of Kif20a exhibited poor prognosis in STS
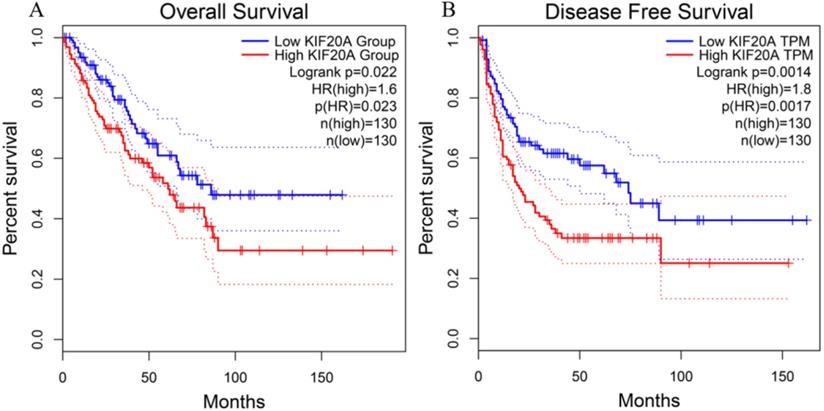
To investigate the relationship between Kif20a transcription level and STS prognosis, the clinical characteristics and expression data of samples collected in the TCGA-SARC project were analyzed. Patient survival and Kif20a expression were analyzed using GEPIA2 (http://gepia2.cancer-pku.cn/) [17]. High Kif20a expression levels compared with median expression levels in all samples exhibited significantly (p<0.05) lower overall survival and disease-free survival rate (Figure 1).
Kif20a predicted protein-protein interaction analysis
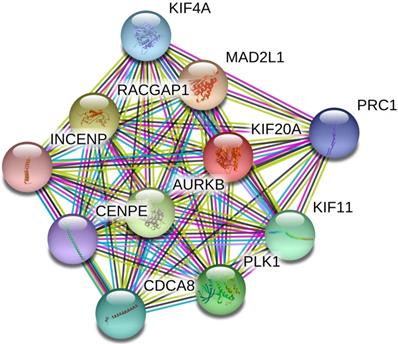
Based on the data from the STRING database, a PPI internetwork (PPI) including 11 nodes and 55 edges was constructed (PPI enrichment p-value: 9.99e-16) and visualized using Cytoscape software (Figure 2). Co-expression analysis revealed that Kif20a was co-expressed with centromere protein E (CENPE), cell division cycle associated 8 (CDCA8), inner centromere protein (INCENP), Rac GTPase-activating protein 1 (RACGAP1), aurora kinase B (AURKB), polo like kinase 1 (PLK1), kinesin family member 11 (KIF11), MAD2 mitotic arrest deficient-like 1 (MAD2L1), kinesin family member 4A (KIF4A) and protein regulator of cytokinesis 1 (PRC1).
GSEA identified the biological functions and pathways associated with Kif20a
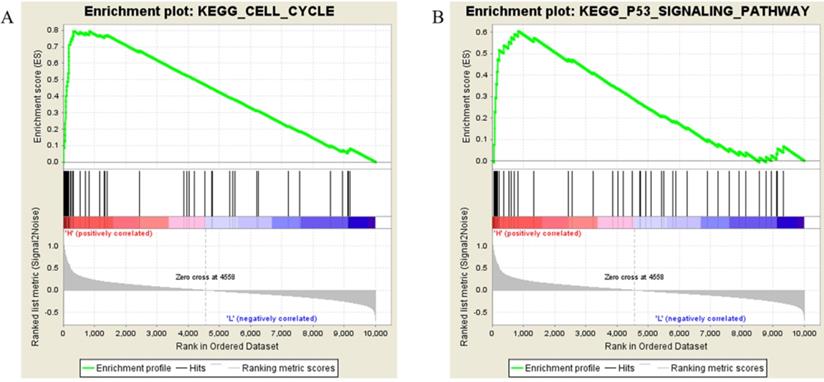
Samples from the TCGA-SARC project were divided into two groups (H vs L), the threshold defined by the median Kif20a expression level. Followed by gene set enrichment, the high Kif20a expression group was enriched in cell cycle function, cell cycle pathway and p53 signaling pathways (Figure 3, Table 2).
The most enriched KEGG pathways of the high KIF20A expression group
| NAME | SIZE | NOM p-value | FDR q-value |
|---|---|---|---|
| KEGG_OOCYTE_MEIOSIS | 46 | 0 | 0 |
| KEGG_CELL_CYCLE | 57 | 0 | 0 |
| KEGG_P53_SIGNALING_PATHWAY | 42 | 0.001912 | 7.40E-04 |
| KEGG_PROGESTERONE_MEDIATED_OOCYTE_MATURATION | 39 | 0.001969 | 0.003036 |
| KEGG_PANCREATIC_CANCER | 35 | 0.00789 | 0.059943 |
| KEGG_SMALL_CELL_LUNG_CANCER | 49 | 0.018145 | 0.147014 |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 23 | 0.02554 | 0.152741 |
| KEGG_PROSTATE_CANCER | 41 | 0.051793 | 0.310258 |
| KEGG_BLADDER_CANCER | 28 | 0.111765 | 0.39316 |
| KEGG_PYRIMIDINE_METABOLISM | 27 | 0.121032 | 0.538301 |
Kif20a is upregulated in STS cell lines
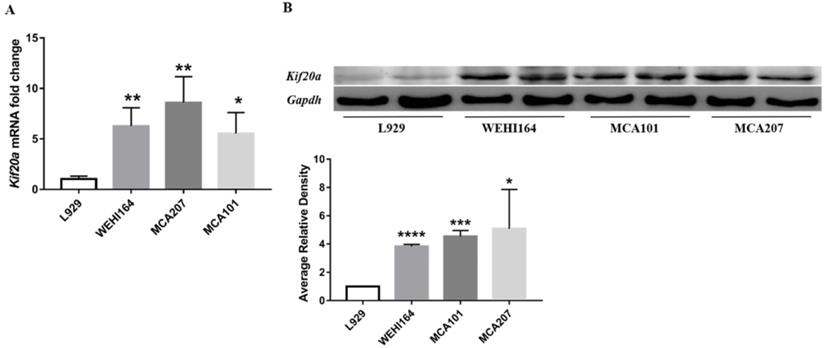
In order to explore Kif20a expression in STS, we assessed mRNA and protein expression levels in three STS cell lines (WEHI164, MCA101, MCA207) and L929 cells by RT-qPCR and Western blotting. There was a significant (p<0.05) higher Kif20a expression level in all STS cell lines (WEHI164, MCA101, MCA207) compared with the L929 control cells (Figure 4).
Higher expression of KIF20A resulted in significantly lower rates of (A) overall survival and (B) disease-free survival.
Protein-protein interaction network (PPI) centered on KIF20A.
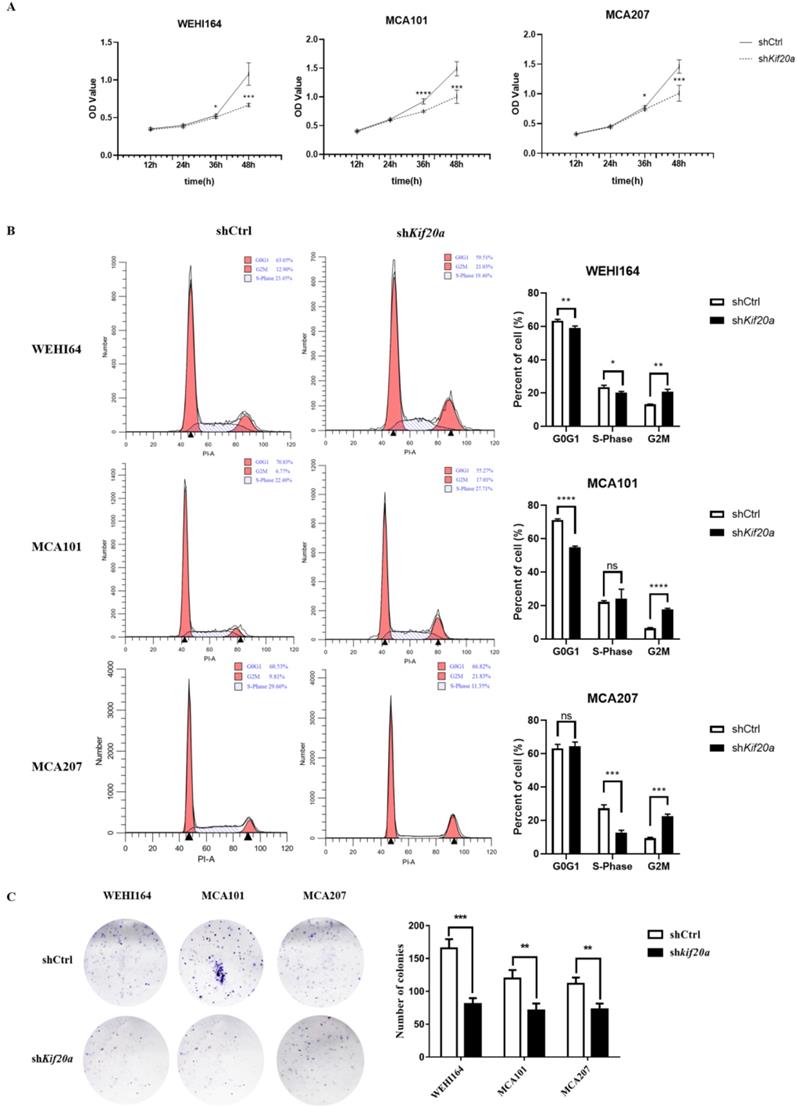
Downregulation of Kif20a inhibits proliferation in vitro
To investigate the biological function of Kif20a, three STS cell lines (WEHI164, MCA101, MCA207) were transfected with a Kif20a shRNA plasmid (shKif20a cells) and control plasmid (shCtrl cells). Transfection efficiency was determined by fluorescence microscopy, RT-qPCR and Western blotting analysis. We found that almost all cells were transfected by plasmids (Fig. S1A) and Kif20a was successfully knocked down at both mRNA (Fig. S1B) and protein expression levels (Fig. S1C) in WEHI164, MCA101 and MCA207 cell lines. These data indicate that Kif20a shRNA plasmids specifically and significantly inhibited Kif20a expression in STS cells. To determine the potential effects of Kif20a on cell proliferation, CCK-8 analysis was performed. Cell viability was found to decrease in shKif20a cells (Figure 5A). FACS analysis was used to explore cell cycle in shCtrl and shKif20a cells. The results demonstrated that G2 phase arrest occurred in all three STS cell lines after Kif20a gene knockdown (Figure 5B). A clonogenic assay further confirmed that in Kif20a knockout cells, colony formation was also significantly (P<0.05) inhibited in the STS cell lines (Figure 5C). These results suggest that downregulation of Kif20a inhibited cell proliferation in STS cells by regulating cell cycle distribution.
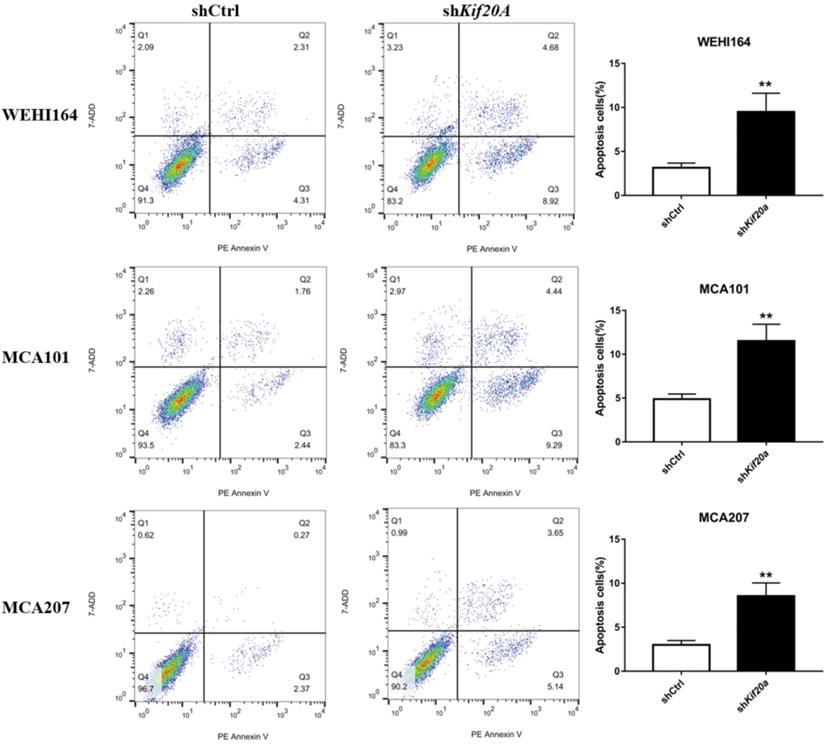
Downregulation of Kif20a promote cell apoptosis in vitro
After Kif20a was downregulated, the proportion of cells in the second (late apoptosis and necrosis) and third quadrants (early apoptosis) increased. Apoptosis in WEHI164 cells increased from 6% to 13%, MCA101 from 4% to 13%, and MCA207 from 2% to 8% (Figure 6).
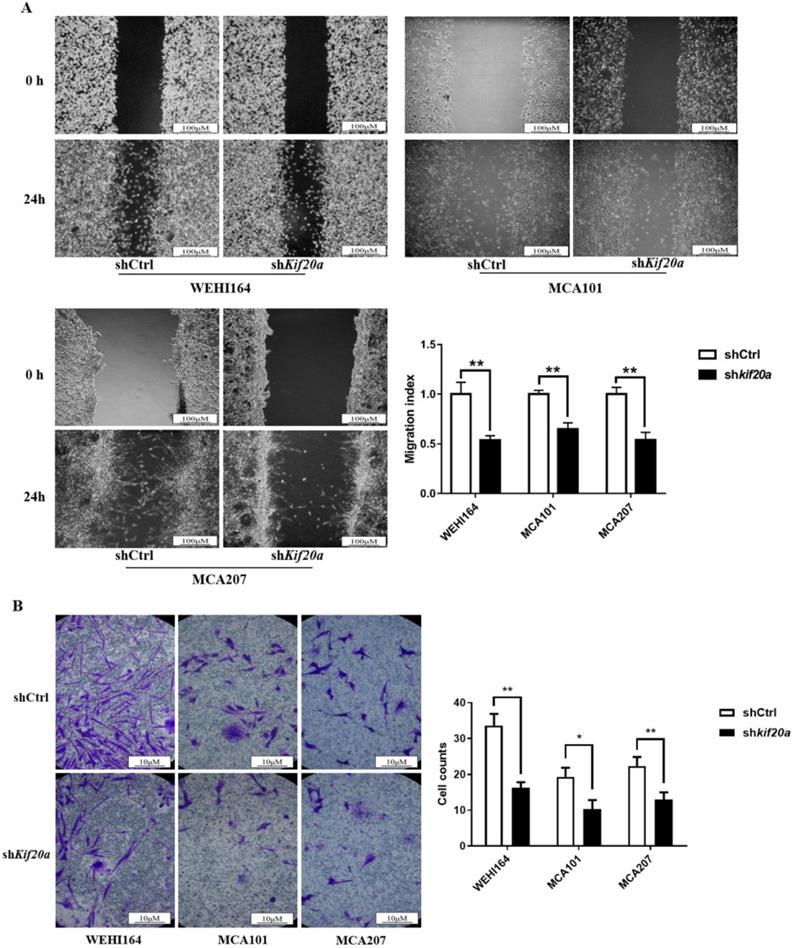
Downregulation of Kif20a inhibits cell migration and cell invasion in vitro
To determine the effect of Kif20a on the metastasis capability of STS cells, migration (Figure 7A) and invasion (Figure 7B) assays were performed in shCtrl and shKif20a cells using wound healing and transwell assays. These tests indicated that the capability of the cells in both respects were significantly (P<0.05) reduced after Kif20a knockdown.
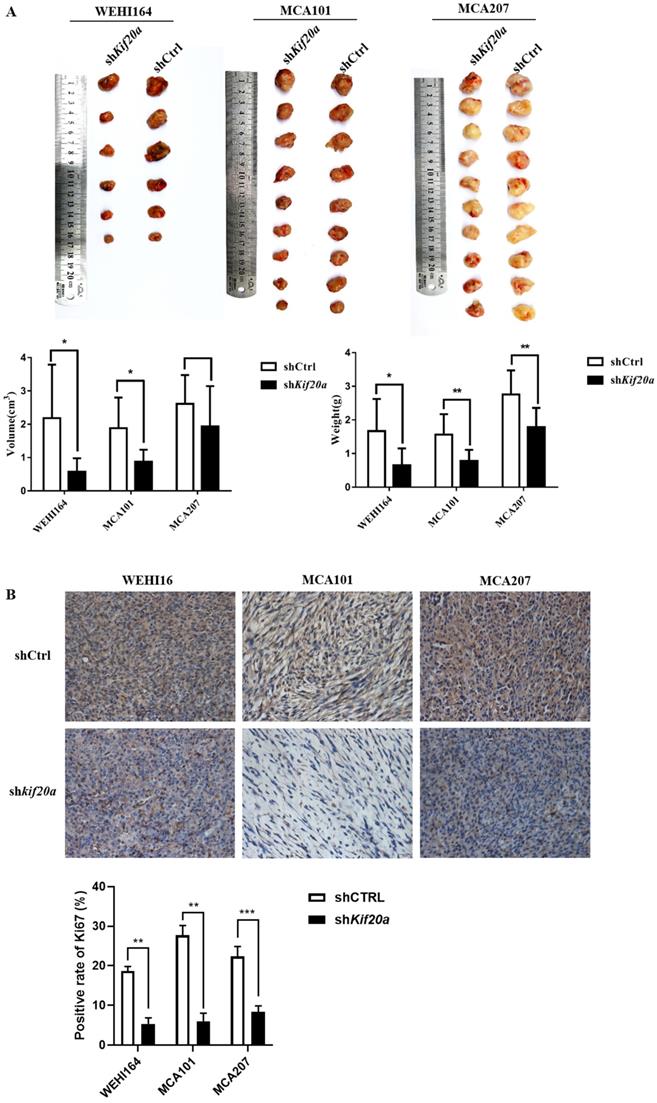
Downregulation of Kif20a suppresses tumor growth in vivo
To investigate the effect of Kif20a on tumor growth, we established a tumor-bearing mouse model. In vivo animal experiments demonstrated that tumor growth was slower after knock down (Figure 8A). Analysis of Ki67 by IHC indicated a reduction in tumor proliferation (Figure 8B). We further measured metastasis of the tumor and found no metastasis in the heart, liver, spleen, lung, kidney or brain (Fig. S2). These results reveal that downregulation of Kif20a suppressed tumor growth, but not tumor metastasis, in vivo.
Discussion
In this study, we found that high expression of Kif20a indicated poor prognosis in STSs. Further analysis indicated that Kif20a is related to cell cycle and the p53 signaling pathway. We also confirmed that Kif20a is upregulated in STS cell lines. Downregulation of Kif20a inhibited proliferation and colony formation in vitro and suppressed tumor growth in vivo.
Results of gene set enrichment analysis. Samples with higher expression of KIF20A were significant enriched in (A) cell cycle and (B) p53 pathways.
Kif20a was upregulated in STS cell lines. (A) Relative Kif20a mRNA expression was determined in three STS cell lines (WEHI164, MCA207, MCA101) and immortalized L929 cells by qRT-PCR and (B) by Western blotting. qRT-PCR and Western blot assays were normalized to GAPDH. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Downregulation of Kif20a inhibited proliferation and colony formation in vitro. (A) Cell viability was determined by CCK8 assay. (B) Cell cycle distribution was determined by flow cytometry. (C) Cell colony formation was determined by colony forming assay. ***P<0.001, ****P<0.0001.
Downregulation of Kif20a promote cell apoptosis in vitro. Q2 indicating cells in late apoptosis and necrosis, Q3 indicating cells in early apoptosis.
STS has a low incidence but poor prognosis. The capillary morphogenesis gene 2 (CMG2) has been demonstrated to promote endothelial cell proliferation and exhibits angiogenic properties. In STSs, low CMG2 expression was correlated with a worsened prognosis [18]. In this study, we found that high Kif20a expression predicted poor prognosis, which is a new finding.
Kif20a is a member of the kinesin superfamily of proteins. It consists of three domains: an N-terminal motor domain, a coiled coil domain and a C-terminal tail domain. The N-terminal domain mediates motor activity, the coiled coil domain interacts with partner proteins and participates in dimerization. The C-terminal tail domain can also interact with partner proteins and contributes to vesicle transport [19]. The present study found that Kif20a is isolated in the nucleus during G2 phase, thus preventing the cross-linking and depolymerization of cytoplasmic microtubules [20]. After the knock-out of Kif20a, it was shown that STS cells became stuck in the G2 phase in this study. In tumors, Kif20a expression increased significantly, it promoted the growth of tumors and tumor cell lines, and was related to carcinogenesis and aggressiveness [19]. The present study demonstrated that Kif20a was upregulated in STSs, promoted the growth of STSs, similar to most tumors. In pancreatic cells, Kif20a was shown to promote cell motility and invasiveness by transport of RNA-binding proteins [21]. We found that Kif20a regulated cell division and contributed to tumor growth both in vitro and in vivo and in STSs. In order to explore the mechanism further, PPI network analysis was conducted, which predicted that Kif20a co-expressed with PLK1, INCENP and AURKB. PLK1 is a serine/threonine-protein-like kinase. It can form a complex with Kif20a which it phosphorylates, regulating its activity [22]. INCENP, AURKB, Survivin and Borealin contain chromosomal passenger complexes (CPCs), which play an important role in regulating the accuracy of the mitotic process. Previous studies have revealed that Kif20a participates in the recruitment of the CPC to the spindle [23]. We hypothesize that Kif20a, along with PLK1, INCENP and AURKB regulate STS, but further evidence is required to confirm this speculation.
Downregulation of KIF20A inhibited cell migration and cell invasion in vitro. (A) Cell migration was determined by wound healing assay. (B) Cell invasion was determined by transwell invasion assay. *P<0.05, **P<0.01.
Downregulation of KIF20A suppressed tumor growth in vivo. (A) Tumor tissues from different treatment groups demonstrating differences in volume and weight of tumors (Left to right: WEHI164, MCA101, MCA207). (B) Ki-67 expression in tumors determined by IHC.
It should be noted that due to the lack of normal control samples and sufficient cell lines in this study, the level of Kif20a expression in STSs needs to be further confirmed.
In summary, our findings revealed that Kif20a performs an important role in STS, indicating that it is a potential prognostic biomarker and potentially representing a therapeutic target for the disease.
Supplementary Material
Supplementary figures.
Acknowledgements
This study was supported by the Special Projects of Health in Jilin Province (3D5148273428) and the Key R&D Program of Guangdong Province (2018B090906001).
Competing Interests
The authors have declared that no competing interest exists.
References
1. van der Graaf WTA, Orbach D, Judson IR, Ferrari A. Soft tissue sarcomas in adolescents and young adults: a comparison with their paediatric and adult counterparts. Lancet Oncol. 2017;18:e166-e75
2. Hoefkens F, Dehandschutter C, Somville J, Meijnders P, Van Gestel D. Soft tissue sarcoma of the extremities: pending questions on surgery and radiotherapy. Radiat Oncol. 2016;11:136
3. Linch M, Miah AB, Thway K, Judson IR, Benson C. Systemic treatment of soft-tissue sarcoma-gold standard and novel therapies. Nat Rev Clin Oncol. 2014;11:187-202
4. Lai F, Fernald AA, Zhao N, Le Beau MM. cDNA cloning, expression pattern, genomic structure and chromosomal location of RAB6KIFL, a human kinesin-like gene. Gene. 2000;248:117-25
5. Hill E, Clarke M, Barr FA. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. Embo j. 2000;19:5711-9
6. Echard A, Jollivet F, Martinez O, Lacapere JJ, Rousselet A, Janoueix-Lerosey I. et al. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279:580-5
7. Sheng Y, Wang W, Hong B, Jiang X, Sun R, Yan Q. et al. Upregulation of KIF20A correlates with poor prognosis in gastric cancer. Cancer Manag Res. 2018;10:6205-16
8. Duan J, Huang W, Shi H. Positive expression of KIF20A indicates poor prognosis of glioma patients. Onco Targets Ther. 2016;9:6741-9
9. Kawai Y, Shibata K, Sakata J, Suzuki S, Utsumi F, Niimi K. et al. KIF20A expression as a prognostic indicator and its possible involvement in the proliferation of ovarian clearcell carcinoma cells. Oncol Rep. 2018;40:195-205
10. Liu SL, Lin HX, Qiu F, Zhang WJ, Niu CH, Wen W. et al. Overexpression of Kinesin Family Member 20A Correlates with Disease Progression and Poor Prognosis in Human Nasopharyngeal Cancer: A Retrospective Analysis of 105 Patients. PLoS One. 2017;12:e0169280
11. Stangel D, Erkan M, Buchholz M, Gress T, Michalski C, Raulefs S. et al. Kif20a inhibition reduces migration and invasion of pancreatic cancer cells. J Surg Res. 2015;197:91-100
12. Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F. et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35:990-1002
13. Goldman M, Craft B, Hastie M, Repečka K, Kamath A, McDade F. et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2019 p: 326470
14. Crosara KTB, Moffa EB, Xiao YZ, Siqueira WL. Merging in-silico and in vitro salivary protein complex partners using the STRING database: A tutorial. J Proteomics. 2018;171:87-94
15. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-50
16. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267-73
17. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W60
18. Greither T, Wedler A, Rot S, Kessler J, Kehlen A, Holzhausen HJ. et al. CMG2 Expression Is an Independent Prognostic Factor for Soft Tissue Sarcoma Patients. Int J Mol Sci. 2017 p:18
19. Wu WD, Yu KW, Zhong N, Xiao Y, She ZY. Roles and mechanisms of Kinesin-6 KIF20A in spindle organization during cell division. Eur J Cell Biol. 2019;98:74-80
20. Fontijn RD, Goud B, Echard A, Jollivet F, van Marle J, Pannekoek H. et al. The Human Kinesin-Like Protein RB6K Is under Tight Cell Cycle Control and Is Essential for Cytokinesis. Mol Cell Biol. 2001;21:2944-55
21. Taniuchi K, Furihata M, Saibara T. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia. 2014;16:1082-93
22. Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU. et al. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. The Journal of cell biology. 2003;162:863-75
23. Hummer S, Mayer TU. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Current biology: CB. 2009;19:607-12
Author contact
![]() Corresponding authors: Mei Zhang, Professor, College of Chemistry, Jilin University, Changchun, China. E-mail: zhangmeiedu.cn, Work phone: 0431-81875991; Dahui Sun, Professor, Department of Orthopaedic Trauma, The First Hospital of Jilin University, Changchun, China. E-mail: sundahui1971com, Work phone: 0431-81875777.
Corresponding authors: Mei Zhang, Professor, College of Chemistry, Jilin University, Changchun, China. E-mail: zhangmeiedu.cn, Work phone: 0431-81875991; Dahui Sun, Professor, Department of Orthopaedic Trauma, The First Hospital of Jilin University, Changchun, China. E-mail: sundahui1971com, Work phone: 0431-81875777.

 Global reach, higher impact
Global reach, higher impact