Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(24):7237-7245. doi:10.7150/jca.46055 This issue Cite
Review
The role of Transfer RNA-Derived Small RNAs (tsRNAs) in Digestive System Tumors
1. Department 1 of General Surgery, the First Hospital of China Medical University, Shenyang 110001, China
2. Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang 110001, China
#, Equal contributor.
Received 2020-3-16; Accepted 2020-9-30; Published 2020-10-18
Abstract
Transfer RNA-derived small RNA(tsRNA) is a type of non-coding tRNA undergoing cleavage by specific nucleases such as Dicer. TsRNAs comprise of tRNA-derived fragments (tRFs) and tRNA halves (tiRNAs). Based on the splicing site within the tRNA, tRFs can be classified into tRF-1, tRF-2, tRF-3, tRF-5, and i-tRF. TiRNAs can be classified into 5′-tiRNA and 3′-tiRNA. Both tRFs and tiRNAs have important roles in carcinogenesis, especially cancer of digestive system. TRFs and tiRNAs can promote cell proliferation and cell cycle progression by regulating the expression of oncogenes, combining with RNA binding proteins such as Y-box binding protein 1 (YBX1) to prevent transcription. Despite many reviews on the basic biological function of tRFs and tiRNAs, few have described their correlation with tumors especially gastrointestinal tumor. This review focused on the relationship of tRFs and tiRNAs with the biological behavior, clinicopathological characteristics, diagnosis, treatment and prognosis of digestive system tumors, and would provide novel insights for the early detection and treatment of digestive system tumors.
Keywords: tsRNAs, tRFs, tiRNAs, tRNAs, digestive system tumors, cancer
Introduction
Transfer RNAs (tRNAs) have important roles in protein biosynthesis, which has a secondary cloverleaf structure containing three hairpin loops: a D loop, an anti-codon loop, and a TψC loop. TRNA may recognize codons of messenger RNA (mRNA) via its anti-codon loop and then the corresponding amino acid is transferred to the polypeptide synthesized in the ribosome(1). Recently, in-depth analyses of new-generation data have revealed a large number of non-coding RNAs with unique function, including microRNA (miRNA), piwi-interacting RNA (piRNA), long non-coding RNA (lncRNA) and circle RNA (circRNA). Among them, transfer RNA-derived small RNAs (tsRNAs) have been paid increasing attention. TsRNAs can be found in a variety of organisms (2, 3). Numerous researches have demonstrated that tsRNAs could regulate cell proliferation, invasion, metastasis and gene expression, and thus play a crucial role in human cancer (4, 5). Aberrant regulation of tsRNAs has been reported in multiple cancers. Five tsRNAs were down-regulated in chronic lymphocytic leukemia and lung cancer, including ts-46, ts-47, ts-49, ts-53 and ts-101, while ts-4 was up-regulated in them (6). Ts-86 was down-regulated in breast cancer whereas ts-66 was up-regulated (6). Differentially expressed tsRNAs have the potential to be diagnostic markers and therapeutic targets of cancer (7, 8).
The main classification of tRNA-derived fragments (tRFs) and tRNA halves(tiRNAs). In this figure, the tRFs or tiRNAs were shown the source from tRNAs, according to the color which is the same with tRNAs'.
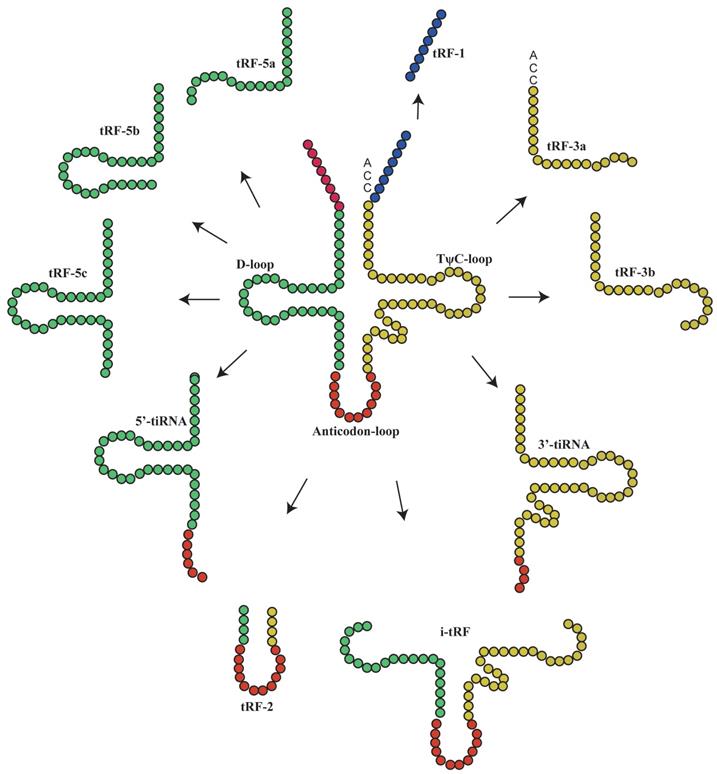
Basic biological functions
Classification of tRFs and tiRNAs
Under special conditions such as hypoxia, stress or UV radiation, tRNA is cleaved into small tsRNA fragments by specific nucleases(9), which can be classified into tRFs and tiRNAs(10, 11). To date, more detailed studies have been performed on tRFs than tiRNAs, as evidenced by the establishment of tRF databases(12). TRFs are non-coding RNAs with 14-30 nucleotides in length. They are derived from the end of pre-tRNAs or mature tRNAs and can be classified into different types based on their original site in these molecules: tRF-1, tRF-3, tRF-5(13), i-tRFs and tRF-2(14, 15). TRF-3 (13-22Nucleotide[nt]) can be further classified into tRF-3 and tRF-3b, while t-RF-5 can be classified into three subtypes: tRF-5a (14-16nt), tRF-5b (22-24nt) and tRF-5c (28-30nt)(16). TiRNAs are non-coding small RNAs with 31-40 nucleotides in length, which can be classified into 3'-tiRNAs and 5'-tiRNAs(11, 17-21). The type of tsRNAs was shown in Figure 1.
It has been suggested that each tRF and tiRNA are produced by different enzymes. TRF-5 and tRF-3 are produced by endonucleolytic and exonucleolytic digestion of mature tRNAs, respectively. TRF-5 is mainly produced by Dicer cleavage of tRNA D loop or the stem loop between D loop and anti-codon(4, 11, 17, 22-25); tRF-3 is produced through TψC loop cleavage by Dicer and Angiogenin (ANG), a member of ribonuclease A superfamily(4, 11, 17, 26-29). TRF-1 is derived from the 3'-end of pre-tRNA mainly via digestion by endonuclease Z (RNase Z/ELAC2)(11, 17-21). TRF-2 is derived from the cleavage of tRNA anti-codon(11, 17, 21). The i-tRFs are mainly derived from the internal region (between D loop and T loop) of mature tRNA, which is across the anti-codon region(6, 11, 21). TiRNAs are derived from mature tRNA anti-codons through digestion by ANG. The 3'-tiRNAs are generated from the 3'-end of anti-codon, while 5'-tiRNAs are from the 5'-end of anti-codon(11, 17, 21, 30). The naming rules of tsRNA are listed as Figure 2 and Table 1.
Data from the tRFdb*
| tRF ID | Organism | Type | tRNA Gene Co-ordinates | tRNA Name |
|---|---|---|---|---|
| 1001 | human | tRF-1 | chr10-69524261-69524342 | chr10.tRNA2-SerTGA |
| 1003 | human | tRF-1 | chr17-8090184-8090265 | chr17.tRNA7-SerGCT |
| 1004 | human | tRF-1 | chr12-98897281-98897352 | chr12.tRNA5-AspGTC |
| 1042 | human | tRF-1 | chr19-33667963-33668036 | chr19.tRNA4-ThrAGT |
| 3001a | human | tRF-3 | chr17-8023713-8023632 | chr17.tRNA42-LeuTAG |
| 3001a | human | tRF-3 | chr16-22207113-22207032 | chr16.tRNA27-LeuTAG |
| 3001a | human | tRF-3 | chr5-180528840-180528921 | chr5.tRNA3-LeuAAG |
| 3001a | human | tRF-3 | chr5-180524555-180524474 | chr5.tRNA19-LeuAAG |
| 3001a | human | tRF-3 | chr5-180601125-180601044 | chr5.tRNA16-LeuAAG |
| 3001a | human | tRF-3 | chr6-28911480-28911399 | chr6.tRNA98-LeuAAG |
| 3001a | human | tRF-3 | chr5-180614701-180614782 | chr5.tRNA7-LeuAAG |
| 3001a | human | tRF-3 | chr16-22308461-22308542 | chr16.tRNA16-LeuAAG |
| 3001a | human | tRF-3 | chr14-21078291-21078372 | chr14.tRNA1-LeuAAG |
| 3001a | human | tRF-3 | chr6-28956779-28956860 | chr6.tRNA78-LeuAAG |
| 3001a | human | tRF-3 | chr6-28446481-28446400 | chr6.tRNA126-LeuAAG |
| 3033a | human | tRF-3 | chr1-16847153-16847080 | chr1.tRNA136-AsnGTT |
| 5001a | mouse | tRF-5 | chr13-23609561-23609632 | chr13.tRNA111-MetCAT |
| 5001a | mouse | tRF-5 | chr15-69227124-69227053 | chr15.tRNA876-MetCAT |
| 5001a | mouse | tRF-5 | chr13-21998980-21999051 | chr13.tRNA82-MetCAT |
| 5001a | human | tRF-5 | chr8-67026223-67026311 | chr8.tRNA5-TyrGTA |
| 5001a | human | tRF-5 | chr8-67025602-67025694 | chr8.tRNA4-TyrGTA |
| 5001a | human | tRF-5 | chr2-27273650-27273738 | chr2.tRNA2-TyrGTA |
| 5001a | human | tRF-5 | chr6-26595102-26595190 | chr6.tRNA17-TyrGTA |
| 5001a | human | tRF-5 | chr6-26577332-26577420 | chr6.tRNA16-TyrGTA |
| 5001a | human | tRF-5 | chr6-26569086-26569176 | chr6.tRNA14-TyrGTA |
| 5001a | human | tRF-5 | chr14-21151432-21151520 | chr14.tRNA5-TyrGTA |
| 5001a | human | tRF-5 | chr14-21131444-21131351 | chr14.tRNA16-TyrGTA |
| 5001a | human | tRF-5 | chr14-21128210-21128117 | chr14.tRNA17-TyrGTA |
| 5001a | human | tRF-5 | chr14-21125716-21125623 | chr14.tRNA18-TyrGTA |
| 5001a | human | tRF-5 | chr14-21121351-21121258 | chr14.tRNA19-TyrGTA |
| 5001b | human | tRF-5 | chr8-67026223-67026311 | chr8.tRNA5-TyrGTA |
| 5001b | human | tRF-5 | chr8-67025602-67025694 | chr8.tRNA4-TyrGTA |
| 5001b | human | tRF-5 | chr6-26595102-26595190 | chr6.tRNA17-TyrGTA |
| 5001b | human | tRF-5 | chr14-21151432-21151520 | chr14.tRNA5-TyrGTA |
| 5001b | human | tRF-5 | chr14-21131444-21131351 | chr14.tRNA16-TyrGTA |
| 5001b | human | tRF-5 | chr14-21128210-21128117 | chr14.tRNA17-TyrGTA |
| 5001b | human | tRF-5 | chr14-21125716-21125623 | chr14.tRNA18-TyrGTA |
| 5001b | human | tRF-5 | chr14-21121351-21121258 | chr14.tRNA19-TyrGTA |
| 5001b | mouse | tRF-5 | chr3-90279853-90279782 | chr3.tRNA792-MetCAT |
| 5001b | mouse | tRF-5 | chr13-22023109-22023038 | chr13.tRNA983-MetCAT |
| 5032c | human | tRF-5 | chr1-161440276-161440205 | chr1.tRNA69-AspGTC |
*, This data is from tRFdb, which shows tRFID from 1001-1042 belongs to tRF-1; 3001-3033 belongs to tRF-3 and 5001-5032 belongs to tRF-5. And the naming rule is always “chrX.tRNA XXX-Amino acid + anticodon”. The anticodon is made up of three nucleotide bases.
Differences and correlation between tRFs and tiRNAs
Under specific conditions, tRFs and tiRNAs are similar but not identical in terms of their structure and function. First, both of them are derived from tRNA. However, due to different enzymes and cleavage sites involved, the products differ in terms of their length and structure(17). Second, tRFs and tiRNAs are mostly located within cells except for a small part in peripheral circulation; however, their intracellular original sites and distribution are different. TRF-5s are produced in cytoplasm and transported to nucleus via a similar mechanism for tRNA transportation(10, 17, 30). TRF-1 might be generated in cytoplasm or nucleus. Some scholars reported that tRF-1s were generated in cytoplasm(4). However, others believed that tRF-1s was likely to be produced in nucleus and transported to cytoplasm via specific mechanism(17). TRF-3s are mainly distributed in cytoplasm (10, 17). TiRNAs are mainly located in cytoplasm with a small part in nucleus and mitochondria, while tiny levels are found in circulatory system(10). However, whether the location of tRFs and tiRNAs is associated with their diverse function remains unknown. Third, both tRFs and tiRNAs have important roles in cellular transcription, gene expression and viral infection(11, 31), with various mechanisms. For instance, regarding transcriptional regulation, tRF-5s were suggested to mainly inhibit transcriptional elongation, whereas 5'-tiRNAs mainly inhibits transcriptional initiation(6, 32, 33). TRFs and tiRNA have many features in common, though further investigations are required to explore the correlation between their characteristics and disease development, especially tumor development and progression.
Function of tRFs and tiRNAs
Although the function of most tRFs and tiRNAs have not been elucidated, accumulating evidence has suggested that they are involved in regulating cell proliferation and apoptosis, DNA damage response, gene expression and post-transcriptional modifications. TRFs and tiRNAs have the following function (Table 2).
Regulating cellular proliferation and apoptosis
TRFs were found to might inhibit cellular proliferation. TRF-3027 (tRNAGly-GCC) may bind to AGO, an important component of RNA-induced silencing complexes (RISCs), block replication protein A1 (RPA1) to inhibit cellular proliferation, and regulate DNA damage response(6, 9, 11, 17, 21, 30).
TsRNAs were also indicated to induce or inhibit apoptosis. During apoptosis induction, inhibition of a specific tsRNA, 3'-tsRNA-LeuCAG, has been shown to induce apoptosis in rapidly dividing cells in vitro and a patient-derived orthotopic hepatocellular carcinoma model of mice. The tsRNA binds to at least two ribosomal protein mRNAs (RPS28 and RPS15) to enhance their translation. A decrease in the translation of RPS28 mRNA blocks pre-18S ribosomal RNA processing, resulting in a reduction in the number of 40S ribosomal subunits to induce apoptosis(34). During apoptosis inhibition, tsRNAs and tRNAs have similar roles. Early studies of tRNA showed that mature tRNA bind to cytochrome C to inhibit the formation of apoptotic bodies and cysteine-aspartic protease (caspase-9) activity to stimulate cellular survival(35). Recent studies showed that tsRNAs derived from tRNA also bind to cytochrome C(36), which then bind to apoptotic protease activating factor 1 (APAF1) and form apoptotic bodies(36). In response to hyperosmotic stress, ANG mediates the competitive binding of 5'-tiRNA and 3'-tiRNA to cytochrome C generating the ribonucleoprotein complex, inhibits the formation and activity of apoptotic bodies, and stimulates cellular survival(10, 36-38) (Figure 3). However, the complete biological mechanisms remain unclear and require further investigation.
Function similar to miRNA and piRNA
Certain tRFs may act as miRNAs or piRNAs to bind to AGO protein, which is an important RISC(34) with biological function. TRF-1s tend to interact with AGO3 and AGO4(6, 39). TRF-5s and tRF-3s interact with AGO1, AGO3 and AGO4(17, 31, 39). Like miRNAs, they have seed sequences complementary to target mRNAs and can be recruited to AGO complexes to regulate target RNA expression and function(17). TRF-5 and tRF-3 both bind to miRNA via the seed region(16, 40), which has been confirmed in many other tRFs. For instance, ts-53 and ts-101 not only bind to AGO proteins but also PIWIL2 (a molecule similar to piRNA), regulating target gene expression and post-transcriptional modification(6, 30). However, interactions between tiRNA and AGO remain unknown and need further investigation.
The type and function of tsRNAs for human
| Type | Name | tRF ID in tRF database | Function | Reference |
|---|---|---|---|---|
| tRF-5 | tRNA-Gln CTG | tRF-5021 | Inhibit translation | (44) |
| tRF-5 | tRNA-GluCTC | tRF-5030 | Inhibit the target mRNA | (3, 43) |
| tRF-3 | tRNA-GlyGCC | tRF-3027 | Repress the endogenous Replicating Protein A1 | (9) |
| tRF-1 | tRNA-SerTGA | tRF-1001 | Patriciate in G2-M transition | (4) |
| 5'-tiRNA | tiRNA-Ala | - | Inhibit translation | (33) |
| tRF-5 | tRNA-Leu/tRF/miR-1280 | tRF-5019 | Repress colorectal cancer cell proliferation | (5) |
| 5'-tiRNA | Several tsRNAs | - | Associated with hepatitis B-associated HCC | (51) |
The naming rules of tRFs and tiRNAs. The name described the tRFs or tiRNAs should contain the type (tRF-5, tRF-3, tRF-1, 5'-tiRNA, 3'-tiRNA), Chromosome number, tRNA number, the amino acid carried and the anticodon carried. The anticodon is the codon which tRNA carried, then this anticodon will be translated into the codon which is matched amino acid. And every tRFs has a name in tRFdb (tRFdatabase), for example, tRF-1001 is Chr10. Trna2-SerTGA and it belongs to tRF-1 type, and the number underline in tRFdb means the type (tRF-5, tRF-3 or tRF-1).
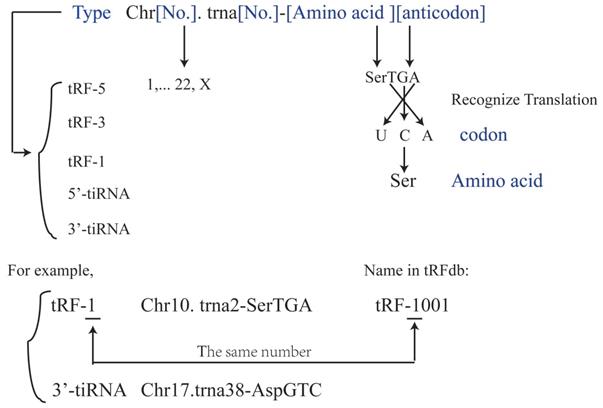
5'-tiRNA could promote the cell survival.ThetsRNAs that were derived from the tRNA also binds to the cytochrome C, which then would bind to the apoptotic protease activating factor 1 (APAF1) to form the apoptotic bodies. And the 5'tiRNA and 3'tiRNA could bind to the cytochrome C to form the ribonucleoprotein complex, inhibit formation and activity of the apoptotic bodies, and stimulate the cellular survival. APAF1, apoptotic protease activating factor 1.
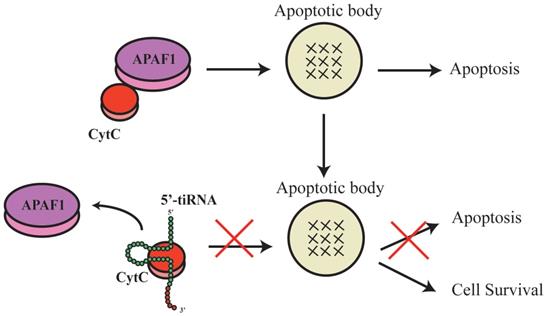
tRF-3 could regulate ribosomal functions. The tRF-3s may specifically bind to TWI12 protein (member of the AGO/PiWi protein family), and recruit Tan1 protein and exoribonuclease XRN2, to form pre-ribosomal RNA splicing complex (TXT), process pre-rRNA during rRNA synthesis, and then regulate translation.
Translation regulation
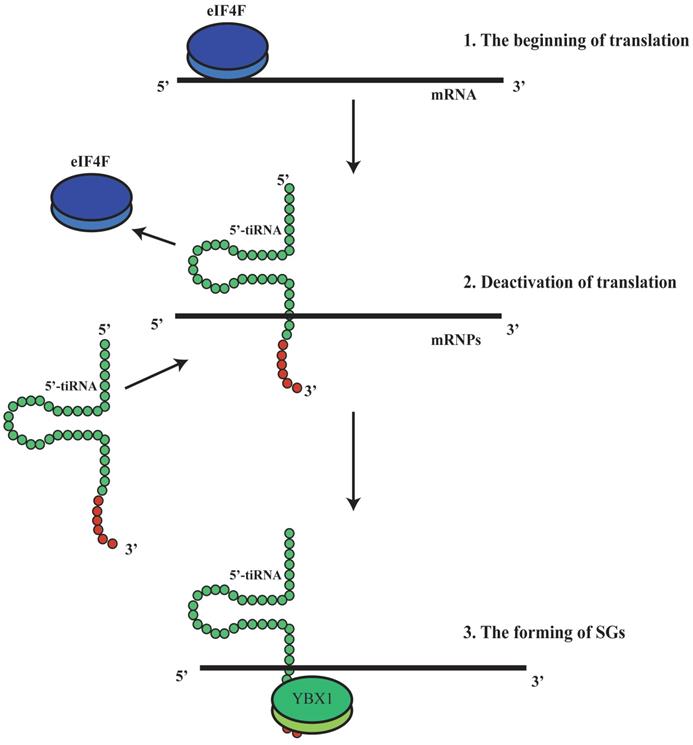
It has been shown that tRFs can regulate ribosomal function. The tRF-3s may specifically bind to TWI12, a member of AGO/PIWI protein family, and recruit TAN1 protein and exoribonuclease XRN2 to form pre-ribosomal RNA splicing complex (TXT), process pre-rRNA during rRNA synthesis, and then regulate translation(41). TRF-3 (e.g. 3'-tRNA-LeuCAG) may bind to two ribosomal protein mRNAs (RPS28 and RPS15) to stimulate translation, ribosomal synthesis and cellular proliferation(10) (Figure 4). However, tiRNAs have not been reported to regulate ribosomal synthesis(10, 11, 17, 41). Some tRFs and tiRNAs may directly regulate cellular translation(33, 42-44). Recent studies have demonstrated that 5'-tiRNAs (5'-tiRNA-Ala and 5'-tiRNA-Cys) contain 5'-terminal oligoguanine motifs (5'-TOGs) and may replace eIF4F, a eukaryotic translation initiation factor, at mRNA m7GTP position to inhibit translation initiation and produce multiple mRNA protein complexes (mRNPs)(45). These tiRNAs may further bind to the cold shock domain (CSD) of YBX-1 RNA binding protein to form 5'-TOG-tiRNA-protein complexes and then stimulate the production of stress granules (SGs)(6, 31, 37, 38, 45, 46) (Figure 5). 5'-tiRNAs may induce SGs assembling via phosphorylated eiF2α(6, 10, 11, 17, 38, 39).
Another interesting phenomenon was found that the functional pattern was distinct from that of canonical miRNA. TRF5-GluCTC, a type of 5'-tRFs, plays a gene-silencing role on the target mRNA with complementary target sequence. Regulation studies showed the 5'-portion of miRNAs is the key determinant in target recognition, while this suggested the 3'-portion of tRF5-GluCTC is critical for its gene silencing function through a trans-silencing mechanism (3).
Compared with miRNAs and piRNAs, tRFs and tiRNAs may directly regulate cellular translation, suggesting that these small RNA molecules have more complicated regulatory potentials to maintain various biological function and thus more important roles.
5'-tiRNA could promotethe formation of stress granules (SGs). 5'-tiRNA may replace eIF4F, which is an eukaryotic translation initiation factor, at the mRNA m7GTP position, to inhibit translation initiation and produce multiple mRNA protein complexes (mRNPs).Those tiRNA may further bind to the cold shock domain (CSD) of the YBX-1 protein (RNA binding protein) to form 5'-TOG-tiRNA-protein complex and then stimulate production of the stress granules (SGs). SGs, stress granules; YBX-1, Y-Box Binding Protein 1; mRNPs, mRNA protein complexes.
Roles in digestive tract (DT) tumors
Roles of tiRNAs and tRFs in the cellular biology of DT tumor
TRFs and tiRNAs have important roles in tumor development and progression. However, the study on the correlation between tRFs/tiRNAs and tumor remains at an early stage. The expression levels of tRFs and tiRNAs were suggested to be abnormal in many tumor cells(47, 48) and correlated with tumor cell proliferation, migration and transformation(6, 48). Recent studies on DT tumor (rectal cancer) showed that tRF/miR-1280 expression levels were decreased(5). Furthermore, tRF/miR-1280 inhibits the 3' UTR of JAG2, reducing JAG2 biosynthesis to inhibit the Notch pathway and directly inhibit proliferation, migration and epithelial-mesenchymal transformation (EMT) of rectal cancer cells(5, 49, 50). TRF/miR-1280 also decreases the expression of CD133+stem cell marker in CRC cells, reduces their activity and migration, and prevents formation of the microenvironment of rectal cancer cell metastasis(5). Moreover, Ts-53 and ts-101 might have important roles in the early transformation stage of rectal cancer(48). Ts-40 might be oncogenic during rectal cancer development(48). Ts-36, correlated with cellular proliferation, may exert roles in the final stage of metastatic transformation of rectal cancer(48). A research applying tiRNA sequencing was performed in 30 HCC patients (including 10 with HBV and 20 with HCV) and 9 controls, and tiRNAs were found to be the most abundant small RNAs in chronically infected liver and the abundance of 5'- tiRNAs was reduced in liver cancer. In addition, in hepatitis B-associated HCC, 5'- tiRNAs abundance was correlated with the expression of tRNA-cleaving ribonuclease, angiogenin (51). Therefore, tRFs and tiRNAs are critical for the biological behavior of DT tumor cells. In-depth investigations would benefit understanding how to exploit these molecules to interfere with tumor progression and how to select molecular targets for therapy.
Correlation between tiRNAs/tRFs and the clinicopathological characteristics of DT tumor
In clinical case, distant metastasis of DT tumor is a major obstacle to optimal patient prognosis(52). Thus, researchers have been exploring the correlation of tiRNAs/tRFs with the cellular invasion and distant metastasis of DT tumor. Recent studies showed that colorectal cancer had elevated ANG levels, increased the levels of 5'-tiRNA-Val, 5'-tiRNA-Cys and 5'-tiRNA-Ala, then promoted cancer cell invasion and migration without affecting cellular proliferation(52). Additionally, 5'-tiRNA-Val levels are positively correlated with lymph node metastasis and distant metastasis of colorectal cancer cells(52). However, the mechanism remains unclear. Further investigations would benefit the early detection of distant tumor metastasis, providing new ideas for the treatment of metastatic tumors and improving prognosis.
Correlation between tiRNAs/tRFs and DT tumor diagnosis
Seminal exosomes were found to contain more tRFs than miRNAs, suggesting that tRFs could be stably expressed and were abundant in body fluids(53). That makes them to be potential markers for early tumor diagnosis. Recent studies showed that 5'tRF-GluCTC and 5'tRF-ValCAC levels were decreased in colorectal cancer cells, indicating that they were involved in the development and progression of colorectal cancer and could serve as potential diagnostic markers(2). Moreover, tiRNA-5034-GluTTC-2 was significantly decreased in gastric cancer (GC) tissue, neoplasm and GC cells, and low levels of tiRNA-5034-GluTTC-2 in GC tissue were correlated with tumor size(54). TiRNA-5034-GluTTC-2 in GC tissue had higher sensitivity and specificity with the large area under receiver operating characteristics (ROC) curve as a tumor marker. All these findings were statistically significant, suggesting its potential as a marker of early GC(54). Recently, it has been shown that the expression of tsRNAs in plasma exosomes of liver cancer patients is significantly higher than that in healthy controls (55). Four tsRNAs were up-regulated including tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5 and tRNA-GluCTC-5, suggesting that these differentially expressed tsRNAs have the potential to be novel diagnostic markers of liver cancer (55). Exploration on the correlation between tRFs/tiRNAs and DT cancer development would benefit identifying markers for the early diagnosis of DT cancer and optimize detection.
Correlation of tiRNAs/tRFs with DT tumor treatment and prognosis
Investigating the role of tiRNAs/tRFs in tumor progression would improve treatment protocols and disease prognosis. Previous reports demonstrated that elevated ANG in rectal cancer increased 5'-tiRNA-Val levels, and thus established an ANG-tiRNAs-cell invasion/metastasis regulatory axis(52). Such findings provided insights on the mechanism of rectal cancer development and revealed new therapeutic targets for colorectal cancer. TRF/miR-1280 could inhibit the Notch pathway and subsequent EMT and distant metastasis of colorectal cancer(5), suggesting that it could affect cancer prognosis. Quantitative PCR (qPCR) illustrated that tRFs and tiRNAs (tRF-3-LeuAAG-1-1[AS-tDR-000064], tRF-3-GlnCTG-1-1[AS-tDR-000069], tRF-3-AlaCGC-1-1[AS-tDR-000102] and tiRNA-5-ProCGG-1-1[AS-tDR-001391]) had abnormally elevated expression in cancerous pancreatic cells, but not normal pancreatic cells(56). KEGG and GO pathway analyses showed that all the four tRFs and tiRNAs were significantly enriched in cancer-related pathways (including RAS signaling pathway, cancer pathways, axon guidance and PI3K/AKT signaling pathway). Therefore, they might be able to serve as markers for early pancreatic cancer diagnosis and provide clues for pancreatic cancer development(56). Furthermore, the survival rate of patients with low expression of tiRNA-5034-GluTTC-2 was much higher than that of individuals with higher expression levels, indicating that tiRNA-5034-GluTTC-2 could be a potential prognostic marker of GC(54). All the findings suggested that tRFs and tiRNAs were crucial in DT cancer treatment and prognosis.
Summary and future directions
Several limitations should be acknowledged. First, although the sequencing has identified many tRFs and tiRNAs, bioinformatic methodology needs to be improved for more identification. Second, current study only focused on the basic biological function of tRFs and tiRNAs without the correlations with diseases, especially DT tumor. Third, the naming roles of molecules remain not unified. Fourth, we urgently need a tiRNA database and a database to integrate the tRFs (tRFdb) and tiRNAs.
In summary, tRFs and tiRNAs have important roles in gene expression, gene translation, and the development and progression of various DT tumors(17). They affect tumor cell proliferation, migration and invasion(48, 57). To date, the origin and function of tRFs and tiRNAs remain to be fully elucidated, and their roles in DT tumor progression also require further investigation. In-depth studies would benefit early detection, prognosis evaluation and DT tumor therapy, further improving prognosis and survival.
Acknowledgements
We thank H. Nikki March, PhD, from LiwenBianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author Contributions
Dr. Xin-ping Zhong conceived and designed this study. Dr. Ben-gang Wang and Dr. Li-rong Yan wrote the paper, Dr. Qian Xu and Xin-ping Zhong revised the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lorenz C, Lunse CE, Morl M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017;7(2):35
2. Soares AR, Fernandes N, Reverendo M, Araujo HR, Oliveira JL, Moura GM. et al. Conserved and highly expressed tRNA derived fragments in zebrafish. BMC molecular biology. 2015;16:22
3. Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Molecular therapy: the journal of the American Society of Gene Therapy. 2013;21(2):368-79
4. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639-49
5. Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W. et al. tRF/miR-1280 Suppresses Stem Cell-like Cells and Metastasis in Colorectal Cancer. Cancer Res. 2017;77(12):3194-206
6. Sun C, Fu Z, Wang S, Li J, Li Y, Zhang Y. et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16-25
7. Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA-novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340(2):201-11
8. Mleczko AM, Celichowski P, Bąkowska-Żywicka K. Ex-translational function of tRNAs and their fragments in cancer. Acta biochimica Polonica. 2014;61(2):211-6
9. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1404-9
10. Li S, Xu Z, Sheng J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes (Basel). 2018;9(5):246
11. Shen Y, Yu X, Zhu L, Li T, Yan Z, Guo J. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med (Berl). 2018;96(11):1167-76
12. Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43(Database issue):D141-5
13. Veneziano D, Di Bella S, Nigita G, Lagana A, Ferro A, Croce CM. Noncoding RNA: Current Deep Sequencing Data Analysis Approaches and Challenges. Hum Mutat. 2016;37(12):1283-98
14. Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG. et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157(3):651-63
15. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161(4):790-802
16. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78
17. Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci. 2016;41(8):679-89
18. Chen H, Xu Z, Liu D. Small non-coding RNA and colorectal cancer. J Cell Mol Med. 2019;23(5):3050-7
19. Li S, Chen Y, Sun D, Bai R, Gao X, Yang Y. et al. Angiogenin Prevents Progranulin A9D Mutation-Induced Neuronal-Like Cell Apoptosis Through Cleaving tRNAs into tiRNAs. Mol Neurobiol. 2018;55(2):1338-51
20. Li F, Kaczor-Urbanowicz KE, Sun J, Majem B, Lo HC, Kim Y. et al. Characterization of Human Salivary Extracellular RNA by Next-generation Sequencing. Clin Chem. 2018;64(7):1085-95
21. Zhu L, Ge J, Li T, Shen Y, Guo J. tRNA-derived fragments and tRNA halves: The new players in cancers. Cancer Lett. 2019;452:31-7
22. Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15(10):1015-23
23. Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW. et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15(12):2147-60
24. Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY. et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151(4):2120-32
25. Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y. et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8(1):158-77
26. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673-95
27. Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S. et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157
28. Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37(19):6575-86
29. Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24(24):2742-7
30. Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-Derived Small RNAs in Cancer: New Potential Biomarkers and Target for Therapy. Adv Cancer Res. 2017;135:173-87
31. Soares AR, Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA. 2017;8(5). [Epub ahead of print. 2017 Jun 12]
32. Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N. et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959-68
33. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613-23
34. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552(7683):57-62
35. Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G. et al. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37(5):668-78
36. Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH. et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34(13):2450-63
37. Saikia M, Hatzoglou M. The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health. J Biol Chem. 2015;290(50):29761-8
38. Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020;235(2):683-690
39. Green D, Fraser WD, Dalmay T. Transfer RNA-derived small RNAs in the cancer transcriptome. Pflugers Arch. 2016;468(6):1041-7
40. Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct. 2015;10:51
41. Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3' fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell. 2012;48(4):509-20
42. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35-42
43. Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T. et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Molecular therapy: the journal of the American Society of Gene Therapy. 2015;23(10):1622-9
44. Sobala A, Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10(4):553-63
45. Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016;44(14):6949-60
46. Suresh PS, Tsutsumi R, Venkatesh T. YBX1 at the crossroads of non-coding transcriptome, exosomal, and cytoplasmic granular signaling. Eur J Cell Biol. 2018;97(3):163-7
47. Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7(17):24766-77
48. Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL. et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114(30):8071-6
49. Van de Walle I, De Smet G, Gartner M, De Smedt M, Waegemans E, Vandekerckhove B. et al. Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions. Blood. 2011;117(17):4449-59
50. Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N. et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121(4):1373-85
51. Selitsky SR, Baran-Gale J, Honda M, Yamane D, Masaki T, Fannin EE. et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep. 2015;5:7675
52. Li S, Shi X, Chen M, Xu N, Sun D, Bai R. et al. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer. 2019;145(5):1395-407
53. Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP. et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42(11):7290-304
54. Zhu L, Li T, Shen Y, Yu X, Xiao B, Guo J. Using tRNA halves as novel biomarkers for the diagnosis of gastric cancer. Cancer Biomark. 2019;25(2):169-76
55. Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D. et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Molecular cancer. 2019;18(1):74
56. Jin L, Zhu C, Qin X. Expression profile of tRNA-derived fragments in pancreatic cancer. Oncol Lett. 2019;18(3):3104-14
57. Mo D, Jiang P, Yang Y, Mao X, Tan X, Tang X. et al. A tRNA fragment, 5'-tiRNA(Val), suppresses the Wnt/beta-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019;457:60-73
Author contact
![]() Corresponding authors: Dr. Xin-ping Zhong, Department 1 of General Surgery, the First Hospital of China Medical University, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83283308; fax: +86-024-83282383. Email: bgwangedu.cn;xpzsubmcom. Dr. Qian Xu, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83282153; fax: +86-024-83282383. Email: qxuedu.cn.
Corresponding authors: Dr. Xin-ping Zhong, Department 1 of General Surgery, the First Hospital of China Medical University, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83283308; fax: +86-024-83282383. Email: bgwangedu.cn;xpzsubmcom. Dr. Qian Xu, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83282153; fax: +86-024-83282383. Email: qxuedu.cn.

 Global reach, higher impact
Global reach, higher impact