Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(24):7276-7282. doi:10.7150/jca.46086 This issue Cite
Research Paper
Risk Stratification Study of Indeterminate Thyroid Nodules with a next-generation Sequencing Assay with Residual ThinPrep® Material
1. Department of Pathology, National Cancer Center/ National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
2. Department of Ultrasound, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
*These authors contributed equally to this work.
Received 2020-3-7; Accepted 2020-10-4; Published 2020-10-21
Abstract
Objective: The management of indeterminate thyroid nodules is challenging. Molecular testing has emerged as a promising method for stratifying this gray area of fine-needle aspiration (FNA) cytology. Next-generation sequencing (NGS) can be used to test a large variety of genetic changes with very small amounts of nucleic acids obtained from FNA samples.
Methods: Thyroid FNA assays were classified according to the Bethesda System for Reporting Thyroid Cytopathology after routine ThinPrep® slide preparation. Indeterminate nodules with surgical outcomes were assayed with an 18-gene NGS panel with the residual ThinPrep® material, including nodules categorized as atypia of undetermined significance (AUS)/follicular lesions of undetermined significance (FLUS) or follicular neoplasm (FN)/suspicious for a follicular neoplasm (SFN). We evaluated the diagnostic efficacy of the 18-gene panel for thyroid malignancies and potential malignancies and compared it with a well-accepted examination, ThyroSeq v2 testing.
Results: A total of 36 indeterminate nodules were assayed, seven were categorized as AUS/FLUS and 29 as FN/SFN. All of them had adequate DNA for the NGS procedure. When noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) was considered malignant, the risk of malignancy was 71.4% for AUS/FLUS nodules, and 69.0%for FN/SFN nodules. The 18-gene panel showed 72.0% sensitivity, 72.7% specificity, 85.7% positive predictive value (PPV), and 53.3% negative predictive value (NPV) in identifying malignancies and potential malignancies in the indeterminate nodules. Compared with a multicenter report from ThyroSeq v2 testing, 18-gene panel showed a lower NPV (p=0.005), but a higher PPV (p=0.02).
Conclusions: NGS assays are feasible on residual ThinPrep® material, with the advantage of not requiring additional FNA procedure. The 18-gene panel testing can be used as a 'rule in' test for surgical management based on indeterminate nodules and showed a lower NPV but a higher PPV compared to ThyroSeq v2 testing.
Keywords: fine-needle aspiration, cytology, thyroid nodules, thyroid cancer, molecular diagnosis
Introduction
With the wide application of thyroid ultrasound in physical examinations, thyroid cancer has become the fastest growing type of cancer identified throughout the world, including on the Chinese mainland [1]. Fine-needle aspiration (FNA) is the most effective diagnostic method for thyroid cancer. FNA allows the diagnosis of cancer or a benign nodule in most patients, although about 20% of FNA samples yield an indeterminate diagnosis [2]. These indeterminate nodules include two subcategories of cytological diagnosis: atypia of undetermined significance (AUS)/follicular lesion of undetermined significance (FLUS) and follicular neoplasm (FN)/suspicious for a follicular neoplasm (SFN) [3]. A predictor of indeterminate nodules that may place certain nodule types at higher malignancy rates is required.
Molecular testing is recommended by the 2015 American Thyroid Association guidelines as an adjunct technique to further stratify the risk of cytologically indeterminate nodules [4]. To date, various commercial molecular tests, such as Afirma, ThyroSeq, or ThyGen X, have been approved by the U.S. Food and Drug Administration to evaluate cytologically indeterminate thyroid nodules [5]. However, there is currently no molecular test that can definitively rule malignancy either in or out. More molecular data on indeterminate nodules are required. Next-generation sequencing (NGS) can be used to test a large variety of genetic changes simultaneously with a very small amount of nucleic acids, and allows multiple genes to be tested in FNA samples.
In this study, we used an 18-gene panel based on the NGS technology to test cytologically indeterminate nodules with residual ThinPrep® material. We evaluated the risk of malignancy in patients with a positive mutation detected with the 18-gene panel, and also compared the risk stratification achieved with the 18-gene panel to ThyroSeq v2 testing, which was also based on NGS platform and has been well-accepted.
Materials and Methods
Case selection
The thyroid FNA samples were collected from the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College between November 2017 and June 2019 and analyzed retrospectively. The patients were selected on the basis of the following criteria: 1) cytological diagnosis of AUS/FLUS or FN/SFN according to the Bethesda System for Reporting Thyroid Cytopathology (TBSRTC) [3]; 2) they had undergone thyroid surgery and had correlated cytological-histological results; and 3) an adequate residual specimen was available for DNA extraction after the routine cytological diagnosis. The cytological-histological correlation was performed by matching the locations and sizes of nodules in both the ultrasound and pathology reports. All the patients gave their informed consent before FNA. This study protocol was reviewed and approved by the Ethics Committee of the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital.
Specimen preparation
All FNA biopsies were performed under ultrasound guidance by radiologists. The aspirates were rinsed into a vial of CytoLyt® (Hologic Inc., Marlborough, MA, USA) and prepared as slides with ThinPrep® 2000 (Hologic Inc.). The slides were fixed in 95% alcohol and stained with Papanicolaou stain. They were then interpreted by two cytopathologists with experience ranging from 14 to 19 years. The residues were collected for DNA extraction. The residue selection criterion was defined as ten groups of cells on a slide in 10 ml of PreservCyt® solution [6]. The liquid materials were stored at -20 °C and used for molecular testing within 3 months.
DNA extraction
After centrifugation, the cells were incubated in 500 μl of DNA lysis solution (1 mg/ml proteinase K, 10 mmol/l Tris-HCl (pH 8.0), 0.1 mol/l EDTA (pH 8.0), 0.5% (w/v) SDS) at 55 °C for approximately 12 h. The DNA was then extracted with the phenol-chloroform method and stored at -20 °C for future use. The concentration and purity of the DNA were measured with a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA) spectrophotometer.
Targeted DNA sequencing
Targeted DNA sequencing was performed for all patients with available DNA. The DNA was profiled with a capture-based targeted sequencing panel (Burning Rock Biotech, Guangzhou, People's Republic of China) that targets 18 genes (BRAF, NRAS, HRAS, KRAS, RET, NTRK1, ETV6, ALK, PPARG, TERT, EIF1AX, PTEN, AKT1, PIK3CA, TP53, CTNNB1, TSHR, and GNAS) and spans 140 kb of the human genome. In this way, we detected all single-nucleotide variants in these 18 genes and any gene fusions involving RET, NTRK1, ETV6, ALK, and PPARG. The design of 18-gene panel is based on data from public database and previous study in histology specimens [7,8]. bioanalyzer high-sensitivity DNA assay was performed to assess the quality and size of the fragments. The available indexed samples were sequenced on a NextSeq 500 sequencer (Illumina Inc., San Diego, CA, USA]) as pair-end reads.
Sequence data analysis
The sequence data were aligned to the human genome (hg19) with Burrows-Wheeler Aligner 0.7.10. Local alignment optimization and variant calling were performed with GATK v3.2-2. Both TopHat2 and Factera 1.4.3 were used for the DNA translocation analysis. To assess the level of DNA degradation, the insert size distribution and library complexity of each sample were computed. To avoid false positive mutation calls arising from DNA damage, different mutation calling thresholds were applied to DNA samples of different quality. Variants with population frequencies > 0.1% in the ExAC, 1000 Genomes, dbSNP, and ESP6500SI-V2 databases were grouped as common single-nucleotide polymorphisms and removed. Integrative Genomics Viewer (Broad Institute, USA) was used to visualize the variants aligned against the reference genome to confirm the accuracy of the variant calls by checking for possible strand bias and sequencing errors. Copy number variation was assessed by normalizing the read depth in each region to the total read number and region size, and correcting for GC bias using the LOESS algorithm.
Statistical analysis
The cytological and molecular results were correlated with the histopathological results. A χ2 test was used to assess the differences in categorical variables. All statistical analyses were performed with SPSS 17.0, and p < 0.05 was considered statistically significant.
Results
Baseline characteristics of patients and nodules
Between November 2017 and June 2019, 434 thyroid nodules showed indeterminate cytology. Thirty-six indeterminate nodules with correlated surgical outcomes and residual ThinPrep® materials were enrolled. The patient and nodule characteristics are listed in Table 1. The mean age of patients was 49 years, and the ratio of females to males was 3:1. The median nodule size was 1.3 cm. Surgical pathology showed that 23 nodules were malignant, and the follicular variant of papillary thyroid carcinoma was the commonest malignant histopathology (47.8%). Benign pathologies included two adenomas and nine nodular hyperplasias. Two nodules were classified as noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP).
Baseline characteristics of patients and nodules
| Age, Mean (SD) | 49 (13) |
|---|---|
| Sex ratio, F:M | 3:1 |
| Size (median) | 1.3 cm |
| Histopathological diagnosis of study population (n=36) | |
| Benign (n=11) | 11 (30.6%) |
| Nodular hyperplasia / with adenomatous hyperplasia | 9 (81.8%) |
| Adenoma | 2 (18.2%) |
| Malignant | 23 (63.9%) |
| PTC-classical | 1 (4.3%) |
| PTC-classial and follicular variants | 2 (8.7%) |
| PTC-follicular variants | 11 (47.8%) |
| PTC-cribriform-morular variants | 1 (4.3%) |
| PTC-oncocytic variant | 1 (4.3) |
| Follicular thyroid carcinoma | 6 (26.1%) |
| Hürthle cell carcinoma | 1 (4.3%) |
| NIFTP | 2 (5.5%) |
SD, standard deviation; NIFTP, non-invasive follicular thyroid neoplasm with papillary-like nuclear features; PTC, papillary thyroid carcinoma.
Risk of malignancy in indeterminate nodules
Among the 36 indeterminate nodules, seven were categorized as AUS/FLUS and 29 as FN/SFN. The ROM was 71.4% in the AUS/FLUS nodules and 69.0% in the FN/SFN nodules when NIFTP was considered malignant. If NIFTP was classified as benign, the prevalence of malignancy was 71.4% and 62.1%, respectively (Table 2).
Risk of malignancy and histopathology outcomes in indeterminate cytologic diagnoses
| Cytologic diagnosis | Benign | NIFTP | Malignancy | ROMa | ROMb |
|---|---|---|---|---|---|
| AUS/FLUS | 2 | 0 | 5 | 71.4% | 71.4% |
| FN/SFN | 9 | 2 | 18 | 69.0% | 62.1% |
AUS/FLUS, atypia of undetermined significance/follicular lesion of undetermined significance; FN/SFN, follicular neoplasm/suspicious for a follicular neoplasm; NIFTP, non-invasive follicular thyroid neoplasm with papillary-like nuclear features; ROM, risk of malignancy; a NIFTP considered malignant; b NIFTP considered benign.
Diagnostic capacity of 18-gene panel and comparison with ThyroSeq v2 assay
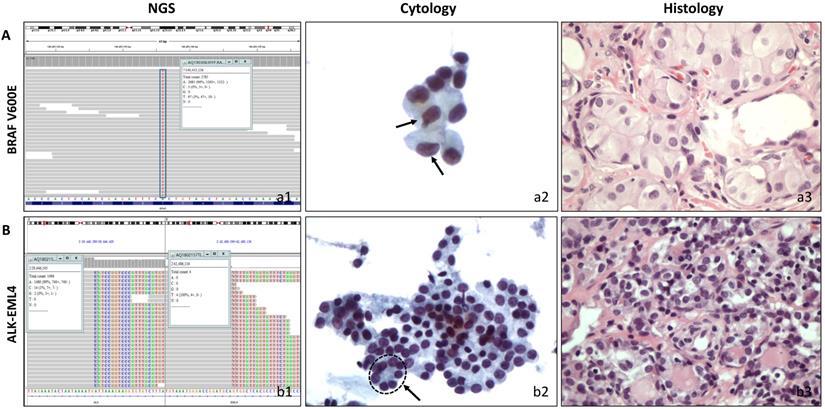
The performance characteristics of the 18-gene panel testing for the diagnosis of thyroid malignancy including their sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) are shown in Table 3. When NIFTP was considered malignant, the 18-gene panel had 72.0% sensitivity, 72.7% specificity, 85.7% PPV, and 53.3% NPV. If NIFTP was considered benign, the PPV decreased to 76.2%, NPV maintained a value of 53.3%. Table 3 also shows the comparison between our 18-gene panel and the previous reports from ThyroSeq v2. A statistical analysis was performed between present study and a multicenter report from ThyroSeq v2 in 2019 and 18-gene panel showed a lower NPV (p=0.005), but a higher PPV (p=0.02) with ThyroSeq v2. Two representative cases with positive molecular results are shown in Figure 1.
Correlation of cytological and histological morphology and molecular results. A, A follicular variant of papillary thyroid carcinoma with cytological diagnosis of AUS/FLUS. (a1) Molecular testing showed BRAF mutation. (a2) Cytopathology, a low cellular specimen composed of cells with enlarged nuclei. Two cells showed slightly irregular nuclei (arrow). The atypia cells were rare and insufficient for a diagnosis of suspicious malignant. (ThinPrep®, Papanicolaou stain, 400). (a3). Histopathology, thyroidectomy section (HE stain, 400). B, A follicular variant of thyroid papillary carcinoma with cytological diagnosis of FN/SFN. (b1) Molecular testing showed ALK/EML4 fusion. (b2) Cytopathology, a moderate cellular smear composed of uniform follicular cells with microfollicular arrangement (circle with arrow). (ThinPrep®, Papanicolaou stain, 400) (b3). Histopathology, thyroidectomy section (HE stain, 400).
Comparison of published experiences with ThyroSeq v2 assay to the present study
| Author | Panel | Material | Cytologic category | No. of surgery | Diagnostic performance | |
|---|---|---|---|---|---|---|
| Present study | 18-gene | Residual liquid-based FNA sample | AUF/FLUS and FN/SFN | 36 | NIFTP = malignant | NIFTP = benign |
| SN = 72.0% | SN = 69.6% | |||||
| SP = 72.7% | SP = 61.5% | |||||
| PPV = 85.7% | PPV = 76.2% | |||||
| NPV = 53.3% | NPV = 53.3% | |||||
| Nikiforov [2014] | ThyroSeq v2 | 1dedicated FNA | FN/SFN | 143 | NIFTP = malignant | |
| SN = 90% | ||||||
| SP = 93% | ||||||
| PPV = 83% | ||||||
| NPV = 96% | ||||||
| Valderrabano [2017] | ThyroSeq v2 | 1 dedicated FNA | AUF/FLUS and FN/SFN | 102 | NIFTP = malignant | NIFTP = benign |
| SN = 70% | SN = 73% | |||||
| SP = 77% | SP = 75% | |||||
| PPV = 42% | PPV = 33% | |||||
| NPV = 91% | NPV = 94% | |||||
| Taye [2017] | ThyroSeq v2 | 1 dedicated FNA | AUS/FLUS and FN/SFN | 60 | NIFTP = malignant | |
| SN = 89% | ||||||
| SP = 43% | ||||||
| PPV = 22% | ||||||
| NPV = 96% | ||||||
| Marcadis [2019] (multicenter) | ThyroSeq v2 | 1 dedicated FNA | AUS/FLUS and FN/SFN | 273 | NIFTP = malignant | NIFTP = benign |
| SN = 85% | SN = 87% | |||||
| SP = 62% | SP = 52% | |||||
| PPV = 59% | PPV = 35% | |||||
| NPV = 86% | NPV = 93% | |||||
AUS/FLUS, atypia of undetermined significance/follicular lesion of undetermined significance; FN/SFN, follicular neoplasm/suspicious for a follicular neoplasm; SN, sensitivity; SP, specificity; PPV, positive predictive value; NPV, negative predictive value; NIFTP, non-invasive follicular thyroid neoplasm with papillary-like nuclear features; FNA, fine needle aspiration.
Detected gene changes and the associated risk of malignancy
| Detected gene changes | Positive | ROMa | ROMb |
|---|---|---|---|
| Mutations | |||
| BRAF V600E | 2 | 100% (2/2) | 100% (2/2) |
| NRAS | 7 | 71% (5/7) | 57% (4/7) |
| NRAS Q61K | 3 | 67% (2/3) | 67% (2/3) |
| NRAS Q61R | 3 | 67% (2/3) | 33% (1/3) |
| NRAS G13R | 1 | 100% (1/1) | 100% (1/1) |
| HRAS | 1 | 100% (1/1) | 100% (1/1) |
| HRAS Q61R | 1 | 100% (1/1) | 100% (1/1) |
| TERT promoter | 1 | 100% (1/1) | 100% (1/1) |
| PTEN | 2 | 0% (0/2) | 0% (0/2) |
| PTEN A126D | 1 | 0% (0/1) | 0% (0/1) |
| PTEN K144 | 1 | 0% (0/1) | 0% (0/1) |
| AKT1 E17K | 1 | 100% (1/1) | 100% (1/1) |
| RET R475W | 2 | 100% (2/2) | 100% (2/2) |
| PIK3CA H1047R | 1 | 100% (1/1) | 100% (1/1) |
| CTNNB1 S33C | 1 | 100% (1/1) | 100% (1/1) |
| GNAS | 4 | 100% (4/4) | 100% (4/4) |
| GNAS D448A | 1 | 100% (1/1) | 100% (1/1) |
| GNAS S455A | 1 | 100% (1/1) | 100% (1/1) |
| GNAS R844G | 1 | 100% (1/1) | 100% (1/1) |
| GNAS A368T | 1 | 100% (1/1) | 100% (1/1) |
| Fusions | |||
| NCOA4/RET | 2 | 100% (2/2) | 100% (2/2) |
| ETV6/NTRK3 | 3 | 100% (3/3) | 100% (3/3) |
| ELM4/ALK | 2 | 100% (2/2) | 50% (1/2) |
| CNTN4/ALK | 1 | 100% (1/1) | 0% (0/1) |
ROM, risk of malignancy; a NIFTP considered malignant; b NIFTP considered benign.
Gene changes detected and the associated risk of malignancy
Table 4 shows the gene changes detected with the 18-gene panel and the associated risk of malignancy. Among the gene mutations, the most commonly affected gene was RAS (HRAS Q61R, NRAS Q61K, Q61R, or G13R), which was mutated in eight nodules, followed by GNAS (D448A, S455A, R844G, or A368T), which was mutated in three nodules (GNAS D448A and S455A were mutated simultaneously in one nodule). The identified gene fusions involved RET (RET/NCOA4) and ALK (ALK/ELM4 or ALK/CNTN4) in two nodules, ETV6 (ETV6/NTRK3) in three nodules. The ALK/ELM4 and ALK/CNTN4 fusions were encountered simultaneously in one NIFTP nodule. Another NIFTP nodule analyzed in our series carried the NRAS Q61R mutation. When NIFTP was considered malignant, mutations BRAFV600E, HRAS Q61R, NRAS G13R, the TERT promoter, AKT1 E17K, RET R475W, PIK3CA H1047R, CTNNBI S33C, or GNAS (D448A, S455A, R844G or A368T) and a fusion of RET/NCOA4, ALK/ELM4, ALK/CNTN4or ETV6/NTRK3 all carried a 100% risk of malignancy. However, mutations NRAS (Q61R, Q61K) and PTEN (A126D, K144) were not specifically associated with malignancy or potential malignancy. The malignancy rates (NIFTP considered malignant) for the NRAS and PTEN genes were 71% and 0%, respectively.
Discussion
Over the past decade, molecular testing has emerged as a promising method for stratifying indeterminate thyroid FNAs. Several molecular testing panels, such as the Afirma Gene Expression Classifier, ThyroSeq v2, and ThyGenX/ThyraMIR, are commercially available in the USA [5, 9]. However, none of them has been approved or is available in mainland China. Ultrasound characteristics and clinical features are currently the main criteria used to stratify indeterminate nodules. The decision to operate is rarely made with reference to molecular results.
We used NGS that targeted 18 genes to retrospectively analyze 36 cytologically indeterminate samples of thyroid lesions. All the samples were diagnosed as AUS/FLUS (7 cases) or FN/SFN (29 cases) with cytology, and the patients had undergone surgery based on their clinical and ultrasound features. The molecular analysis was performed after surgery with the residual liquid cytology samples after routine ThinPrep® slide preparation, which had been stored at -20 °C. To guarantee enough DNA for analysis, the selection criterion for the residues was defined as ten groups of cells on the slide in 10 ml of PreservCyt solution, as in our previous study [6]. The DNA quality of all 36 samples fulfilled the requirements for NGS. Molecular testing based on residual ThinPrep® material has the advantage of not requiring an additional FNA.
In this study, the rates of malignancy for AUS/FLUS regardless of whether NIFTP was considered malignant or not were both 71.4%. The ROM of the FN/SFN nodules was 69.0% when NIFTP was considered malignant and fell to 62.1% when NIFTP was reclassified as benign. ROM was higher than the idealized ROMs described by TBSRTC [10]. This discrepancy is mainly attributable to the surgical cohort selected. Our hospital is the national cancer center of China. The experiences of surgeons and radiologist make this selection more effective. The various differences in ROMs described in the present and previous studies emphasize the need for surgeons to understand their individual data, rather than rely on TBSRTC predictions [11-13].
When NIFTP was considered malignant, 18-gene panel showed 85.7% PPV and 53.3% NPV for the diagnosis of thyroid malignancy in indeterminate nodules. If NIFTP was considered benign, PPV decreased to 76.2%, NPV maintained a value of 53.3%. As previously reported, we considered NIFTP malignant when evaluating molecular tests in clinical practice because the recommended treatment fort NIFTP is surgical excision [14,15]. Our 18-gene panel showed moderate NPV and high PPV when NIFTP was considered malignant, and may serve as a 'rule in' test for surgery.
Our 18-gene panel involves next-generation sequencing that detects gene mutations and fusions. The design of our panel is similar to a well-accepted test designated 'ThyroSeq v2', which was initially suggested to be both a “rule in” and “rule out” test because both its PPV (83%) and NPV (96%) were high [16]. However, validation in the real world has suggested that its PPV may be lower than initially reported [17-19]. Recently, a multicenter study reported a PPV of 59% and an NPV of 86% when NIFTP was considered malignant [19]. Compared with that multicenter study, our 18-gene showed a lower NPV (p = 0.005), but a higher PPV (p = 0.02).
As in previous reports, RAS was the most frequent mutation identified in the indeterminate nodules in this study, and was not specifically associated with malignant or potentially malignant outcomes [20-22]. Another false positive molecular result in our study was the mutation of PTEN. Two nodules with mutations in PTEN were both shown to be hyperplasia nodules. False PTEN mutations have rarely been reported in other studies [15, 17, 18, 23]. This highlights the need for larger clinical studies to evaluate each mutation individually and to better characterize the risk of malignancy. The surgeon's familiarity with this information will allow more-appropriate clinical practices.
This study was not without limitations. Because residual ThinPrep® FNA samples were collected for molecular testing, more nodules categorized as AUS/FLUS were excluded than those categorized as FN/SFN because there were fewer cells in the AUS/FLUS residues. Fewer samples in AUS/FLUS subcategory than that in FN/SFN subcategory may have weakened the power to demonstrate the diagnostic capacities of the 18-gene test in AUS/FLUS nodules. The small sample size may have also limited our understanding of the malignant risk associated with specific gene changes.
Overall, residual ThinPrep® samples are suitable for NGS, and our 18-gene panel showed high PPV and moderate NPV for malignancy and potential malignancy. Therefore, it can be used as a 'rule in' test for stratifying indeterminate nodules. A lower NPV but a higher PPV was found with the use of 18-gene panel testing compared to the well-accepted ThyroSeq v2 testing.
Abbreviations
FNA: fine-needle aspiration; AUS: atypia of undetermined significance; FLUS: follicular lesions of undetermined significance; FN: follicular neoplasm; SFN: suspicious for a follicular neoplasm; NIFTP: noninvasive follicular thyroid neoplasms with papillary-like nuclear features; PPV: positive predictive value; NPV: negative predictive value; PTC: papillary thyroid carcinoma; TBSRTC: The Bethesda System for Reporting Thyroid Cytopathology; NGS: next-generation sequencing.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant no. 81972804] and Beijing Hope Run Special Fund of the Cancer Foundation of China [grant no. LC2017A12].
Ethics Committee Approval and Patient Consent
This study protocol was reviewed and approved by the Ethics Committee of the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College and all the patients gave their informed consent before FNA.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
2. Bongiovanni M, Spitale A, Faquin WC. et al. The Bethesda system for reporting thyroid cytopathology: a meta-analysis. Acta Cytol. 2012;56:333-9
3. Ali SZ, Cibas ES. The Bethesda system for reporting thyroid cytopathology: definitions, criteria and explanatory notes, 2nd ed. Cham Switzerland: Springer. 2017
4. Haugen BR, Alexander EK, Bible KC. et al. 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the American Thyroid Association Guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1-133
5. Bose S, Sacks W, Walts AE. Update on molecular testing for cytologically indeterminate thyroid nodules. Adv Anat Pathol. 2019;26:114-23
6. Zhao H, Zhang ZH, Zhou B. et al. Detection of BRAF c.1799T > A [p.V600E] mutation using residual routine fine-needle aspiration specimens of papillary thyroid carcinoma. Diagn Cytopathol. 2015;43:786-90
7. Duan H, Li Y, Hu P. et al. Mutational profiling of poorly differentiated and anaplastic thyroid carcinoma by the use of targeted next-generation sequencing. Histopathology. 2019;75:890-9
8. Teng L, Deng W, Lu J. et al. Hobnail variant of papillary thyroid carcinoma: molecular profiling and comparison to classical papillary thyroid carcinoma, poorly differentiated thyroid carcinoma and anaplastic thyroid carcinoma. Oncotarget. 2017;8:22023-33
9. Onenerk AM, Pusztaszeri MP, Canberk S. et al. Triage of the indeterminate thyroid aspirate: What are the options for the practicing cytopathologist? Cancer Cytopathol. 2017;125:477-85
10. Cibas ES, Ali SZ. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid. 2017;27:1341-46
11. Kakudo K, Higuchi M, Hirokawa M. et al. Thyroid FNA cytology in Asian practice-active surveillance for indeterminate thyroid nodules reduces overtreatment of thyroid carcinomas. Cytopathology. 2017;28:455-66
12. Shrestha RT, Evasovich MR, Amin K. et al. Correlation between histological diagnosis and mutational panel testing of thyroid nodules: a two-year institutional experience. Thyroid. 2016;26:1068-76
13. Lim JXY, Nga ME, Chan DKH. et al. Subclassification of Bethesda atypical and follicular neoplasm categories according to nuclear and architectural atypia improves discrimination of thyroid malignancy risk. Thyroid. 2018;28:511-21
14. Haugen BR, Sawka AM, Alexander EK. et al. American Thyroid Association Guidelines on the Management of Thyroid Nodules and Differentiated Thyroid Cancer Task Force Review and Recommendation on the Proposed Renaming of Encapsulated Follicular Variant Papillary Thyroid Carcinoma Without Invasion to Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features. Thyroid. 2017;27:481-3
15. Livhits MJ, Kuo EJ, Leung AM. et al. Gene Expression Classifier vs Targeted Next-Generation Sequencing in the Management of Indeterminate Thyroid Nodules. J Clin Endocrinol Metab. 2018;103:2261-8
16. Nikiforov YE, Carty SE, Chiosea SI. et al. Highly accurate diagnosis of cancer in thyroid nodules with follicular neoplasm/suspicious for a follicular neoplasm cytology by ThyroSeq v2 next-generation sequencing assay. Cancer. 2014;120:3627-34
17. Valderrabano P, Khazai L, Leon ME. et al. Evaluation of ThyroSeq v2 performance in thyroid nodules with indeterminate cytology. Endocr Relat Cancer. 2017;24:127-36
18. Taye A, Gurciullo D, Miles BA. et al. Clinical performance of a next-generation sequencing assay (ThyroSeq v2) in the evaluation of indeterminate thyroid nodules. Surgery. 2018;163:97-103
19. Marcadis AR, Valderrabano P, Ho AS. et al. Interinstitutional variation in predictive value of the ThyroSeq v2 genomic classifier for cytologically indeterminate thyroid nodules. Surgery. 2019;165:17-24
20. Bellevicine C, Sgariglia R, Migliatico I. et al. Different qualifiers of AUS/FLUS thyroid FNA have distinct BRAF, RAS, RET/PTC, and PAX8/PPARg alterations. Cancer Cytopathol. 2018;126:317-25
21. Young NA, Win KK, Pomo L. et al. An academic community hospital experience using commercially available molecular testing in the management of indeterminate thyroid nodules. J Am Soc Cytopathol. 2018;7:92-8
22. Cohen DS, Tongson-Ignacio JE, Lolachi CM. et al. Rethinking malignancy risk in indeterminate thyroid nodules with positive molecular studies: southern california permanente experience. Otolaryngol Head Neck Surg. 2019;161:419-23
23. Nikiforova MN, Wald AI, Roy S. et al. Targeted next-generation sequencing panel [ThyroSeq] for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1852-60
Author contact
![]() Corresponding authors: Huiqin Guo MD, Department of Pathology, National Cancer Center /National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Chaoyang District, Panjiayuan 17 in the South, Beijing, 100021, China. Tel: +86 10-8778-8549, Fax: +86 10-6771-3648. E-mail: guohuiqinac.cn; Yong Wang MD, Department of Ultrasound, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. Chaoyang District, Panjiayuan 17 in the South, Beijing, 100021, China. Tel: +86 10-8778-8551, Fax: +86 10-6771-3648, E-mail: drwangyong77com.
Corresponding authors: Huiqin Guo MD, Department of Pathology, National Cancer Center /National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Chaoyang District, Panjiayuan 17 in the South, Beijing, 100021, China. Tel: +86 10-8778-8549, Fax: +86 10-6771-3648. E-mail: guohuiqinac.cn; Yong Wang MD, Department of Ultrasound, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. Chaoyang District, Panjiayuan 17 in the South, Beijing, 100021, China. Tel: +86 10-8778-8551, Fax: +86 10-6771-3648, E-mail: drwangyong77com.

 Global reach, higher impact
Global reach, higher impact