Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(1):198-206. doi:10.7150/jca.48769 This issue Cite
Research Paper
Emerging roles of MiR-133a in human cancers
1. Department of Gastroenterology, Wuxi People's Hospital Affiliated to Nanjing Medical University, 299 Qingyang Road, Wuxi, Jiangsu 214023, China.
2. Department of General Surgery, the First Affiliated Hospital of Nanjing Medical University, Nanjing 210029, P.R. China.
#Equal contributors.
Received 2020-5-27; Accepted 2020-10-23; Published 2021-1-1
Abstract
MicroRNAs (miRNAs) can post-transcriptionally regulate the expression of cancer-relevant genes via binding to the 3'-untranslated region (3'-UTR) of the target mRNAs. MiR-133a, as a miRNA, participate in tumorigenesis, progression, autophagy and drug-resistance in various malignancies. Based on the recent insights, we discuss the functions of miR-133a in physiological and pathological processes and its potential effects on cancer diagnosis, prognosis and therapy.
Keywords: cancer, miRNA, MiR-133a
Introduction
Cancer is a worldwide health concern due to its high mortality. Treatments of cancer, especially that in advanced stage, lay a tremendous economic burden for both patients and the society [1].Therefore, it is urgent to explore novel, highly specific biomarkers and targeted treatments for cancer. MiRNAs, a class of noncoding RNA (ncRNA) about 22-nucleotide RNAs in size, are highly conserved molecules regulating gene expression post-transcriptionally by binding to the 3'-untranslated region (3'-UTR) of target message RNAs (mRNAs) [2] and participating in tumorigenesis, proliferation, invasion and drug resistance in cancer [3]. Among the miRNAs, miR-133a has been considered as a tumor suppressor and a biomarker for prognosis of various cancers, such as osteosarcoma [4], esophageal cancer (EC) [5], colorectal cancer (CRC) [6], non-small cell lung cancer (NSCLC) [7], bladder cancer [8], breast cancer [9] and gastric cancer (GC) [10].
The miR-133 family (miR-133a, miR-133b), also classified as myomiRNAs for its role in skeletal and cardiac muscle development, is located on the 18th (miR-133a-1), 20th (miR-133a-2), and 6th (miR-133b) chromosomes, and transcribed as bicistronic transcripts with miR-1-2, miR-1-1, or miR-206. MiR-133 is involved in a variety of diseases, including cardiac hypertrophy, heart failure, cardiac arrhythmia, muscular dystrophy and cancers [11, 12].
The biochemical and molecular properties of MiR-133a
MiR-133a was first experimentally characterized in mice and found highly conserved in mice, flies and human. MiR-133a-1 and miR-133a-2 possess identical nucleotide sequences, while miR-133b differs from miR-133a by a single nucleotide at the 3' end (GU→A) [13].
The methods for the detection of MiR-133a
It is well known that the ways of detecting microRNAs mainly include high throughput sequencing, quantitative real time polymerase chain reaction (RT-qPCR) and microarrays[14]. RT-qPCR technology is the most frequently applied to detect the expression of miR-133a in cancer tissues and cell lines. As a circulating microRNA, miR-133a can also be deteced in body fluid, like blood and gastric juice[15], suggesting its value as a non-invasive biomarker.
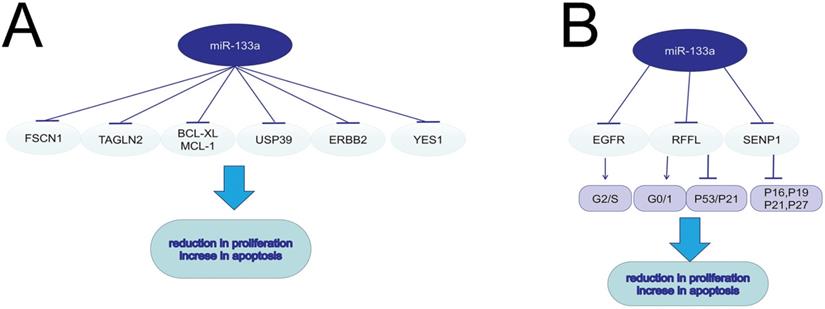
The role of miR-133a in cell circle, proliferation and apoptosis.
Target genes of miR-133a in various cancers
| MiR-133 | Cancers | Target genes |
|---|---|---|
| Downregulated | CRC | SOX4, FSCN1, RFFL, SENP1, Eif4A1, Rho A |
| Bladder cancer | FSCN1, TAGLN2, EGFR, GSTP1 | |
| GC | FSCN1, USP39, ERBB2/HER2, GABARAPL1, FOXP3, DSEN1, TAGLN2, Sp1 | |
| Nasopharyngeal cancer | EGFR | |
| ESCC | FSCN1, EGFR, COL1A1, SOX4 | |
| RCC | TAGLN2 | |
| Breast cancer | EGFR, MAML1, LASP1, UCP2 | |
| NSCLC | EGFR, YES1, CORO1C | |
| Osteosarcoma | BCL-xl, Mcl-1, RBPJ, PTHR1 | |
| PC | EGFR, USP39, IGF1R | |
| Ovaria cancer | PYGB, PVT1 | |
| OSCC | COL1A1 | |
| Prostate Cancer | EGFR, FGFR, IGF1R, MET | |
| HCC | FOSL2, MMP9, FSCN1, ATP7B, ABCC1 | |
| GBC | RBPJ | |
| Glioma | MMP9, MT1-MMP | |
| Endometrial cancer | PDE7A | |
| Cervical cancer | EGFR, SOX4 |
CRC: colorectal cancer; GC: gastric cancer; ESCC: esophageal squamous cancer; RCC: renal cell carcinoma; NSCLC: non-small cell lung cancer; PC: pancreatic cancer; HCC: hepatocellular carcinoma; GBC: gallbladder carcinoma.
Summarization of lncRNA-miR-133a interactions in human cancers
| Types of cancer | LncRNA | Expression | Reference |
|---|---|---|---|
| osteosarcoma | LINC01278 | upregulated in tumor tissues and cells | [4] |
| PC | FEZF1-AS1 | upregulated in tumor tissues and cells | [22] |
| Bladder cancer, PC, CRC | XIST | upregulated in tumor tissues and cells | [30,31,106] |
| PC | AFAP1-AS1 | upregulated in tumor tissues and cells | [39] |
| NSCLC, ovarian cancer | HOXD-AS1 | upregulated in tumor tissues and cells | [42,43] |
| CRC | ABHD11-AS1 | upregulated in tumor tissues and cells | [46] |
| cervical cancer | NEAT1 | upregulated in tumor tissues and cells | [50] |
CRC: colorectal cancer; NSCLC: non-small cell lung cancer; PC: pancreatic cancer.
The interactions of MiR-133a with other molecules
MiR-133a plays the tumor suppressive role via tageting and regulating the genes like SOX4, EGFR, FSCN1, COL1A1 and so on (Table 1). Accumulating evidence demonstrated that long non-coding RNAs (lncRNAs) could function as endogenous miRNA sponges or competing endogenous RNA by binding to miRNAs and regulating their function. We summarized the lncRNA-miR-133a interactions in Table 2. Whether circular RNA (circRNA) is involved in miR-133a-mediated tumor inhibition is worth studying.
MiR-133a in cell proliferation and apoptosis
Malignant tumors are manifested as activated cell proliferation and reduced cell apoptosis. Low-frequent ultrasound-targeted microbubble destruction (UTMD) of miR-133a injected in mice with breast cancer significantly suppressed tumor growth [9]. MiR-133a could negatively regulate the cell cycle and further suppress cell proliferation in breast cancer via targeting EGFR. The over-expression of miR-133a in MCF-7 and MDA-MB-231 cells could decrease G2/S phase and interfere DNA synthesis through the EGFR/Akt signaling pathway [16]. MiRNA-133a induced NSCLC cell apoptosis and decreased cell proliferation via the EGFR/AKT/ERK signaling pathway [17]. In esophageal cancer, miR-133a could targeted EGFR, and thus promote cell apoptosis and radio-sensitivity via downregulating the MEK/ERK pathway [5]. MiR-133a also served a tumor suppressor by modulating the ERalpha and AhR signaling pathways in breast cancer cell line MCF-7 [18]. MiR-133a increased apoptosis and inhibited proliferation in colorectal cancer by regulating the RFFL expression, inducing a G0/G1-phase arrest and activating p53/p21 signaling [19]. In human osteosarcoma, miR-133a suppressed cell proliferation and promoted cell apoptosis by repressing Bcl-xL and Mcl-1 mRNA [20]. MiR-133a-3p was sponged by LINC01278 and the decreased miR-133a-3p could lead to upregulation of parathyroid hormone type 1 receptor (PTHR1), consequently promoting cell proliferation and restraining apoptosis of osteosarcoma cells [4]. MiR-133a could suppress cell proliferation in non-small cell lung cancer by binding to the 3'-UTR of YES1 mRNA. YES1, a member of Src family tyrosinekinases (SFKs), could play a prominent role in tumor growth [7]. Fused in Sarcoma (FUS) could function as an AR-interacting protein that enhances AR transcriptional activity in prostate cancer. MiR-133a-5p inhibited cell proliferation in AR-positive prostate cancer cell line VCaP and LNCaP by targeting both FUS and AR, which further decreased the resistance to androgen ablation therapies[21]. In pancreatic cancer (PC) cell lines, FEZF1-AS1 served as an oncogene to induce cell proliferation and invasion through miR-133a/EGFR axis under normoxic condition [22]. In colorectal cancer, miR-133a-3p inhibited SENP1 expression, and then upregulated CDK inhibitors such as p16, p19, p21, and p27, resulting in the reduction in cell proliferation [6]. In GC and PC, ubiquitin-specific protease 39 (USP39), which was negatively regulated by miR-133a, was verified to induce cell proliferation and suppress cell apoptosis through modulating the AKT signaling pathway [10, 23]. MiR-133a inhibits the proliferation of gastric cancer cell SNU-1 and promotes SNU-1 cell apoptosis by modulating downstream ERBB2 (also called HER2, a member of EGFR family), and reducing the p-ERK1/2 and p-AKT expression [24].
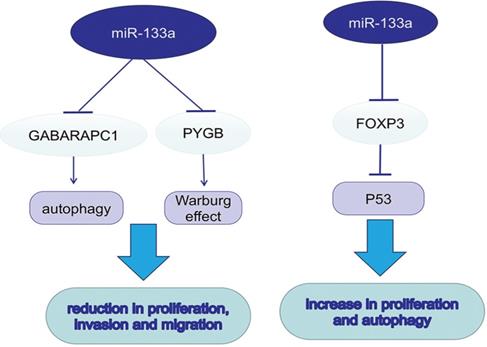
MiR-133a in autophagy and metabolism
Autophagy promotes tumor development in multiple cancers via consuming intracellular particles and providing sufficient energy for aggressive behaviors. MicroRNA-133a-3p could target GABARAPL1 to block autophagy-mediated glutaminolysis, further repressing gastric cancer growth and metastasis [25]. Forkhead protein 3 (FOXP3), a forkhead transcription factor and specific marker for T regulatory (Treg) cells, can downregulate TP53 by directly targeting its promoter and inhibiting its transcription. In contrast to previous studies, miR-133a-3p could promote the proliferation and autophagy in different gastric cancer cell lines by binding to the 3′-UTR of FOXP3 [26]. In dedifferentiated liposarcoma (DDLPS), loss of miR-133a could reduce oxidative metabolism, supporting the tumor Warburg effect. Braggio, D. first found that overexpression of miR-133a could increase glycolysis and OXPHOS both in vitro and in vivo; however, the metabolic change was not sufficient to suppress DDLPS cell proliferation and progression [27]. It was known that glycogen metabolism contributed to cancer development. The upregulation of glycogen phosphorylase B (PYGB) was detected in ovarian cancer tissues, while PYGB knockdown suppressed ovarian cancer cells proliferation, invasion and migration. The same procedure was also observed when miR-133a-3p was overexpressed, suggesting its role as a negative regulator in PYGB expression [28].
miR-133a promotes autophagy and the Warburg effect, which further promotes tumor progression.
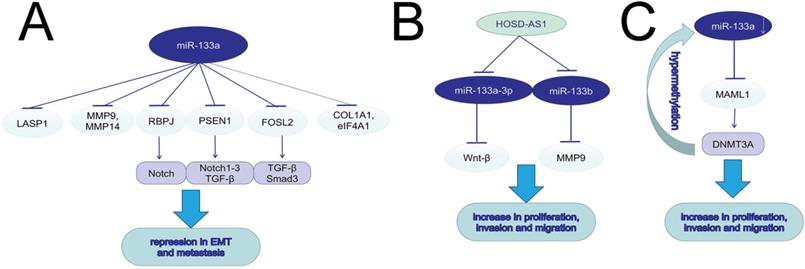
miR-133a induces EMT, invasion and migration in various tumors.
MiR-133a in cell migration and invasion
The increased migration and invasion ability accounts for the high mortality rate of cancers. In both in vitro and in vivo experiments, silencing miR-133a-3p by DNA hypermethylation in breast cancer cell lines and tissues led to increased cell migration, invasion and proliferation. MiR-133a-3p could target and downregulate the mastermind-like transcriptional coactivator 1 (MAML1) mRNA, and then up-regulated DNA methyltransferase 3A (DNMT3A), promoting hypermethylation of the miR-133a-3p promoter [29]. The long non-coding RNA X inactive specific transcript (XIST) could function as an oncogene, and promote tumor progression in bladder and pancreatic cancer via regulating miR-133a/EGFR axis [30, 31]. MiR-133a-3p inhibited cell invasion and migration in esophageal squamous cancer (ESCC) and oral squamous cell carcinoma (OSCC) via negatively regulating COL1A1, an important factor in extracellular matrix [32, 33]. It was hypothesized that epithelial mesenchymal transition (EMT) was vital to cancer migration, with evidence of upregulating expression of E-cadherin, downregulating N-cadherin, vimentin and Slug. Wang, T. et al found higher circP4HB expression in NSCLC tumor tissues than in normal ones, and the role of circP4HB in promoting EMT depended on the suppression of miR-133a-5p [34]. Also in GC line SGC-7901 and BGC-823, miR-133a played an antigrowth and antimetastasis role by inhibiting transforming growth factor-beta1 (TGF-beta1)-induced EMT via targeting PSEN1. Moreover, the decrease of PSEN1 could further downregulate Notch 1, Notch 2, and Notch 3 [35]. MiR-133a could not only suppress cell proliferation, induce cell cycle arrest at G0/G1 stage and accelerate cell apoptosis, but also inhibit cell migration and invasion in vivo and in vitro via targeting IGF-1R and negatively regulating downstream AKT and ERK signal pathway [36-38]. In PC, AFAP1-AS1 was reported to induce IGF1R transcription and activate the AKT/ERK pathways to promote EMT and cell metasis by sponging miR-133a [39]. Downregulation of miR-133a-3p functioned as a supportive factor in bone metastasis of prostate cancer (PCa) possibly by targeting EGFR, FGFR1, IGF1R and MET, and inactivating PI3K/AKT signaling pathway [40]. MiR-133a could suppress hepatocellular carcinoma (HCC) by targeting Fos-related antigen 2 (FOSL2) and inactivating TGF-β/Smad3 signaling pathway. FOSL2, a member of the AP-1 family, included rich isoforms of Fos and Jun [41]. LncRNA HOXD-AS1 could induce NSCLC cell proliferation and migration by targeting miR-133b/MMP-9 axis and promote epithelial ovarian cancer (EOC) cell invasion and EMT by regulating miR-133a-3p/Wnt/β-catenin pathway [42, 43]. MiR-133a could suppress breast cancer via targeting LASP1, a actin-binding protein reacting to cAMP and cGMP signaling [44]. In ovarian cancer, miR-133a influenced cancer progression and prognosis as a downstream target of plasmacytoma variant translocation 1 (PVT1) [45]. The ABHD11-AS1/miR-133a/SOX4 axis played a significant role in the progression in CRC [46]. MiR-133a exerted inhibitory effects on gallbladder carcinoma (GBC) via negative regulation of RBPJ, an important transcriptional regulator in the Notch signaling pathway [47]. The same modulation found in osteosarcoma contributed to DCs maturation and activation [48]. Astragalus polysaccharides (APS) could not only repress osteosarcoma cells proliferation and invasion but also induce cell apoptosis by inducing miR-133a and further inactivating JNK pathway [49]. In cervical cancer, miR-133a was also significantlly downregulated. Upregulating miR-133a can restrain the progression of cervical cancer via targeting EGFR and SOX4. NEAT1, a oncogene in various cancers, was reported to sponge to miR-133a and inhibit miR-133a expression, further regulate the cervial cancer progression via the NEAT1/miR-133a/SOX4 axis[50]. Matrine was found to suppress the invasion and metastasis of NCI-H1299 cells by upregulating miR-133a, which further repressed the EGFR/Akt/MMP-9 signal pathway [51]. MiR-133a could act as a tumor-suppressor by targeting and downregulating eukaryotic translation initiation factor 4A1 (eIF4A1) [52], matrix metallopeptidase 9 (MMP9) [53, 54], MMP14 [55], membrane-type 1 matrix metalloproteinase (MT1-MMP) [56], Fascin1 (FSCN1) [57-59], phosphodiesterase 7A (PDE7A) [60], Coronin-1C (CORO1C) [61] and Sox4 [62].
The dual role of miR-133a in drug resistance.
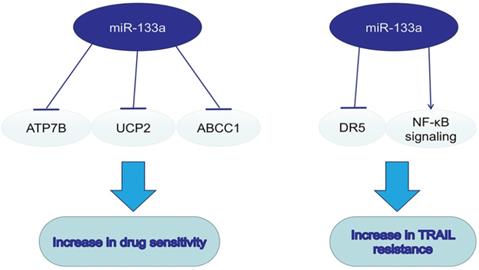
MiR-133a in drugs and drug sensitivity
On the basis of dysregulation of miR-133a and its tumorsupressive role in various cancers, miR-133a was also a pivotal regulator in drug resistance. Combining drugs targeting miR-133a and traditional chemotherapy may enhance the efficacy of antitumor therapy. MiR-133a could enhance the sensitivity to cisplatin via decreasing the ATP7B expression in Hep-2v cells [63]. Upregulating miR133a could reverse the resistance to doxorubicin via decreasing the expression of mitochondria uncoupling protein 2 (UCP-2) in doxorubicin-resistant breast cancer cell line MCF-7/Dox [64]. MiR-133a overexpression could enhance the sensitivity to ADM in HepG2 cells by inhibiting multidrug resistance-associated protein 1 (ABCC1), a ATP-binding cassette (ABC) transporter of cellular drug disposition [65]. TNF-related apoptosis-inducing ligand (TRAIL), a new anti-cancer drug, could specifically induce cancer cell apoptosis. MiR-133a was found to promote TRAIL resistance in glioblastoma via down-regulating DR5 expression and activating NF-κB signaling [66].
MiR-133a in cancer prognosis
Accumulating evidence has suggested the association between miR-133a and poor overall survival (OS) of patients with solid cancer [67, 68]. A meta-analysis revealed that high expression of miR-133a was related to better prognosis and ameliorated clinicopathological features in digestive system cancers[69]. A research in 110 patients showed that the reduced serum miR-133a contributed to poor prognosis of pancreatic cancer (PC), indicating that miR-133a might function as a specific diagnostic indicator for PC [70]. The terminal differentiation induced ncRNA (TINCR), a target of miR-133a, was associated with shorter disease-free survival (DFS) and OS of HCC patients [71]. MiR-133a was also found to be associated with the prognosis of bladder cancer, as well as better survival and less resistance to treatment in prostate cancer [72, 73]. The expression of miR-133a was negatively correlated with lymphatic metastasis, clinical stage and MMP-14, EGFR levels of NSCLC [74, 75].
MiR-133a as a potential biomarker
Plenty of efforts have been devoted to evaluating the efficacy of miR-133a as a diagnostic marker. By conducting a case-control investigation in 50 pairs of breast cancer and normal tissues, Bitaraf, A. et al. considered miR-133a-3p, along with miR-127-3p, miR-155-5p, miR-199b-5p, and miR-342-5p, were promising biomarkers for BC [76]. MiR-133a-3p was significantly modulated in triple-negative breast cancer (TNBC), suggesting that miR-133a-3p might be a predictor of cancer invasion and prognosis [77]. Early stage renal cell carcinoma is often failed to be diagnosed due to its asymptomatic feature. Luckily, miR-133a-2 was a newly found biomarker of renal cell carcinoma (RCC) [78]. At present, it is still hard to estimate the metastatic potential of gastrointestinal neuroendocrine neoplasms (GI-NENs), yet a lower level of miR-133a was detected in 51 primary GI-NENs with liver metastases, and a higher level of miR-133a was found in appendiceal carcinoids without metastases compared to the primary tumors [79]. A multicenter, retrospective research proposed that circulating microRNAs like miR-133a were more sensitive in diagnosing preclinical and early-stage hepatocellular carcinoma than AFP20 [80]. Interestingly, the methylation-silencing of miR-133a could be reversed by H. pylori eradication, suggesting the potential role of miR-133a in H. pylori-induced gastric carcinogenesis [81]. Combined with gastroscopy, miR-133a in gastric juice could also be a novel test target for screening gastric cancer [15, 82]. The combination of miR-133a and FOBT could be a potential detection mode in colorectal cancer screening [83]. Based on the information from The Cancer Genome Atlas (TCGA) database and Meta-analysis results, along with bioinformatics analysis, miR-133a was recognized as a potential biomarker of NSCLC [84-86], oral cancer [87, 88], osteosarcoma [89], cervical cancer [90], prostate cancer [91], digestive tumors [92] including EC [93-95], GC [96, 97], pancreatic ductal adenocarcinoma (PDAC) [98] and CRC [99]. A study of 8006 tumors including 19 tumor types suggested that hypoxia contributed to genomic instability. Among the dysregulated hypoxia-associated microRNAs, miR-133a-3 was considered to be a novel prognosis biomarker for hypoxic tumors with elevated risk for distant metastasis [100]. MiR-133a-3p dysregulation was also found in smoking-induced HPV (+) oropharyngeal cancer patients[101]. Taken together, the dysregulation of miR-133a occurs at the early stage of tumorigensis, and low expression of miR-133a frequently indicates poor prognosis, which is consistent with its tumorsuppressive role in cancer development and progression. MiR-133a might serve as a promising biomarker for cancer early diagnosis, monitoring cancer progression and treatment response.
MiR-133a mediating different signaling pathways
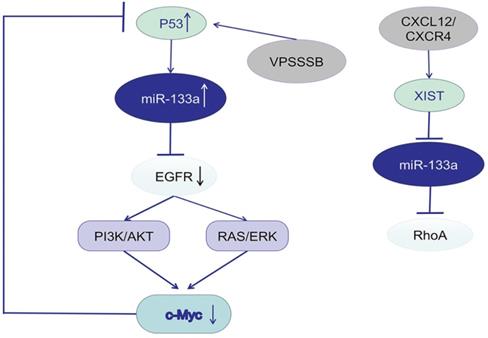
Studies focusing on the potential mechanism of miR-133a described EGFR as a promising target gene and the downstream signaling pathway. P53 was predicted to bind to the promoter of miR-133a-1, a precursor of miR-133a-3p, and thus activate miR-133a-3p expression. C-Myc, an oncogene, could influence p53 transcription, suppress P53 expression, and further downregulate miR-133a-3p expression. MiR-133a-3p could directly target the mRNA of EGFR [16, 102], inactivate the EGFR/RAS/ERK/c-Myc pathway or EGFR/PI3K/AKT/c-Myc pathway and increase p53 expression. VPS33B, a member of Sec-1 domain family, could activate miR-133a-3p by inducing p53 nuclear translocation, further regulate the EGFR/RAS/ERK/c-Myc/p53/miR-133a-3p feedback loop, and thus suppress CRC progression [103]. VPS 33B could also inhibit cell proliferation and chemoresistance to fluorouracil (5-FU) both in vivo and in vitro by regulating EGFR/PI3K/AKT/c-Myc/P53/miR-133a-3p signaling loop [104]. In nasopharyngeal carcinoma (NPC), cinobufotalin (CB) induced FOXO1-mediated cisplatin sensitivity by down-regulating MYH9 which was colocalized with FOXO1 in the cytoplasm via the PI3K/AKT/c-Myc/P53/miR-133a-3p pathway [105]. Activation of CXCL12/CXCR4 axis upregulated LncRNA XIST, which acted as a ceRNA to sponge miR-133a-3p, and promote malignant progression of CRC cells via lncRNA XIST/ miR-133a-3p/RhoA axis. RhoA, a potential target of miR-133a-3p, could regulate cell motility through cytoskeletal reorganization by promoting actin polymerization and actomyosin contractility via the ROCK/p-MLC pathway [106].
miR-133a involves in EGFR/c-Myc/P53 axis and the associated signaling pathway.
Conclusions
MiR-133a is significantly downregulated in malignant cancers and often accompanied by poor prognosis. It also participates in various biological processes, including proliferation, apoptosis, autophagy, migration, invasion and drug resistance, which rely on the regulation of downstream target genes and signaling pathways, with the EGFR/c-Myc/P53 axis as the commonly seen one. Accumulating studies have shown that miRNA may serve as an effective molecular targeted drug for the treatment of cancers because the recovery of abnormally expressed miRNA levels can affect the onset, development and metastasis of multiple cancers [107]. We hereby suggest that miR-133a might be a potential biomarker and therapeutic target in maligant tumors, and new attempts like combining miR-133a with traditional cancer therapy should be made.
Acknowledgements
Funding
This work was supported by Wuxi Municipal Health Committee (Z201713).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
2. Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. The international journal of biochemistry & cell biology. 2010;42:1273-81
3. Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS letters. 2011;585:2087-99
4. Qu Z, Li S. Long noncoding RNA LINC01278 favors the progression of osteosarcoma via modulating miR-133a-3p/PTHR1 signaling. Journal of cellular physiology. 2020 [Epub ahead of print]
5. Yang QS, Jiang LP, He CY, Tong YN, Liu YY. Up-Regulation of MicroRNA-133a Inhibits the MEK/ERK Signaling Pathway to Promote Cell Apoptosis and Enhance Radio-Sensitivity by Targeting EGFR in Esophageal Cancer In vivo and In vitro. Journal of cellular biochemistry. 2017;118:2625-34
6. Zhou GQ, Han F, Shi ZL, Yu L, Li XF, Yu C. et al. miR-133a-3p Targets SUMO-Specific Protease 1 to Inhibit Cell Proliferation and Cell Cycle Progress in Colorectal Cancer. Oncology research. 2018;26:795-800
7. Shen Y, Chen F, Liang Y. MicroRNA-133a inhibits the proliferation of non-small cell lung cancer by targeting YES1. Oncology letters. 2019;18:6759-65
8. Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K. et al. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urologic oncology. 2013;31:115-23
9. Ji Y, Han Z, Shao L, Zhao Y. Evaluation of in vivo antitumor effects of low-frequency ultrasound-mediated miRNA-133a microbubble delivery in breast cancer. Cancer medicine. 2016;5:2534-43
10. Dong X, Su H, Jiang F, Li H, Shi G, Fan L. miR-133a, directly targeted USP39, suppresses cell proliferation and predicts prognosis of gastric cancer. Oncology letters. 2018;15:8311-8
11. Yu H, Lu Y, Li Z, Wang Q. microRNA-133: expression, function and therapeutic potential in muscle diseases and cancer. Current drug targets. 2014;15:817-28
12. Li N, Zhou H, Tang Q. miR-133: A Suppressor of Cardiac Remodeling? Frontiers in pharmacology. 2018;9:903
13. Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3(1):9-21
14. Kang K, Peng X, Luo J, Gou D. Identification of circulating miRNA biomarkers based on global quantitative real-time PCR profiling. J Anim Sci Biotechnol. 2012;3(1):4
15. Shao J, Fang PH, He B, Guo LL, Shi MY, Zhu Y. et al. Downregulated MicroRNA-133a in Gastric Juice as a Clinicopathological Biomarker for Gastric Cancer Screening. Asian Pacific journal of cancer prevention: APJCP. 2016;17:2719-22
16. Cui W, Zhang S, Shan C, Zhou L, Zhou Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. The FEBS journal. 2013;280:3962-74
17. Guo N, Zhao Y, Zhang W, Li S, Li S, Yu J. MicroRNA-133a downregulated EGFR expression in human non-small cell lung cancer cells via AKT/ERK signaling. Oncology letters. 2018;16:6045-50
18. Mobini K, Tamaddon G, Fardid R, Keshavarzi M, Mohammadi-Bardbori A. Aryl hydrocarbon-estrogen alpha receptor-dependent expression of miR-206, miR-27b, and miR-133a suppress cell proliferation and migration in MCF-7 cells. Journal of biochemical and molecular toxicology. 2019: e22304.
19. Dong Y, Zhao J, Wu CW, Zhang L, Liu X, Kang W. et al. Tumor suppressor functions of miR-133a in colorectal cancer. Molecular cancer research: MCR. 2013;11:1051-60
20. Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y. et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56:220-6
21. Zheng L, Kang Y, Zhang L, Zou W. MiR-133a-5p inhibits androgen receptor (AR)-induced proliferation in prostate cancer cells via targeting FUsed in Sarcoma (FUS) and AR. Cancer biology & therapy. 2020;21:34-42
22. Ou Z-L, Zhang M, Ji L-D, Luo Z, Han T, Lu Y-B. et al. Long noncoding RNA FEZF1-AS1 predicts poor prognosis and modulates pancreatic cancer cell proliferation and invasion through miR-142/HIF-1α and miR-133a/EGFR upon hypoxia/normoxia. Journal of cellular physiology. 2019 [Epub ahead of print]
23. Cai J, Liu T, Huang P, Yan W, Guo C, Xiong L. et al. USP39, a direct target of microRNA-133a, promotes progression of pancreatic cancer via the AKT pathway. Biochemical and biophysical research communications. 2017;486:184-90
24. Li C, Li X, Gao S, Li C, Ma L. MicroRNA-133a Inhibits Proliferation of Gastric Cancer Cells by Downregulating ERBB2 Expression. Oncology research. 2017;25:1169-76
25. Zhang X, Li Z, Xuan Z, Xu P, Wang W, Chen Z. et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. Journal of experimental & clinical cancer research: CR. 2018;37:320
26. Li J-P, Zhang H-M, Liu M-J, Xiang Y, Li H, Huang F. et al. miR-133a-3p/FOXP3 axis regulates cell proliferation and autophagy in gastric cancer. Journal of cellular biochemistry. 2020;121:3392-405
27. Yu PY, Lopez G, Braggio D, Koller D, Bill KLJ, Prudner BC. et al. miR-133a function in the pathogenesis of dedifferentiated liposarcoma. Cancer cell international. 2018;18:89
28. Zhou Y, Jin Z, Wang C. Glycogen phosphorylase B promotes ovarian cancer progression via Wnt/beta-catenin signaling and is regulated by miR-133a-3p. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;120:109449
29. Shi W, Tang T, Li X, Deng S, Li R, Wang Y. et al. Methylation-mediated silencing of miR-133a-3p promotes breast cancer cell migration and stemness via miR-133a-3p/MAML1/DNMT3A positive feedback loop. Journal of experimental & clinical cancer research: CR. 2019;38:429
30. Zhou K, Yang J, Li X, Chen W. Long non-coding RNA XIST promotes cell proliferation and migration through targeting miR-133a in bladder cancer. Experimental and therapeutic medicine. 2019;18:3475-83
31. Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. Journal of cellular biochemistry. 2017;118:3349-58
32. Yin Y, Du L, Li X, Zhang X, Gao Y. miR-133a-3p suppresses cell proliferation, migration, and invasion and promotes apoptosis in esophageal squamous cell carcinoma. Journal of cellular physiology. 2019;234:12757-70
33. He B, Lin X, Tian F, Yu W, Qiao B. MiR-133a-3p Inhibits Oral Squamous Cell Carcinoma (OSCC) Proliferation and Invasion by Suppressing COL1A1. Journal of cellular biochemistry. 2018;119:338-46
34. Wang T, Wang X, Du Q, Wu N, Liu X, Chen Y. et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochemical and biophysical research communications. 2019;513:904-11
35. Chen XB, Li W, Chu AX. MicroRNA-133a inhibits gastric cancer cells growth, migration, and epithelial-mesenchymal transition process by targeting presenilin 1. Journal of cellular biochemistry. 2019;120:470-80
36. Chen G, Fang T, Huang Z, Qi Y, Du S, Di T. et al. MicroRNA-133a Inhibits Osteosarcoma Cells Proliferation and Invasion via Targeting IGF-1R. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;38:598-608
37. Zhang W, Liu K, Liu S, Ji B, Wang Y, Liu Y. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:9779-88
38. Gong Y, Ren J, Liu K, Tang LM. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World journal of gastroenterology. 2015;21:2949-58
39. Chen B, Li Q, Zhou Y, Wang X, Zhang Q, Wang Y. et al. The long coding RNA AFAP1-AS1 promotes tumor cell growth and invasion in pancreatic cancer through upregulating the IGF1R oncogene via sequestration of miR-133a. Cell cycle (Georgetown, Tex). 2018;17:1949-66
40. Tang Y, Pan J, Huang S, Peng X, Zou X, Luo Y. et al. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. Journal of experimental & clinical cancer research: CR. 2018;37:160
41. Sun L, Guo Z, Sun J, Li J, Dong Z, Zhang Y. et al. MiR-133a acts as an anti-oncogene in Hepatocellular carcinoma by inhibiting FOSL2 through TGF-beta/Smad3 signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;107:168-76
42. Xia H, Jing H, Li Y, Lv X. Long noncoding RNA HOXD-AS1 promotes non-small cell lung cancer migration and invasion through regulating miR-133b/MMP9 axis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;106:156-62
43. Zhang Y, Dun Y, Zhou S, Huang XH. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/beta-catenin signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;96:1216-21
44. Sui Y, Zhang X, Yang H, Wei W, Wang M. MicroRNA-133a acts as a tumour suppressor in breast cancer through targeting LASP1. Oncology reports. 2018;39:473-82
45. Yang Q, Yu Y, Sun Z, Pan Y. Long non-coding RNA PVT1 promotes cell proliferation and invasion through regulating miR-133a in ovarian cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;106:61-7
46. Lei X, Li L, Duan X. Long non-coding RNA ABHD11-AS1 promotes colorectal cancer development through regulation of miR-133a/SOX4 axis. Bioscience reports. 2018;38(6):BSR20181386
47. Huang Y, Wu Y, Dong J, Han D, Yang S, Jiang L. MicroRNA-133a-3p exerts inhibitory effects on gallbladder carcinoma via targeting RBPJ. American journal of cancer research. 2016;6:2448-62
48. Gao X, Han D, Fan W. Down-regulation of RBP-J mediated by microRNA-133a suppresses dendritic cells and functions as a potential tumor suppressor in osteosarcoma. Experimental cell research. 2016;349:264-72
49. Chu Y, Fang Y, Chi J, Li J, Zhang D, Zou Y. et al. Astragalus polysaccharides decrease proliferation, migration, and invasion but increase apoptosis of human osteosarcoma cells by up-regulation of microRNA-133a. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas. 2018;51:e7665
50. Yuan LY, Zhou M, Lv H, Qin X, Zhou J, Mao X. et al. Involvement of NEAT1/miR-133a axis in promoting cervical cancer progression via targeting SOX4. Journal of cellular physiology. 2019;234:18985-93
51. Liao H, Zhao X, Qu J, Zhang J, Cai H. Matrine suppresses invasion and metastasis of NCI-H1299 cells by enhancing microRNA-133a expression. International journal of clinical and experimental medicine. 2015;8:10714-22
52. Li W, Chen A, Xiong L, Chen T, Tao F, Lu Y. et al. miR-133a acts as a tumor suppressor in colorectal cancer by targeting eIF4A1. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39:1010428317698389
53. Wang J, Li J, Guo F, Yan Y. MicroRNA-133a inhibits the malignant behavior of glioma via downregulation of matrix metallopeptidase 9. Molecular medicine reports. 2016;13:3220-6
54. Chen X, Bo L, Zhao X, Chen Q. MicroRNA-133a inhibits cell proliferation, colony formation ability, migration and invasion by targeting matrix metallopeptidase 9 in hepatocellular carcinoma. Molecular medicine reports. 2015;11:3900-7
55. Wang H, Zhang Y, Zhang Y, Liu W, Wang J. Cryptotanshinone inhibits lung cancer invasion via microRNA-133a/matrix metalloproteinase 14 regulation. Oncology letters. 2019;18:2554-9
56. Sakr M, Takino T, Sabit H, Nakada M, Li Z, Sato H. miR-150-5p and miR-133a suppress glioma cell proliferation and migration through targeting membrane-type-1 matrix metalloproteinase. Gene. 2016;587:155-62
57. Zheng K, Liu W, Liu Y, Jiang C, Qian Q. MicroRNA-133a suppresses colorectal cancer cell invasion by targeting Fascin1. Oncology letters. 2015;9:869-74
58. Wang G, Zhu S, Gu Y, Chen Q, Liu X, Fu H. MicroRNA-145 and MicroRNA-133a Inhibited Proliferation, Migration, and Invasion, While Promoted Apoptosis in Hepatocellular Carcinoma Cells Via Targeting FSCN1. Digestive diseases and sciences. 2015;60:3044-52
59. Lai C, Chen Z, Li R. MicroRNA-133a inhibits proliferation and invasion, and induces apoptosis in gastric carcinoma cells via targeting fascin actin-bundling protein 1. Molecular medicine reports. 2015;12:1473-8
60. Yamamoto N, Nishikawa R, Chiyomaru T, Goto Y, Fukumoto I, Usui H. et al. The tumor-suppressive microRNA-1/133a cluster targets PDE7A and inhibits cancer cell migration and invasion in endometrial cancer. International journal of oncology. 2015;47:325-34
61. Mataki H, Enokida H, Chiyomaru T, Mizuno K, Matsushita R, Goto Y. et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. Journal of human genetics. 2015;60:53-61
62. Li S, Qin X, Li Y, Zhang X, Niu R, Zhang H. et al. MiR-133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. American journal of translational research. 2015;7:1390-403
63. Wang X, Zhu W, Zhao X, Wang P. miR-133a enhances the sensitivity of Hep-2 cells and vincristine-resistant Hep-2v cells to cisplatin by downregulating ATP7B expression. International journal of molecular medicine. 2016;37:1636-42
64. Yuan Y, Yao YF, Hu SN, Gao J, Zhang LL. MiR-133a Is Functionally Involved in Doxorubicin-Resistance in Breast Cancer Cells MCF-7 via Its Regulation of the Expression of Uncoupling Protein 2. PloS one. 2015;10:e0129843
65. Ma J, Wang T, Guo R, Yang X, Yin J, Yu J. et al. Involvement of miR-133a and miR-326 in ADM resistance of HepG2 through modulating expression of ABCC1. Journal of drug targeting. 2015;23:519-24
66. Wang SS, Feng L, Hu BG, Lu YF, Wang WM, Guo W. et al. miR-133a Promotes TRAIL Resistance in Glioblastoma via Suppressing Death Receptor 5 and Activating NF-kappaB Signaling. Molecular therapy Nucleic acids. 2017;8:482-92
67. Gao S, Zhao ZY, Zhang ZY, Zhang Y, Wu R. Prognostic Value of MicroRNAs in Esophageal Carcinoma: A Meta-Analysis. Clinical and translational gastroenterology. 2018;9:203
68. Xiao J, Zou Y, Lu X, Xie B, Yu Q, He B. et al. Prognostic value of decreased microRNA-133a in solid cancers: a meta-analysis. OncoTargets and therapy. 2016;9:5771-9
69. Zhu W, Ji X. The Impact of MicroRNA-133a on Prognosis and Clinicopathological Parameters for Digestive System Cancers: a Comprehensive Study Based on Meta-Analysis and TCGA Database. Pathology oncology research: POR. 2020;26:771-81
70. Wang Z. Diagnostic performance for declined microRNA-133a in pancreatic cancer. Journal of cellular biochemistry. 2019 [Epub ahead of print]
71. Tian F, Xu J, Xue F, Guan E, Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Bioscience reports. 2017;37(4):BSR20170301
72. Yin XH, Jin YH, Cao Y, Wong Y, Weng H, Sun C. et al. Development of a 21-miRNA Signature Associated With the Prognosis of Patients With Bladder Cancer. Frontiers in oncology. 2019;9:729
73. Foj L, Filella X. Identification of Potential miRNAs Biomarkers for High-Grade Prostate Cancer by Integrated Bioinformatics Analysis. Pathology oncology research: POR. 2019;25:1445-56
74. Wang Y, Li J, Chen H, Mo Y, Ye H, Luo Y. et al. Down-regulation of miR-133a as a poor prognosticator in non-small cell lung cancer. Gene. 2016;591:333-7
75. Lan D, Zhang X, He R, Tang R, Li P, He Q. et al. MiR-133a is downregulated in non-small cell lung cancer: a study of clinical significance. European journal of medical research. 2015;20:50
76. Bitaraf A, Babashah S, Garshasbi M. Aberrant expression of a five-microRNA signature in breast carcinoma as a promising biomarker for diagnosis. Journal of clinical laboratory analysis. 2019: e23063.
77. Song X, Zhang C, Liu Z, Liu Q, He K, Yu Z. Characterization of ceRNA network to reveal potential prognostic biomarkers in triple-negative breast cancer. PeerJ. 2019;7:e7522
78. Tang T, Du X, Zhang X, Niu W, Li C, Tan J. Computational identification and analysis of early diagnostic biomarkers for kidney cancer. Journal of human genetics. 2019;64:1015-22
79. Mandal R, Hardin H, Baus R, Rehrauer W, Lloyd RV. Analysis of miR-96 and miR-133a Expression in Gastrointestinal Neuroendocrine Neoplasms. Endocrine pathology. 2017;28:345-50
80. Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q. et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. The Lancet Oncology. 2015;16:804-15
81. Lim JH, Kim SG, Choi JM, Yang HJ, Kim JS, Jung HC. Helicobacter pylori Is Associated with miR-133a Expression through Promoter Methylation in Gastric Carcinogenesis. Gut and liver. 2018;12:58-66
82. Virgilio E, Giarnieri E, Giovagnoli MR, Montagnini M, Proietti A, D'Urso R. et al. Gastric Juice MicroRNAs as Potential Biomarkers for Screening Gastric Cancer: A Systematic Review. Anticancer research. 2018;38:613-6
83. Luo X, Wu Y, Ji M, Zhang S. Combined Plasma MicroRNA and Fecal Occult Blood Tests in Early Detection of Colorectal Cancer. Clinical laboratory. 2019 65(5)
84. Yang ZQ, Wu CA, Cheng YX. Prognostic Value of microRNA-133a Expression and Its Clinicopathologic Significance in Non-Small Cell Lung Cancer: A Comprehensive Study Based on Meta-Analysis and the TCGA Database. Oncology research and treatment. 2018;41:762-8
85. Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World journal of surgical oncology. 2017;15:175
86. Su L, Li N, Huo X. Mining featured micro ribonucleic acids associated with lung cancer based on bioinformatics. Thoracic cancer. 2015;6:501-7
87. Zeljic K, Jovanovic I, Jovanovic J, Magic Z, Stankovic A, Supic G. MicroRNA meta-signature of oral cancer: evidence from a meta-analysis. Upsala journal of medical sciences. 2018;123:43-9
88. Yan ZY, Luo ZQ, Zhang LJ, Li J, Liu JQ. Integrated Analysis and MicroRNA Expression Profiling Identified Seven miRNAs Associated With Progression of Oral Squamous Cell Carcinoma. Journal of cellular physiology. 2017;232:2178-85
89. Mirghasemi A, Taheriazam A, Karbasy SH, Torkaman A, Shakeri M, Yahaghi E. et al. Down-regulation of miR-133a and miR-539 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Cancer cell international. 2015;15:86
90. Servin-Gonzalez LS, Granados-Lopez AJ, Lopez JA. Families of microRNAs Expressed in Clusters Regulate Cell Signaling in Cervical Cancer. International journal of molecular sciences. 2015;16:12773-90
91. Goto Y, Kurozumi A, Enokida H, Ichikawa T, Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. International journal of urology: official journal of the Japanese Urological Association. 2015;22:242-52
92. Lai CH, Liang XZ, Liang XY, Ma SJ, Li JG, Shi MF. et al. Study on miRNAs in Pan-Cancer of the Digestive Tract Based on the Illumina HiSeq System Data Sequencing. BioMed research international. 2019;2019:8016120
93. Mei LL, Qiu YT, Zhang B, Shi ZZ. MicroRNAs in esophageal squamous cell carcinoma: Potential biomarkers and therapeutic targets. Cancer biomarkers: section A of Disease markers. 2017;19:1-9
94. Gao SH, Liu J, Zhang HJ, Zhao N, Zhang J. Low miR-133a expression is a predictor of outcome in patients with esophageal squamous cell cancer. European review for medical and pharmacological sciences. 2016;20:3788-92
95. Bus P, Kestens C, Ten Kate FJ, Peters W, Drenth JP, Roodhart JM. et al. Profiling of circulating microRNAs in patients with Barrett's esophagus and esophageal adenocarcinoma. Journal of gastroenterology. 2016;51:560-70
96. Li CY, Liang GY, Yao WZ, Sui J, Shen X, Zhang YQ. et al. Identification and functional characterization of microRNAs reveal a potential role in gastric cancer progression. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2017;19:162-72
97. Chen S, Zhu J, Yu F, Tian Y, Ma S, Liu X. Combination of miRNA and RNA functions as potential biomarkers for gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:9909-18
98. Liao X, Wang X, Huang K, Yang C, Yu T, Han C. et al. Genome-scale analysis to identify prognostic microRNA biomarkers in patients with early stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Cancer management and research. 2018;10:2537-51
99. Weber D, Amar L, Godde D, Prinz C. Extensive screening of microRNA populations identifies hsa-miR-375 and hsa-miR-133a-3p as selective markers for human rectal and colon cancer. Oncotarget. 2018;9:27256-67
100. Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, Livingstone J. et al. Molecular landmarks of tumor hypoxia across cancer types. Nature genetics. 2019;51:308-18
101. House R, Majumder M, Janakiraman H, Ogretmen B, Kato M, Erkul E. et al. Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer. PloS one. 2018;13:e0205077
102. Song X, Shi B, Huang K, Zhang W. miR-133a inhibits cervical cancer growth by targeting EGFR. Oncology reports. 2015;34:1573-80
103. Chen Y, Liu Z, Wang H, Tang Z, Liu Y, Liang Z. et al. VPS33B negatively modulated by nicotine functions as a tumor suppressor in colorectal cancer. International journal of cancer. 2020;146:496-509
104. Liang Z, Liu Z, Cheng C, Wang H, Deng X, Liu J. et al. VPS33B interacts with NESG1 to modulate EGFR/PI3K/AKT/c-Myc/P53/miR-133a-3p signaling and induce 5-fluorouracil sensitivity in nasopharyngeal carcinoma. Cell death & disease. 2019;10:305
105. Li Y, Liu X, Lin X, Zhao M, Xiao Y, Liu C. et al. Chemical compound cinobufotalin potently induces FOXO1-stimulated cisplatin sensitivity by antagonizing its binding partner MYH9. Signal transduction and targeted therapy. 2019;4:48
106. Yu X, Wang D, Wang X, Sun S, Zhang Y, Wang S. et al. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. Journal of experimental & clinical cancer research: CR. 2019;38:32
107. Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F. et al. Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano. 2015;9:9731-40
Author contact
![]() Corresponding author: Min Xia, Department of Gastroenterology, Wuxi People's Hospital Affiliated to Nanjing Medical University, 299 Qingyang Road, Wuxi, Jiangsu 214023, China; E-mail address: xmzb1013com.
Corresponding author: Min Xia, Department of Gastroenterology, Wuxi People's Hospital Affiliated to Nanjing Medical University, 299 Qingyang Road, Wuxi, Jiangsu 214023, China; E-mail address: xmzb1013com.

 Global reach, higher impact
Global reach, higher impact