Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(2):387-396. doi:10.7150/jca.47863 This issue Cite
Research Paper
Silencing of CASC8 inhibits non-small cell lung cancer cells function and promotes sensitivity to osimertinib via FOXM1
1. Department of Pathology, First Affiliated Hospital and College of Basic Medical Sciences, China Medical University, Shenyang, China.
2. Department of Pathology, the First Bethune Hospital of Jilin University, Changchun, Jilin, China.
3. Department of Pathology, Basic Medical Sciences, Xinjiang Medical University, Urumqi, China.
Received 2020-5-7; Accepted 2020-10-25; Published 2021-1-1
Abstract
In a meta-analysis, the long noncoding RNA cancer susceptibility candidate 8 (CASC8) was found to be a cancer susceptibility gene closely related to lung cancer, but its functions in lung cancer are unknown. In the Cancer Genome Atlas database, the expression of CASC8 was significantly higher in non-small cell lung cancer than in adjacent normal tissues, and high expression of CASC8 was associated with poor prognosis in patients with lung adenocarcinoma. Silencing CASC8 inhibited proliferation, migration, and invasion in non-small cell lung cancer cell lines. Silencing CASC8 also promoted sensitivity to osimertinib through Forkhead box M1 (FOXM1). Therefore, this pathway can be exploited in patients with lung cancer resistant to targeted therapies. Our study revealed for the first time that silencing CASC8 inhibited the proliferation, migration, and invasion of non-small cell lung cancer cells and promoted their sensitivity to osimertinib, suggesting that CASC8 is closely related to the occurrence and development of non-small cell lung cancer. This may provide insight into mechanisms of treatment for non-small cell lung cancer.
Keywords: Cancer susceptibility candidate 8, Long noncoding RNAs, Non-small Cell Lung, Forkhead Box Protein M1, Osimertinib
Introduction
Nearly 80% of lung cancers are non-small cell lung cancer (NSCLC) [1], and NSCLC is the leading cause of cancer-related deaths in the United States and China [2-4]. Although surgery is the preferred method of treatment, only a portion of early lung cancers can be cured by surgical resection. Further, more than 60% of patients with lung cancer are diagnosed with locally advanced disease or have lymph node metastasis (stage III or IV), and surgery is not the primary treatment option. Patients with good physical condition usually receive chemotherapy first, and then choose whether surgery is needed according to their response [5]. Although recent years have shown the discovery of many genetic drivers in NSCLC, such as epidermal growth factor receptor (EGFR) mutations, anaplastic lymphoma kinase fusion, and human telomerase reverse transcriptase overexpression, these drivers are at present the only therapeutic target genes for NSCLC [6-8]. Studying the molecular mechanisms of NSCLC, identifying new therapeutic targets and improving the treatment of patients with lung cancer are necessary.
Long noncoding RNAs (lncRNAs) are RNAs longer than 200 nucleotides that lack a coding region [9]. Although lncRNA is ubiquitously transcribed in humans and has a mature structure similar to that of mRNA, including its 5' capped architecture and 3' polyadenylation, lncRNA does not have the ability to be translated into protein [10,11]. However, it has various regulatory functions, including cis-acting and trans-acting elements, transcriptional regulation, and signal transduction [12-14]. LncRNAs can also act as a specific sponge of certain microRNAs (miRNAs), and adsorption of miRNAs inhibits their abilities [15-17]. The lncRNA cancer susceptibility candidate 8 (CASC8) has been shown to be a tumor susceptibility gene [18], but its mechanism of action is not clear.
The aim of this study was to investigate how CASC8 affects NSCLC through FOXM1 and examine the effect of CASC8 on osimertinib drug sensitivity.
Materials and Methods
Patients and specimens
Tumors and corresponding normal tissues were collected from patients seen between 2015 and 2019 at the First Hospital Affiliated of China Medical University; the corresponding normal tissue samples were obtained from a location at least 5 cm away from the tumor tissue. The Chinese Medical University Institutional Review Committee approved the study, and all patients provided informed consent. Patients who participated in the study did not receive radiation therapy or chemotherapy before surgery. The clinical prognostic data involved in this study were provided by the Kaplan Meier-plotter database (https://kmplot.com/analysis/).
Cell culture
The cell lines used in the study were all obtained from the Shanghai Cell Bank (Shanghai, China). Human bronchial epithelial (HBE) cells were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM); A549, H1299, and H460 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 media, these cell lines were all cultured in 10% qualified fetal bovine serum (FBS, FB15015; Clark Biosciences, Richmond, VA, USA); the H1975 and HCC827 cells were cultured in RPMI 1640 with 20% FBS.
Plasmid construction and transfection
CASC8 short hairpin RNA (113F078, CASC8-RNAi) and scramble small hairpin RNA-green fluorescent protein shRNA-GFP (negative control) were purchased from GENE (Shanghai, China). Transfection of shRNA was performed using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. shCASC8 or its negative control (NC) were transfected into selected cell lines, and the efficiency was tested after 24 hours. shRNA was also stably transfected into cell lines. Forty-eight hours after transient transfection, G418 (Thermo Fisher Scientific, Waltham, MA, USA) was added, and monoclonal stable-transfected cell lines were screened by limiting dilution. The cells were cultured in a 25T cell culture flask for subsequent experiments.
Western blotting
Total protein was extracted with lysis buffer (P0013; Beyotime Biosciences, Shanghai, China); the volume depended on the number of cells collected. Appropriate amounts of protease inhibitor and phosphatase inhibitor were added to the lysis buffer (B14002 and B15002; Biotool, Shanghai, China). Thirty-five micrograms of each sample were separated by SDS-PAGE and the protein was transferred to a PVDF membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% skim milk (232100; BD Co., Franklin Lakes, NJ, USA) for at least 2 hours, followed by gentle washing with Tween Tris buffered saline (TTBS) and overnight incubation at 4°C with the following primary antibodies: anti-Forkhead box M1 (FOXM1; 20459, 1:500 dilution, Cell Signaling Technology, Danvers, MA, USA), anti-E-cadherin (14472, 1:1000 dilution, Cell Signaling Technology), anti-Snail (2879, 1:1000 dilution, Cell Signaling Technology), anti-cyclin B1 (12231, 1:1000 dilution, Cell Signaling Technology), and anti-glyceraldehyde 3-phospate dehydrogenase (GAPDH) (TA319654, 1:1000 dilution, Origene). The next day, the membrane was incubated with peroxidase-conjugated secondary antibody for 2 hours at 37°C. Densitometry was quantitatively evaluated using ImageJ software.
Quantitative real-time polymerase chain reaction (qPCR)
Ribonucleic acid (RNA) was extracted using TRIzol reagent (15596026, Thermo Fisher Scientific) and used to generate cDNA. After the complementary deoxynucleic acid (cDNA) was generated, an appropriate amount was taken for qPCR using the PrimeScript RT Master Mix (RR036A, TaKaRa, Japan). The reaction volume was 20 µL. The qPCR conditions were 95°C for 30 seconds and 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. A dissociation step was used to generate a dissolution profile and confirm whether the amplification was specific. Relative gene expression was calculated using the 2-ΔΔCt method using GAPDH or U1 as controls. The primer sequences were as follows: CASC8 forward, 5'-TGGCATGGACCAGGAGCACTAG-3′, and reverse, 5′-GGACTTGCCGGTAACCTAGATTGG-3′; U1 small nuclear 1 (RNU1-1) forward, 5'-ACCCCTGCGATTTCCCCAAA-3′, and reverse, 5′-CAGGGGAAAGCGCGAACG-3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT -3′, and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′; and FOXM1 forward, 5′-GATCTGCGAGATTTTGGTACAC-3′, and reverse, 5′-CTGCAGAAGAAAGAGGAGCTAT-3′.
MTS proliferation assay
shCASC8 and NC cells were plated in 96-well culture plates at 3000 cells/well and cultured with cell strain-adapted FBS for 5 days in a sterile incubator at 37°C. A volume of 20 μL 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazole (MTS, G3580, Promega, Madison, Wisconsin, USA USA) was added to each well to test cell viability. After 1 hour incubation in a sterile incubator at 37°C in the absence of light, the absorbance at 490 nm was measured using a microplate reader to measure cell proliferation.
Colony formation assay
A total of 500 cells/well of shCASC8 and NC cells were plated in 6-well culture plates. Four milliliters of RPMI 1640 were added to each well, and cells were incubated in a sterile incubator at 37°C for 10-14 days, at which point colonies were visible to the naked eye. For the colony formation experiment with osimertinib, 500 corresponding cells were plated in each well, the IC50 concentration of the drug was added for 48 hours, and cells were incubated in a sterile incubator at 37°C for 10-14 days. Cells were washed three times with PBS and then fixed with pre-cooled methanol. After fixation, the cells were again washed three times with PBS and then stained with crystal violet. The crystal violet was removed, and the cells were dried and photographed. Cell colonies were counted for statistical analysis.
Invasion and migration analyses
Invasion assays were performed by adding 100 μL of Matrigel (1:9 dilution; BD Biosciences, San Jose, CA, USA) to an insert with a pore size of 8 μm (Costar, Washington, DC, USA). shCASC8 and NC cells were added to the transwell chamber and 600 μL RPMI 1640 with 20% FBS was added to the lower chamber. After 48 hours in a 37°C sterile incubator, cells in the upper chamber were removed with a cotton swab. Cells in the lower chamber were fixed with pre-cooled methanol, then stained with crystal violet. Cells were imaged and the number of cells that had passed through the transwell membrane was counted. Migration experiments were performed in the same fashion but without the use of Matrigel.
Wound-healing assay
shCASC8 and NC cells were plated in a single layer in a 6-well culture plate. When the cells had reached 100% confluence, they were incubated with mitomycin (1758-9327, Inalco, Beijing, China) for 2 hours in a light-free environment, after which the monolayers were gently scraped with a 100-μL pipette tip under sterile conditions. After gently removing the scraped cells and washing the well with PBS, the scratch was imaged. Images were taken at the stated time points, and the distance was measured using PS software.
Statistical analysis
All numerical data were analyzed by SPSS version 24.0 and GraphPad Prism 5. The χ2 test was used to assess the correlation between each indicator and CASC8 expression. At least three independent replicates were performed in each experiment, and P < 0.05 was considered statistically significant.
Results
CASC8 is upregulated in non-small cell lung cancer
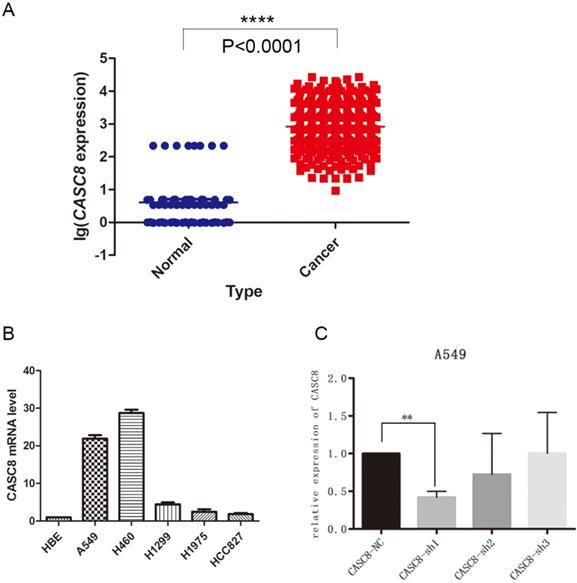
The lncRNA CASC8 is a susceptibility factor for cancer, but its specific role in NSCLC is not well understood. In this study, we analyzed the expression of CASC8 in NSCLC tissues. We downloaded expression data from The Cancer Genome Atlas (TCGA) database and analyzed CASC8 expression in 535 cancer tissues and 59 adjacent normal tissues. The expression of CASC8 was significantly higher in NSCLC tissues than in normal tissues adjacent to the tumor (P < 0.0001, Fig. 1A). We also assessed CASC8 expression in cell lines. We used the normal bronchial epithelial cell line HBE and five common NSCLC cell lines for qPCR. The level of CASC8 was significantly higher in the NSCLC cell lines than in HBE cells (Fig. 1B). We created cells with a stable knockdown of CASC8, which showed reduced expression of CASC8 (Fig. 1C).
Silencing of CASC8 inhibits proliferation, migration, and invasion in NSCLC cells
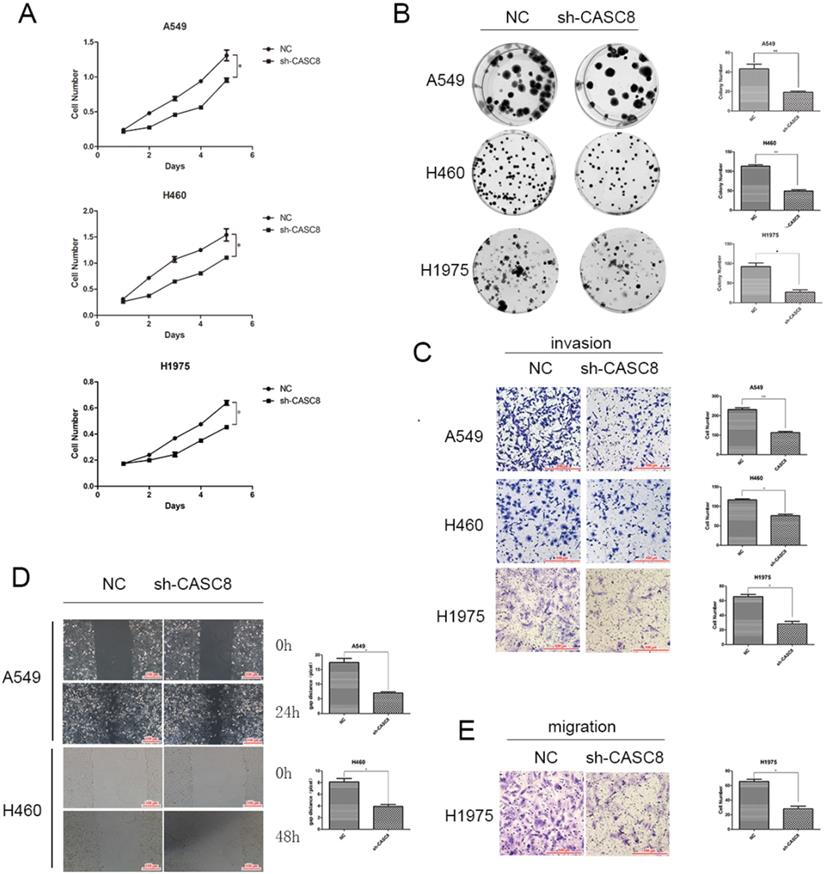
Based on the results of the qPCR and TCGA analyses, we examined the role of CASC8 in cells. Transfection of shCASC8 in A549, H460, and H1975 cells reduced the expression of CASC8. These cells had decreased proliferation (Fig. 2A) and colony-forming ability (Fig. 2B). Invasion was attenuated in cells with CASC8 knockdown (Fig. 2C), as was cell migration, as determined by the wound-healing assay (Fig. 2D) and the transwell assay without Matrigel (Fig. 2E). These results were statistically significant (*P < 0.05, **P < 0.01). These findings indicate that, as the level of CASC8 decreases, the migration distance and migration ability of cells decrease. This result corresponds to our previous results, and it can be concluded that decreasing CASC8 can inhibit vital functions in NSCLC cells.
CASC8 is upregulated in non-small cell lung cancer. A. The expression of CASC8 was analyzed in 535 lung cancer tissues and 59 normal lung tissues from TCGA database. The level of CASC8 was higher in cancer tissues than in normal tissues. ***P < 0.0001. B. Expression of CASC8 mRNA in the HBE (normal bronchial epithelial), A549 (NSCLC), H460 (NSCLC), H1299 (NSCLC), HCC827 (NSCLC), and H1975 (NSCLC) cell lines. C. Validation of CASC8 knockdown cells. **P < 0.001. Abbreviations: CASC8: cancer susceptibility candidate 8; TCGA: The Cancer Genome Atlas; NSCLC: non-small cell lung cancer.
CASC8 regulates genes involved in epithelial-to-mesenchymal transition (EMT) via FOXM1, and FOXM1 expression is associated with poor prognosis in NSCLC
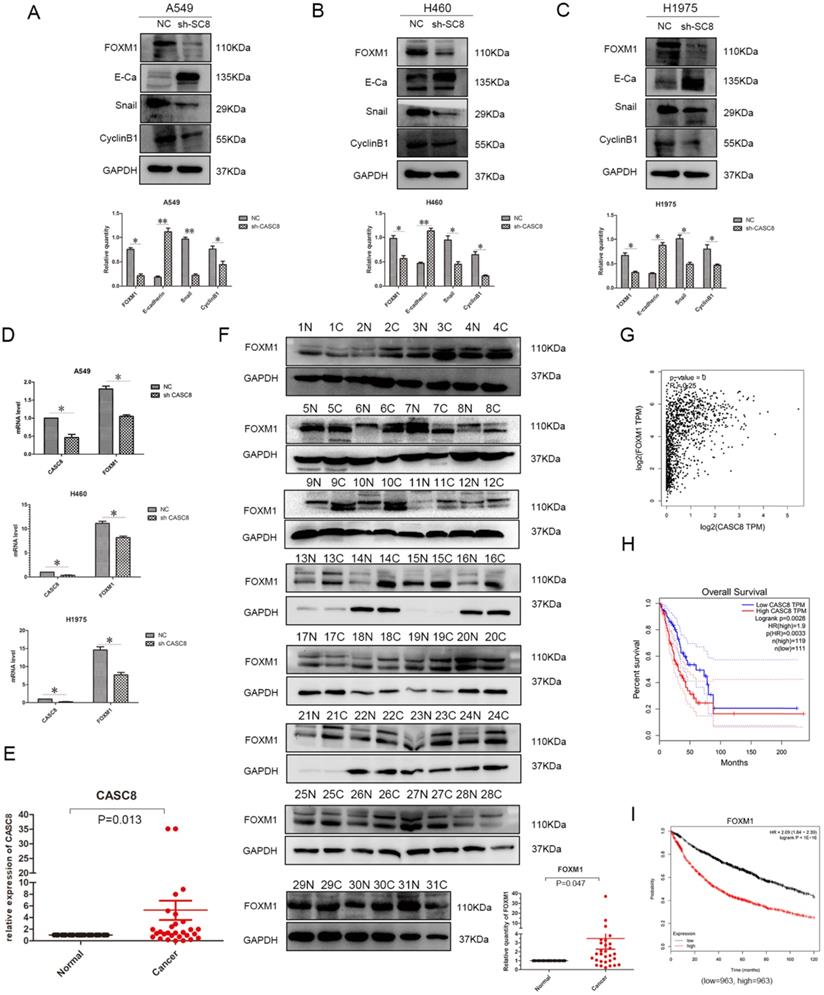
Understanding the effects of CASC8 on the in vitro behavior of NSCLC cells, we further explored its mechanism of action. We assessed the relationship between CASC8 and FOXM1 using western blotting. When CASC8 was knocked down, the protein level of FOXM1 decreased, and this was observed in A549 (Fig. 3A), H460 (Fig. 3B), and H1975 (Fig. 3C) cells. CASC8 silencing increased expression of E-cadherin and decreased expression of Snail. These findings suggest that CASC8 silencing inhibits EMT. Further, the expression of cyclin B1, a FOXM1 target, was also decreased, which may explain the results of the MTS and colony formation assays. In order to investigate whether CASC8 regulates FOXM1 at the RNA level, qPCR was performed. The mRNA level of FOXM1 decreased with CASC8 knockdown, indicating that CASC8 regulates FOXM1 at the transcriptional level. We also selected 31 paired cancer and adjacent tissues for qPCR (Fig. 3E) and western blot (Fig. 3F). Both the levels of CASC8 and the levels of FOXM1 were higher in the cancer tissues than in the adjacent normal tissues. Through the Gene Expression Profiling Interactive Analysis (GEPIA) database, we obtained the same results as the experimental results, that is, CASC8 and FOXM1 are positively correlated, and the relationship between the two is statistically significant (Fig. 3G, P < 0.0001, R=0.25). In addition, we evaluated the relationship between CASC8 expression and FOXM1 expression and patient prognosis using the GEPIA database and the Kaplan-Meier plotter database. CASC8 expression (Fig. 3H, P =0.0028) and FOXM1 expression (Fig. 3I, P < 0.0001) were associated with poor prognosis in NSCLC.
Silencing of CASC8 promotes sensitivity to osimertinib in NSCLC cells
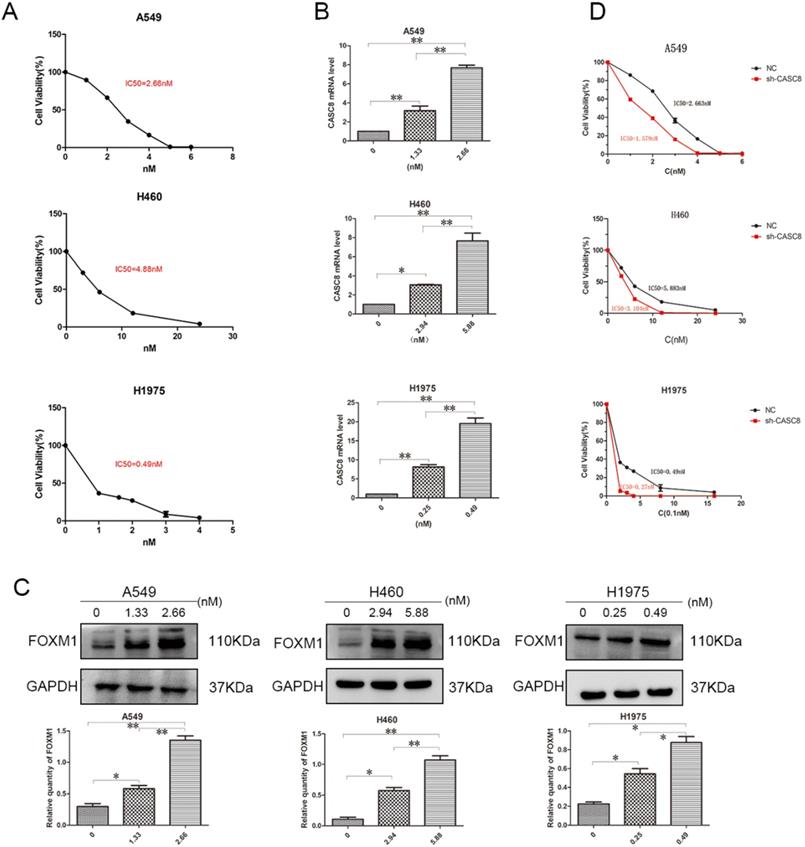
Targeted drugs are very important in the treatment of NSCLC, but resistance is an issue. Therefore, we examined the ability of CASC8 to act as a mechanism of resistance to the targeted drug osimertinib. First, to test the IC50 of osimertinib in A549, H460, and H1975 cells, a concentration gradient of osimertinib was added to the cells. After 48 hours, the IC50s of osimertinib in A549, H460, and H1975 cells were 2.663 nM, 5.838 nM, and 0.49 nM, respectively (Fig. 4A). Next, concentrations of osimertinib corresponding to the individual IC50s and half the concentration of the IC50s were added, and the CASC8 and FOXM1 levels were assessed. In all three cell lines, the level of CASC8 increased with increasing drug concentration, which proved that the increase in CASC8 was osimertinib-dependent (Fig. 4B). Similar results were obtained in western blot experiments. FOXM1 was also increased in an osimertinib-dependent manner (Fig. 4C). These results indicate that both CASC8 and FOXM1 have a role in resistance to osimertinib. After silencing CASC8, the IC50s of A549, H460, and H1975 cells decreased to 1.579 nM, 3.104 nM, and 0.27 nM, respectively (Fig. 4D). A colony formation experiment was conducted and it was found that when adding the IC50 concentration of osimertinib to the A549 cells, H460 cells and H1975 cells, the cells with lower CASC8 expression had the worst colony formation ability (Supplementary A). These experiments confirm our hypothesis that silencing CASC8 can promote sensitivity to osimertinib in NSCLC cells, which provides new insights for its clinical application.
Discussion
NSCLC has a huge burden on human health and life worldwide, and the treatment of NSCLC has caused enormous economic burdens for individuals and society. Research and treatment into the development of NSCLC is very important. In this study, the role of the lncRNA CASC8 in NSCLC was identified.
First, we showed that the expression of CASC8 was significantly higher in tumors than in adjacent tissues using the TCGA database, which provides a rationale for the study of CASC8. The A549 and H460 NSCLC cell lines, which have relatively high levels of CASC8, and the H1975 NSCLC cell line, which harbors an EGFR T790M mutation, were selected as experimental models for this study, and a series of in vitro experiments were performed.
In vitro behavioral experiments, we learned that silencing of CASC8 inhibits the proliferation, migration, and invasion of NSCLC cells. In addition, the expression of cyclin B1, which is associated with the G2/M transition, decreased with CASC8 knockdown, which corresponds to the results of the MTS assay and the colony formation assay. We were intrigued to find that FOXM1 also decreased with CASC8 knockdown. FOXM1 is a transcription factor that is required for cell proliferation, promotes tumor development, and is an oncogene [19]. FOXM1 promotes proliferation in NSCLC cells, and it can promote the transition from G2 to M phase via transcriptional regulation of cyclin B1 [20-22]. This result further underscored the role of CASC8 in NSCLC cells. However, how CASC8 affects FOXM1 is still unknown. The miRTarBase indicates that CASC8 can interact with miR-671-5P, and miR-761-5P can inhibit the transcription of FOXM1 [23]. Therefore, we hypothesize that CASC8 can act as a sponge of miR-671-5P. However, the exact role of CASC8 and miR-671-5P was not explored in this study, and this should be further verified. But through the GEPIA database and our experiments, we can prove that there is a positive regulatory relationship between CASC8 and FOXM1, and the relationship between the two is statistically significant. At the same time, the high expression of both is related to the poor prognosis of patients (from GEPIA and Kaplan Meier-Plotter).
FOXM1 is also essential for pulmonary fibrosis and EMT [24]. FOXM1 regulates the expression of E-cadherin [20,25], and E-cadherin downregulation is a hallmark of EMT. When the E-box sequence in the E-cadherin promoter is inhibited, Snail can bind to the E-box sequence, resulting in decreased expression of E-cadherin [26-28]. Snail plays a vital role in EMT as a procedural switch [29]. When E-cadherin expression is reduced, tumor cells undergo migration and invasion [30]. We observed several potential EMT-related changes with CASC8 knockdown, such as an increase in E-cadherin and a decrease in Snail. This may explain the decreased migration and invasion in NSCLC cell lines after CASC8 silencing.
Only a portion of early lung cancer can be cured by surgical resection. Further, more than 60% of patients with lung cancer are diagnosed with locally advanced lung cancer or have lymph node metastasis (stage III or IV), and surgery is not the primary treatment option [5]. EGFR is a commonly mutated gene in NSCLC. Although some therapeutic targets have been identified, patients inevitably develop resistance, limiting the utility of targeted therapies. Therefore, identifying mechanisms of resistance is vital for improving patient survival.
EGFR-targeted treatment is widely used in the clinic for patients with lung cancer. Early tyrosine kinase inhibitors (TKIs) such as gefitinib and afatinib have good effects in the initial stage of treatment, but the effect cannot be extended beyond 10-14 months, and patients inevitably develop resistance [31-34]. Interestingly, FOXM1 also plays a role in NSCLC resistance to gefitinib, [22] although the most common cause of drug resistance is the T790M mutation [35]. Osimertinib is a third-generation TKI that has been proved to overcome the EGFR T790M mutation [36] and shows better clinical efficacy that gefitinib and erlotinib [37]. Although tumors are more sensitive to later-generation EGFR-TKIs, there is still a drug resistance phenomenon [38,39]. In this study, we demonstrated that silencing of CASC8 can sensitize NSCLC cells to osimertinib, identifying a potential mechanism of osimertinib resistance. However, whether the silencing of CASC8 promotes sensitivity to osimertinib in vivo has not been verified and needs to be confirmed.
Silencing of CASC8 inhibits the proliferation, migration, and invasion of non-small cell lung cancer cells. A. Silencing of CASC8 decreased the proliferation of NSCLC cells. Top panel, A549; middle panel, H460; bottom panel, H1975, *P < 0.05. B. Colony formation assay to detect the effect of CASC8 silencing on the colony-forming ability of NSCLC cells. Right panel, quantification, **P < 0.01, ***P < 0.0001. C. Silencing of CASC8 impairs the invasive ability of NSCLC cells (Transwell invasion assay). Left panel, quantification, *P < 0.05, **P < 0.01. D. Wound-healing assay using A549 and H460 cells after shCASC8 transfection. The cell migration distance was shorter in shCASC8 cells than in control cells. Right panel, quantification, *P < 0.05. E. Transwell migration assay using H1975 cells. Right panel, quantification, *P < 0.05. Abbreviations: CASC8: cancer susceptibility candidate 8; NSCLC: non-small cell lung cancer.
CASC8 induces EMT via FOXM1, and FOXM1 expression is associated with poor prognosis in non-small cell lung cancer. A. Changes in expression levels FOXM1, E-cadherin, Snail, and cyclin B1 after CASC8 knockdown in A549 cells. Left panel, quantification, *P < 0.05, **P < 0.01. B. As in (A) for H460 cells; C. as in (A) for H1975 cells. D. The FOXM1 levels decreased after CASC8 silencing. E. 31 pairs of clinical tissue samples were collected and qPCR was performed. The expression of CASC8 was higher than in NSCLC tissues in adjacent tissues, P < 0.0001. F. Western blot analysis was performed using the same tissue as in (Fig. 3E).The expression of FOXM1 was higher than in NSCLC tissues in adjacent normal tissues. Bottom panel, quantification, *P < 0.05, **P < 0.01. G. Derived from the GEPIA database, indicating that there is a positive correlation between CASC8 and FOXM1 at the gene level and has statistical significance (P = 0, R = 0.25). H. Derived from GEPIA database, high expression of CASC8 is associated with poor patient prognosis in lung adenocarcinoma (P=0.0028). I. High expression of FOXM1 is associated with poor prognosis of NSCLC, P < 0.0001. Abbreviations: CASC8: cancer susceptibility candidate 8; EMT: epithelial-to-mesenchymal transition; FOXM1: Forkhead box M1, NSCLC: non-small cell lung cancer; qPCR: quantitative real-time polymerase chain reaction.
Silencing of CASC8 promotes sensitivity to osimertinib in non-small cell lung cancer cells. A. The half-lethal dose (IC50) of osimertinib was determined in A549, H460, and H1975 cells using an appropriate concentration gradient. After 48 hours of culture, the IC50s of A549, H460, and H1975 cells were 2.663 nM, 5.883 nM, and 0.49 nM, respectively. B. Concentrations of osimertinib corresponding to the appropriate IC50 and half the IC50 were added to the three cell lines. Cells were cultured for 48 hours and RNA was extracted for qPCR. The change in CASC8 expression was osimertinib-dependent. C. Cells were cultured for 48 hours at different drug concentrations and then subjected to western blot analysis. The change in the FOXM1 levels was osimertinib-dependent. Bottom panel, quantification, *P < 0.05, **P < 0.01. D. The IC50s of shCASC8 A549, H460, and H1975 cells were significantly lower than those of their corresponding NC cells, *P < 0.05, **P < 0.01. Abbreviations: CASC8: cancer susceptibility candidate 8; FOXM1: Forkhead box M1, qPCR: quantitative real-time polymerase chain reaction; NC: negative control.
Conclusions
In summary, the lncRNA CASC8 promotes the proliferation, migration, and invasion of NSCLC cells. CASC8 also sensitizes NSCLC cells to osimertinib. However, whether CASC8 can be used as a standard for the diagnosis of NSCLC still requires further experimental verification.
Abbreviations
NSCLC: non-small cell lung cancer; EGFR: epidermal growth factor receptor; lncRNA: long noncoding RNA; miRNA: microRNA; CASC8: cancer susceptibility candidate 8; NC: negative control; FOXM1: Forkhead box M1; qPCR: quantitative real-time polymerase chain reaction; EMT: epithelial-to-mesenchymal transition; MTS: 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazole; TCGA: The Cancer Genome Atlas; TKI: tyrosine kinase inhibitor; shRNA-GFP: scramble small hairpin RNA-green fluorescent protein; FBS: fetal bovine serum; HBE: human bronchial epithelial; DMEM: modified eagle medium; RPMI: Roswell Park Memorial Institute; TTBS: Tween Tris buffered saline; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; RNA: Ribonucleic acid; cDNA: complementary deoxynucleic acid; GEPIA: Gene Expression Profiling Interactive Analysis.
Supplementary Material
Supplementary figure.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Ethics approval and informed consent
The tissues used in this experiment were all approved by the patient.
Data availability
Kaplan Meier-Plotter: https://kmplot.com/analysis/. GEPIA (Gene Expression Profiling Interactive Analysis): http://gepia.cancer-pku.cn/.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Peng W, Wang J, Shan B. et al. Diagnostic and Prognostic Potential of Circulating Long Non-Coding RNAs in Non-Small Cell Lung Cancer. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;49:816-27
2. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
3. Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
4. Global Burden of Disease Cancer C, Fitzmaurice C, Dicker D. et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505-27
5. Osmani L, Askin F, Gabrielson E. et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103-9
6. Maemondo M, Inoue A, Kobayashi K. et al. Gefitinib or Chemotherapy for Non-Small-Cell Lung Cancer with Mutated EGFR. N Engl J Med. 2010;362:2380-88
7. Shaw AT, Kim D-W, Nakagawa K. et al. Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N Engl J Med. 2013;368:2385-94
8. Eldholm V, Haugen A, Zienolddiny S. CTCF mediates the TERT enhancer-promoter interactions in lung cancer cells: Identification of a novel enhancer region involved in the regulation of TERT gene. Int J Cancer. 2014;134:2305-13
9. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491-509
10. Guttman M, Russell P, Ingolia NT. et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240-51
11. Bánfai B, Jia H, Khatun J. et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646-57
12. Nagano T, Mitchell JA, Sanz LA. et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717-20
13. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904-14
14. Kino T, Hurt DE, Ichijo T. et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8
15. Franco-Zorrilla JM, Valli A, Todesco M. et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033-37
16. Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res. 2017;77:3135-39
17. Poliseno L, Salmena L, Zhang J. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-38
18. Cui Z, Gao M, Yin Z. et al. Association between lncRNA CASC8 polymorphisms and the risk of cancer: a meta-analysis. Cancer Manag Res. 2018;10:3141-48
19. Bella L, Zona S, Nestal de Moraes G. et al. FOXM1: A key oncofoetal transcription factor in health and disease. Semin Cancer Biol. 2014;29:32-9
20. Lee JH, Lee HJ, Sim DY. et al. Apoptotic effect of lambertianic acid through AMPK/FOXM1 signaling in MDA-MB231 breast cancer cells. Phytother. Res. 2018;32:1755-63
21. Costa RH, Kalinichenko VV, Major ML. et al. New and unexpected: forkhead meets ARF. Curr Opin Genet Dev. 2005;15:42-8
22. Xu N, Zhang X, Wang X. et al. FoxM1 mediated resistance to gefitinib in non-small-cell lung cancer cells. Acta pharmacologica Sinica. 2012;33:675-81
23. Tan X, Fu Y, Chen L. et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7:293-307
24. Balli D, Ustiyan V, Zhang Y. et al. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 2013;32:231-44
25. Wei P, Zhang N, Wang Y. et al. FOXM1 promotes lung adenocarcinoma invasion and metastasis by upregulating SNAIL. Int J Biol Sci. 2015;11:186-98
26. Hennig G, Lowrick O, Birchmeier W. et al. Mechanisms identified in the transcriptional control of epithelial gene expression. J Biol Chem. 1996;271:595-602
27. Batlle E, Sancho E, Franci C. et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84-9
28. Cano A, Perez-Moreno MA, Rodrigo I. et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76-83
29. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415-28
30. Hou H, Yu X, Cong P. et al. Six2 promotes non-small cell lung cancer cell stemness via transcriptionally and epigenetically regulating E-cadherin. Cell Proliferat. 2019;52:e12617
31. Dearden S, Brown H, Jenkins S. et al. EGFR T790M mutation testing within the osimertinib AURA Phase I study. Lung cancer. 2017;109:9-13
32. Mok TS, Wu YL, Thongprasert S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947-57
33. Wu YL, Zhou C, Hu CP. et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213-22
34. Zhou C, Wu YL, Chen G. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735-42
35. Oxnard GR, Arcila ME, Sima CS. et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin can res. 2011;17:1616-22
36. Tang ZH, Lu JJ. Osimertinib resistance in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Lett. 2018;420:242-6
37. Aguiar PN Jr, Haaland B, Park W. et al. Cost-effectiveness of Osimertinib in the First-Line Treatment of Patients With EGFR-Mutated Advanced Non-Small Cell Lung Cancer. JAMA Oncol. 2018;4:1080-84
38. Oxnard GR, Hu Y, Mileham KF. et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018;4:1527-34
39. Uchibori K, Inase N, Araki M. et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nature comm. 2017;8:14768
Author contact
![]() Corresponding author: Xueshan Qiu, Department of Pathology, College of Basic Medical Sciences, China Medical University, Shenyang, China. E-mail: xsqiuedu.cn; 13386881964.
Corresponding author: Xueshan Qiu, Department of Pathology, College of Basic Medical Sciences, China Medical University, Shenyang, China. E-mail: xsqiuedu.cn; 13386881964.

 Global reach, higher impact
Global reach, higher impact