Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(2):428-437. doi:10.7150/jca.48423 This issue Cite
Research Paper
Long noncoding RNA DLEU2 affects the proliferative and invasive ability of colorectal cancer cells
1. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
2. Department of Pathology, Dezhou People's Hospital, Dezhou 253056, Shandong, China.
3. Department of Orthopedics, The First Affiliated Hospital of Hunan University of Chinese Medicine, Changsha 410007, Hunan, China.
4. Department of Hematology, Xiangya hospital, Central South University, Changsha 410008, Hunan, China.
5. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
Received 2020-5-20; Accepted 2020-10-24; Published 2021-1-1
Abstract
Emerging evidence indicates that long noncoding RNAs (lncRNAs) are closely associated with colorectal cancer (CRC) tumorigenesis. One example is lncRNA Deleted in Lymphocytic Leukemia 2 (DLEU2). However, how DLEU2 contributes to CRC is still poorly understood. This study sought to investigate the effects of DLEU2 on CRC pathogenesis, and the underlying mechanism involved. Using a quantitative real-time polymerase chain reaction (qRT-PCR) assay, we demonstrated that the expression levels of DLEU2 in 45 pairs of CRC tissues were higher than those in the corresponding normal colon mucosal tissues. In addition, CRC patients with high DLEU2 expression levels exhibited poor overall survival (OS) and recurrence-free survival (RFS), as determined by analyses and measurements from the GEO and GEPIA databases. When DLEU2 was silenced using short interfering RNA (siRNA) in CRC cell line, the results demonstrated that DLEU2 silencing suppressed CRC cell tumorigenesis in vitro, which was associated with decreased expression of cyclin dependent kinase 6(CDK6), ZEB1, and ZEB2 as well as enhancing the expression of Cyclin-dependent kinase inhibitor 1A (CDKN1A). Taken together, the results of this study suggested that DLEU2 may play critical roles in the progression of CRC and may serve as a prognostic biomarker for CRC.
Keywords: DLEU2, colorectal cancer, invasion, tumorigenesis, survival
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy and the second leading cause of cancer-related deaths worldwide [1-3]. According to epidemiological investigations, in 2012, there were approximately 1.36 million new cases of CRC, and approximately 690,000 deaths, ranking fourth among all malignant tumors [1]. With the development of therapeutic approaches, substantial progress has been achieved in treating CRC over the past decades. However, the CRC mortality rate has not changed due to tumor cells metastasis [4,5]. CRC progression has been identified as a multistep process that involves inherited and environmental factors [6,7]. With the popularity of high-throughput sequencing and molecular therapy, an increasing number of studies have focused on the molecular pathogenesis of CRC, with the aim of exploring suitable biomarkers for identifying the progression and development of CRC [8-10].
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs (ncRNAs) defined by a transcript length of 200-100,000 nt, and a lack of a complete functional open reading frame (ORF) [11-14]. The molecular functions of lncRNAs at the epigenetic, transcriptional, and post-transcriptional levels are conventionally subdivided as follows: 1) acting as host genes for miRNAs; 2) functioning as decoys; 3) acting as a co-regulator or co-repressor; 4) recruiting and interacting with proteins; and 5) interacting with miRNAs. LncRNAs play important roles in the aspects of tissue differentiation and reproduction, individual development, and immunity [11-14]. Recent studies have shown that lncRNAs play an important role in various cancers, such as nasopharyngeal carcinoma [15], CRC [16], gastric cancer [17], glioblastoma [18], and cervical cancer [19]. These lncRNAs with dysregulated expression may be potential biomarkers valuable in the screening, diagnosis, and therapy of cancers.
In this study, we analyzed two previously published online datasets to analyze the dysregulated expression of lncRNAs in CRC. We found that Deleted in Lymphocytic Leukemia 2 (DLEU2) was significantly overexpressed in the two CRC datasets. DLEU2 is located in the chr13q14.2 region and was originally identified as an important tumor regulator gene [20], whereas little is known concerning the functions and mechanisms of DLEU2 involvement CRC tumorigenesis. In this study, we showed that DLEU2 was overexpressed in CRC tissues and cell lines, and high expression levels in CRC patients were associated with poor overall survival (OS) and recurrence-free survival (RFS). Furthermore, DLEU2 knockdown in CRC cells inhibited malignant proliferation and metastasis. Taken together, these results suggested that DLEU2 may play a critical role in the progression and development of CRC.
Materials and Methods
Tissue samples
A total of 45 CRC lesions and matched adjacent tissues obtained from newly diagnosed CRC patients were collected at the Xiangya Hospital of Central South University. This study was approved by the hospital Research Ethics Board of Xiangya Hospital of Central South University, and all participants provided informed consent for inclusion in the study.
Bioinformatics analysis
All CRC datasets were deposited in the Gene Expression Omnibus (GEO) database: GSE37364 [21], GSE23878 [22], GSE41328 [23] and GSE17538 [24] (these datasets were generated using the Affymetrix Human Genome U133 Plus 2.0 platform). The GSE50760 [25] dataset was obtained using the Illumina HiSeq 2000 platform. The GSE37364 dataset has 56 primary CRC samples and 38 normal colon samples, GSE23878 has 35 primary CRC samples and 24 normal colon samples, GSE41328 has 10 pairs of CRC and adjacent non-tumour tissues, and GSE50760 has 18 pairs of metastasis CRC samples and non-metastatic CRC samples.
GSE17538 contains clinical follow-up data for 231 CRC samples. Based on the results of log-rank tests, GSE17538 was divided into a low DLEU2 expression group (n = 135, i.e. DLEU2low), and a high DLEU2 expression group (n = 96, i.e. DLEU2high). Subsequently, gene set enrichment analysis (GSEA) [26, 27] was used to identify gene set differences between the two groups.
Cell culture and transfection
CRC cell lines (SW480, HT29, LoVo, SW620, and CaCO2) and human normal colon mucosal cell line (NCM460) were obtained from the American Type Culture Collection (ATCC, Manassas, USA). These cell lines were cultured in RPMI-1640 medium with 10% foetal bovine serum, and HCT116 cells were cultured in Dulbecco's modified Eagle's medium with 10% Tris-buffered saline.
When cell densities reached almost 60%, 50 nM siRNA-DLEU2 oligo or a negative control (siRNA-NC) were transfected using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer's instructions. The sequences of the DLEU2 targeting siRNAs were: siRNA-DLEU2-1, 5'- AGUCUACGUUGGAGGUAAA -3'; and siRNA-DLEU2-2, 5'- AAGUAUUCAAUCAAAGAAGUG -3'; Sequences of non-target scramble controls were provided by RiboBio (Guangzhou, China).
Subcellular fractionation analysis
Approximately 1 × 107 HT29 cells were collected in order to determine the cellular localisation of DLEU2. Nuclear and cytoplasmic RNAs were collected using a PARIS Kit (Invitrogen). The computing methods employed were described previously [28].
Quantitative real-time polymerase chain reaction
RNA isolation and amplification and quantitative real-time polymerase chain reaction (qRT-PCR) were performed as described previously [29]. The thermocycling program used was as follows: 95°C for 30 sec, followed by 40 cycles of 60°C for 30 sec and 72°C for 30 sec. Primer sequences for qRT-PCR are shown in Table 1.
Primer sequence for real-time PCR
| Gene | Primer (Forward) | Primer (Reverse) |
|---|---|---|
| DLEU2 | TCCGAGAGTATAGCGCCACT | ACTGCCCTTTGCTCCAAGTA |
| CDK6 | GCTGACCAGCAGTACGAATG | GCACACATCAAACAACCTGACC |
| CDKN1A | CGATGGAACTTCGACTTTGTCA | GCACAAGGGTACAAGACAGTG |
| ZEB1 | GATGATGAATGCGAGTCAGATGC | ACAGCAGTGTCTTGTTGTTGT |
| ZEB2 | GCGATGGTCATGCAGTCAG | CAGGTGGCAGGTCATTTTCTT |
| β-actin | TCACCAACTGGGACGACATG | GTCACCGGAGTCCATCACGAT |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| GAPDH | AACGGATTTGGTCGTATTGG | TTGATTTTGGAGGGATCTCG |
Flow cytometry for cell cycle analysis
After transfection with si-NC or si-DLEU2, approximately 1 × 106 HT29 cells were collected for flow cytometry assays, and DNA content was detected using propidium iodide (Sigma, San Antonio, USA) staining according to the methods described in a previous study [30]. Analysis of cell cycle distribution used cell ModFit software (Beckman Coulter, South Kraemer, USA). Each experiment was repeated three times independently.
CCK8 assay
Cell proliferation was assayed using the Cell Counting Kit-8 (CCK-8) (Dojin Laboratories, Japan) according to the methods described previously [31]. Each experiment was repeated three times independently.
Western blotting
Cell lysis, electrophoresis, and target protein visualisation were performed according to the methods described previously [32].
Transwell Matrigel assay
Transwell Matrigel assay were performed using a 24-well transwell plate (8-μM pore size, Costar, America) to detect the invasiveness of CRC cells according to previously described methods [33]. Briefly, the number of invasive tumor cells was calculated from the total number of cells from three randomly selected 20× fields for each experiment. The histogram represents data from three independent experiments.
Statistical analyses
All statistical analyses were performed using GraphPad Prism v.7.0 software and Statistical Package for the Social Sciences version 18.0. Data are shown as mean ± standard error of the mean (SEM), and the results of analysis were considered significant in log-rank tests if p < 0.05.
Results
DLEU2 is upregulated in CRC
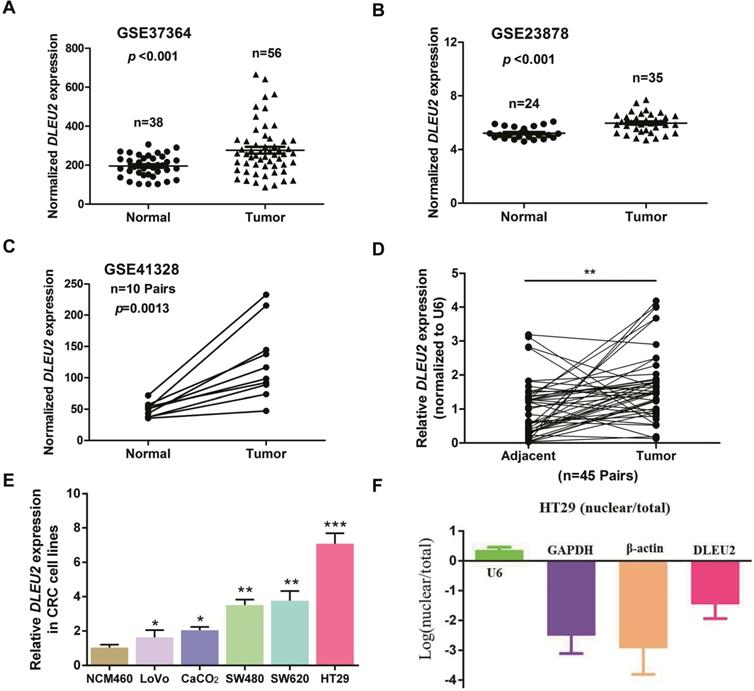
To identify dysregulated lncRNAs in CRC, we analyzed three online GEO datasets (#GSE37364, GSE23878, and GSE41328) based on the Affymetrix Human Genome U133 Plus 2.0 platform. lncRNA DLEU2 was significantly upregulated in CRC tissues compared with non-tumor tissues (p < 0.05, Fig. 1A-C). Furthermore, we detected DLEU2 expression in 45 matched pairs of CRC samples and adjacent non-tumor tissues, and our results indicated that DLEU2 was more highly expressed in CRC tissues than in the adjacent non-tumor tissues (p < 0.05, Fig. 1D). DLEU2 expression levels were also determined by qRT-PCR in five CRC cell lines (SW480, HT29, LoVo, SW620, and CaCO2) and in the normal colon mucosal cell line NCM460. DLEU2 expression was higher in CRC cell lines than in NCM460 cells (all p < 0.05, Fig. 1E), and DLEU2 expression was highest in HT29 cells. Moreover, we analyzed the nuclear and cytoplasmic distribution of DLEU2 and found that the expression of DLEU2 was higher in cytoplasm, indicating that the subcellular localization of DLEU2 in CRC cells was primarily cytoplasmic (Fig. 1F).
Association between DLEU2 expression and clinicopathological features of CRC
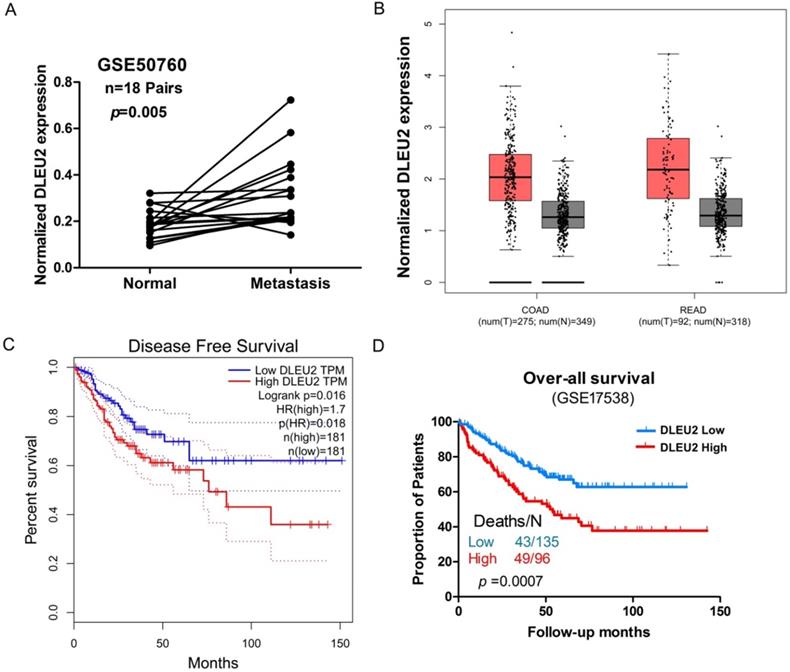
We next assessed potential correlations involving DLEU2 expression with clinicopathological features in CRC. We assessed the correlation between DLEU2 expression and distant metastasis in CRC tissues by analyzing a previously published Affymetrix HG_U133 Plus 2 array dataset (#GSE50760). Elevated expression of DLEU2 was significantly correlated with CRC distant metastasis (p = 0.004, Fig. 2A). Next, by analyzing public CRC datasets in GEPIA [34], we found that DLEU2 was significantly upregulated in colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) samples from TCGA data (p < 0.05, Fig. 2B; Supplemental Fig. 1), and that high DLEU2 expression was associated with poor RFS (p < 0.05, Fig. 2C). We also examined the association between DLEU2 expression levels and OS in the GSE17538 database using Kaplan-Meier analysis with log-rank tests. The results revealed that patients with high DLEU2 expression levels had lower OS (p < 0.05, Fig. 2D). Taken together, these data indicated that high DLEU2 expression is an independent risk factor for CRC patients.
Knockdown of DLEU2 expression in CRC cells inhibits proliferative and metastatic phenotypes
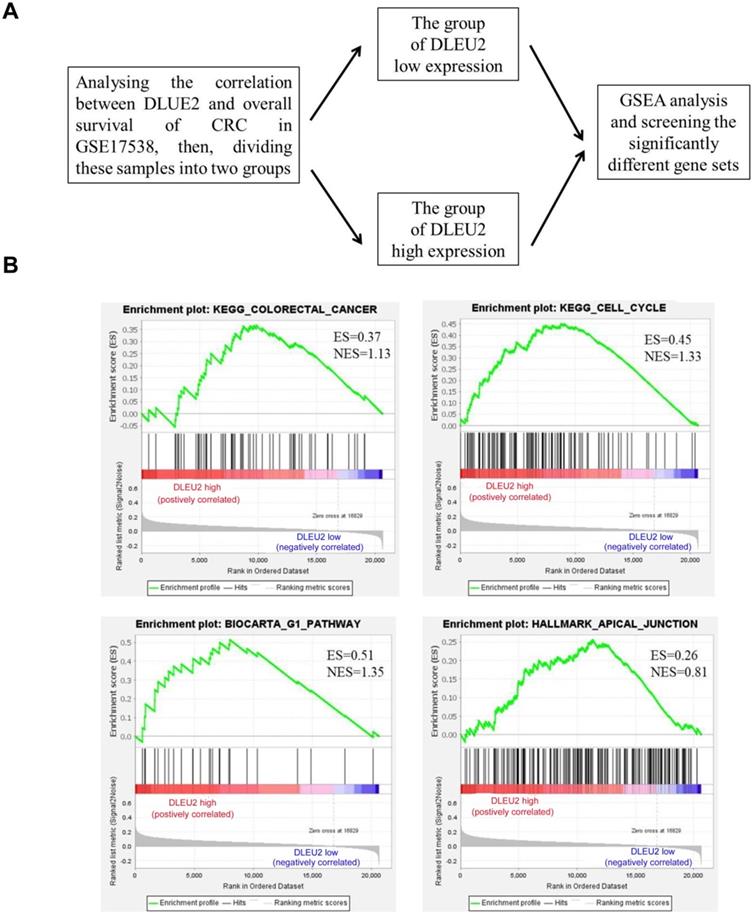
To explore the function of DLEU2 in CRC cells, GSEA was used to analyze the significantly-different gene sets between CRC specimens with high DLEU2 expression (DLEU2high) and low DLEU2 expression (DLEU2low) (Fig. 3A). High expression of DLEU2 was positively correlated with colorectal cancer, cell cycle, G1 pathway, and apical junction signaling sets (Fig. 3B-E; Supplemental Fig. 2-5).
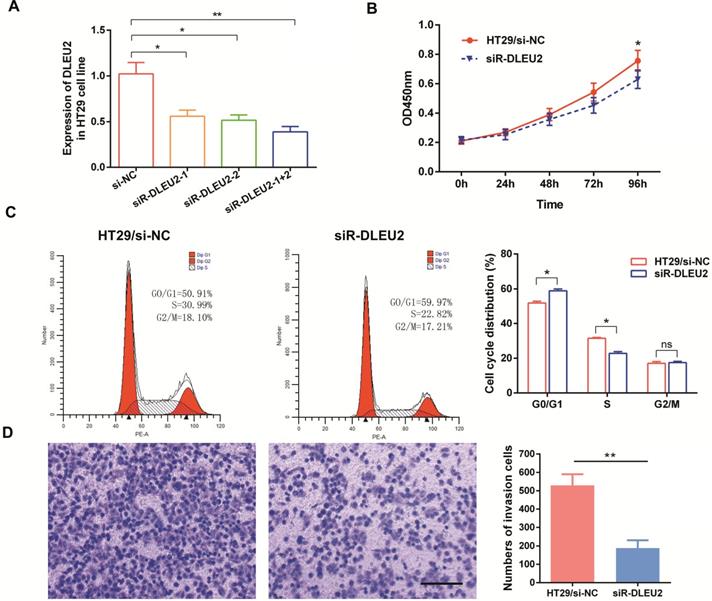
We also measured the efficiency of short interfering RNA (siRNA) siR-DLEU2. DLEU2 expression in both siRNA siR-DLEU2-1 + 2 groups was significantly down-regulated compared to siR-DLEU2-1 and siR-DLEU2-2 groups in HT29 cells (Fig. 4A). After establishing siRNA efficacy, we assessed the effects of DLEU2 knockdown in CRC cells. We found that knocking-down DLEU2 expression significantly inhibited HT29 cell proliferation relative to control cells in 96h (p < 0.05, Fig. 4B). Flow cytometric analysis showed that knocking-down DLEU2 expression in HT29 cells increased the percentage of cells in G1 phase, and decreased the percentage of cells in S phase (p < 0.05, Fig. 4C). Subsequently, we explored the effects of DLEU2 knockdown on the invasiveness of CRC cells using transwell matrigel assays. Knocking-down DLEU2 expression significantly inhibited the invasive capacity of HT29 cells compared to control group cells (p < 0.05, Fig. 4D). These findings suggested that DLEU2 plays a significant role in CRC tumorigenesis.
DLEU2 is upregulated in CRC. (A) #GSE37364 (38 normal colon samples and 56 primary CRC samples), (B) #GSE23878 (24 normal colon samples and 35 primary CRC samples) and (C) #GSE41328 (containing 10 pairs of CRC tissues and corresponding normal colorectal tissues) identified in the GEO database were used to analyze the expression of DLEU2. (D) DLEU2 was highly expressed in the 45 pairs of CRC tissues compared with the corresponding normal colorectal tissues. (E) DLEU2 expression was significantly increased in CRC cell lines (SW480, HT29, LoVo, SW620, and CaCO2) compared with NCM460, a normal colon mucosal cell line. (F) Nuclear and cytoplasmic RNA were analyzed by qRT-PCR to detect the expression levels of DLEU2 in HT29 cells. GPADH and β-Actin were used as cytoplasmic RNA controls; U6 was used as a nuclear RNA control. Data are shown as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control.
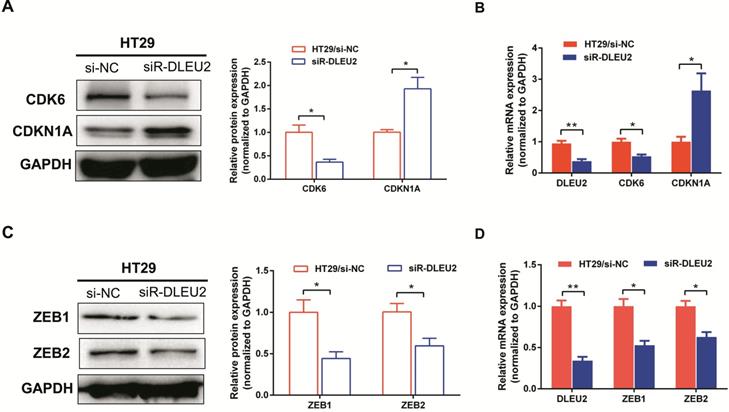
Knockdown of DLEU2 expression in CRC cells influences the expression of proliferation- and EMT-related genes
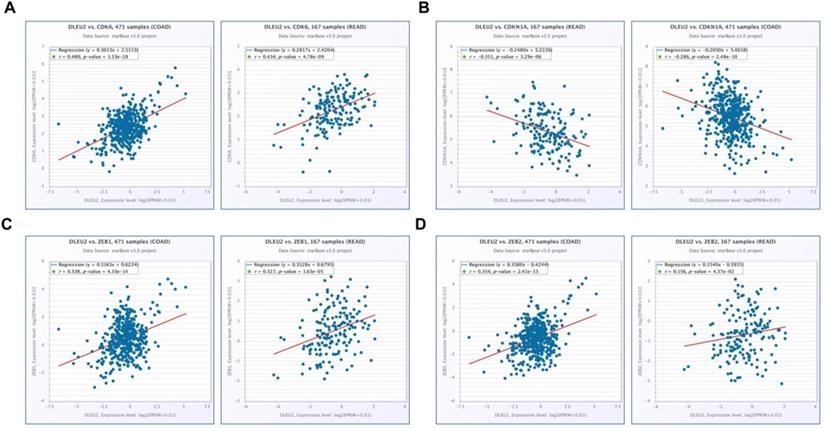
To further elucidate the molecular mechanisms by which knockdown of DLEU2 expression suppressed proliferation and invasion by CRC cells in vitro, we used qRT-PCR and western blotting to assess mRNA and protein levels of the proliferation markers cyclin‑dependent kinase 6 (CDK6) and cyclin-dependent kinase inhibitor 1A (CDKN1A), as well as the epithelial-mesenchymal transition (EMT) markers ZEB1 and ZEB2 in HT29 cells. Knocking-down DLEU2 expression significantly inhibited expression of CDK6, and promoted the expression of CDKN1A (p < 0.05, Fig. 5A-B). Meanwhile, DLEU2 knock-down significantly decreased the expression of the EMT markers ZEB1 and ZEB2 (p < 0.05, Fig. 5C-D). Starbase database (http://starbase.sysu.edu.cn/) analysis showed that DLEU2 expression was positively associated with expression of CDK6, ZEB1, and ZEB2, and negatively associated with expression of CDKN1A in COAD and READ samples (all p < 0.05, Fig. 6). These results indicated that DLEU2 may contribute to the regulation of proliferative and EMT marker expression in CRC cells.
Association between DLEU2 expression and clinicopathological features of CRC. (A) Relative expression of DLEU2 in normal and metastatic CRC tissues was obtained from the GEO database (#GSE50760, 18 matched metastatic CRC samples and adjacent non-metastatic samples). (B) The GEPIA database was used to analyze DLEU2 expression in colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) samples. (C) The GEPIA database was used to analyze the clinic impact of DLEU2 expression patterns on CRC patient recurrence-free survival (RFS). (D) Kaplan-Meier analysis showing overall survival (OS) curves for CRC patients with different expression levels of DLEU2; statistical significance was assessed by log-rank tests (#GSE17538, the specimen was divided into two groups: group 1, DLEU2low, n = 135; group 2, DLEU2high, n = 96).
Discussion
Evidence from recent studies has demonstrated that lncRNAs regulate gene expression at the epigenetic, transcriptional and posttranscriptional levels [12, 35]. LncRNAs are also involved in the pathogenesis of many diseases, especially cancer. Disruption of lncRNA levels is closely correlated with cancer cell proliferation and apoptosis, EMT, and drug resistance [36, 37]. More and more lncRNAs have been reported as being differentially-expressed in CRC and with correlations involving poor prognosis, including MALAT1 [38], LINC01234 [39], AFAP1-AS1 [40], GAS5 [41], LINC00152 [42], NEAT1 [43], among others. lncRNAs are considered ideal biomarkers for tumor diagnosis and monitoring because they can be detected with high specificity and sensitivity, are easy to extract, and are stable in blood and tissues [44].
With the popularity of high-throughput sequencing and RNA-Seq gene microarray technologies, many public databases (for example TCGA, GEO, and Oncomine) have emerged as powerful tools to be used to predict and analyze potentially valuable lncRNAs [45]. In this study, we first analyzed dysregulated lncRNAs in CRC using three GEO datasets (#GSE37364, GSE41328, and GSE23878), and found that lncRNA Deleted in Lymphocytic Leukemia 2 (DLEU2) was significantly overexpressed in two CRC datasets. DLEU2, located in the chr13q14.2 region, was first reported in lymphocytic leukemia [46]. Recent studies have shown that DLEU2 is upregulated in many human cancers, such as non-small cell lung cancer (NSCLC) [47], esophageal adenocarcinoma [48], and osteosarcoma [49]. However, the roles and the mechanisms involving DLEU2 in CRC have rarely been reported. In this study, we found that overexpression of DLEU2 in CRC patients was associated with poorer RFS and OS, independently of other factors by multivariate analysis.
The significantly-different gene sets between CRC specimens with high DLEU2 expression (DLEU2high) and low DLEU2 expression (DLEU2low) were analyzed. (A) Schematic flowchart illustrating the strategy to analyze gene set differences between DLEU2high and DLEU2low cells in CRC specimens from the GSE17538 database revealed by gene set enrichment analysis (GSEA). (B) GSEA identified gene set differences between DLEU2high and DLEU2low. ES: enrichment score; NES: normalized enrichment score.
Knockdown of DLEU2 expression in CRC cells inhibits proliferative and metastatic phenotypes. (A) After transfection of HT29 cells with either si-NC or siR-DLEU2 (1#, 2#, 1+2#) for 48 h, the expression of DLEU2 was analyzed by qRT-PCR. CCK8 assays (B) and flow cytometry (C) were used to detect cell proliferative ability, and transwell matrigel assays (D) were used to detect cell metastatic ability. Data are shown as means ± SEM. *p < 0.05, **p < 0.01 compared with control.
Knockdown of DLEU2 expression suppresses the expression of proliferation and metastatisis markers in CRC. After transfection with si-NC or siR- DLEU2 for 48 h, (A-B) mRNA and protein expression levels of cyclin dependent kinase 6 (CDK6) and cyclin-dependent kinase inhibitor 1A (CDKN1A) in HT29 cells were analyzed by qRT-PCR, western blotting and densitometry; (C-D) the mRNA and protein expressionof ZEB1 and ZEB2 in HT29 cells were analyzed by qRT-PCR, western blotting and densitometry. Data are shown as means ± SEM. *p < 0.05, **p < 0.01 compared with controls.
Scatter plots illustrating the statistical association between DLEU2 expression and levels of proliferative and metastatic markers in CRC. The Starbase database was used to analyze associations involving DLEU2 expression and the expression of CDK6 (A), CDKN1A (B), ZEB1 (C), and ZEB2 (D) in COAD and READ samples.
Recent studies suggest that DLEU2 plays a critical role in the development and progression of human cancers. Xu et al. [50] demonstrated that overexpressed DLEU2 can bind with miR-455 to promote the expression of SMAD2, thereby inducing the proliferation and invasion of pancreatic cancer (PC). Moreover, Guo et al. [51] reported that overexpression of DLEU2 can promote tumorigenesis in hepatocellular carcinoma (HCC) by interacting with the zeste 2 polycomb repressive complex 2 subunit (EZH2). However, the effects of DLEU2 on the tumorigenesis of other malignancies remain far from being understood. In this study, we first identified that the expression of DLEU2 in CRC was positively correlated with colorectal cancer, cell cycle, G1 pathway, and Apical junction signaling sets by GSEA. We demonstrated that knock-down of DLEU2 expression in CRC cells could suppress their proliferative and invasive capabilities by reducing the expression of CDK6, ZEB1, and ZEB2, as well as enhancing the expression of CDKN1A. Furthermore, we showed that the subcellular localization of DLEU2 in CRC cells was mainly cytoplasmic. The subcellular localization of lncRNAs is usually related to their biological functions [52]. lncRNAs, mainly localized in the cytoplasm, can 'sponge' miRNAs to form ceRNA so as to regulate the expression of target genes; whereas nuclear lncRNAs can interact with transcription factors or epigenetic modification-associated proteins to modulate gene expression. Therefore, we speculated that DLEU2 expressed in the cytoplasm of CRC cells may sponge miRNA to form ceRNA, thereby regulating the malignant biological behavior of CRC. In our next study, we will further explore regulatory mechanisms involving DLEU2 in CRC progression.
In summary, our study indicated upregulated expression of lncRNA DLEU2 in CRC, and high DLEU2 expression was positively correlated with poor survival time (RFS and OS) of CRC patients. We also demonstrated that knock-down of DLEU2 could inhibit the proliferative and invasive capabilities of CRC cells. However, there were certain limitations to this study: 1) we did not further investigate the underlying regulatory mechanisms involving DLEU2 in promoting the proliferative and invasive capabilities of CRC cells; 2) our study did not extend to animal experiments to explore the effects of DLEU2 in vivo. Taken together, our study may provide novel targets for candidate CRC therapies in the future.
Supplementary Material
Supplementary figures.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81903032), the Natural Science Foundation of Hunan Province of China (2019JJ40487 and 2019JJ40497), the China Postdoctoral Science Foundation (2020M672520), the Youth Fund of Xiangya Hospital (2018Q011) and the Open Sharing Fund for the Large-scale Instruments and Equipments of Central South University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A. et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-93
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
3. Ou CL, Sun ZQ, Li S, Li GY, Li XY, Ma J. Dual roles of yes-associated protein (YAP) in colorectal cancer. Oncotarget. 2017;8:75727-41
4. Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. JAMA. 2003;289:1288-96
5. Sunkara V, Hebert JR. The colorectal cancer mortality-to-incidence ratio as an indicator of global cancer screening and care. Cancer-Am Ccancer Soc. 2015;121:1563-69
6. Nie H, Wang YT, Liao ZM, Zhou JH, Ou CL. The function and mechanism of circular RNAs in gastrointestinal tumours. Cell Prolif. 2020;53(7):e12815
7. Calvert PM, Frucht H. The genetics of colorectal cancer. Ann Intern Med. 2002;137:603-12
8. Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res. 2016;36:1093-102
9. Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365-85
10. Vaiopoulos AG, Athanasoula K, Papavassiliou AG. Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim Biophys Acta. 2014;1842:971-80
11. Tang Y, He Y, Zhang P, Wang JP, Fan CM, Yang LT. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77
12. He XY, Ou CL, Xiao YH, Han Q, Li H, Zhou SX. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017;8:71325-41
13. Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES. et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. Embo J. 2014;33:981-93
14. Wang YT, Nie H, He XY, Liao ZM, Zhou Y, Zhou JH. et al. The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics. 2020;10:11049-62
15. Fan CM, Tang YY, Wang JJP, Wang Y, Xiong F, Zhang SS. et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2019;40:380-91
16. He X, Li S, Yu B, Kuang GY, Wu YR, Zhang ML. et al. Up-regulation of LINC00467 promotes the tumourigenesis in colorectal cancer. J Cancer. 2019;10:6405-13
17. Song HJ, Sun WL, Ye GL, Ding XY, Liu Z, Zhang SJ. et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225
18. Yan YL, Xu ZJ, Chen X, Wang X, Zeng SS, Zhao ZJ. et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front Cell Dev Biol. 2019;7:217
19. Shang C, Zhu W, Liu T, Wang W, Huang G, Huang J. et al. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol Rep. 2016;35:3185-97
20. Zhou YC, Shi HT, Du YQ, Zhao GQ, Wang XX, Li Q. et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging (Albany NY). 2019;11:7386-401
21. Valcz G, Patai AV, Kalmar A, Péterfia B, Fűri I, Wichmann B. et al. Myofibroblast-derived SFRP1 as potential inhibitor of colorectal carcinoma field effect. Plos One. 2014;9:e106143
22. Uddin S, Ahmed M, Hussain A, Abubaker J, Al-Sanea N, AbdulJabbar A. et al. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol. 2011;178:537-47
23. Lin GX, He XM, Ji H, Shi LM, Davis RW, Zhong S. Reproducibility Probability Score-incorporating measurement variability across laboratories for gene selection. Nat Biotechnol. 2006;24:1476-77
24. Smith JJ, Deane NG, Wu F, Abubaker J, Al-Sanea N, AbdulJabbar A. et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958-68
25. Kim SK, Kim SY, Kim JH, Roh SA, Cho DH, Kim YS. et al. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol. 2014;8:1653-66
26. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267-73
27. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545-50
28. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-19
29. Zhang X, Ai F, Li X, She XL, Li N, Tang AL. et al. Inflammation-induced S100A8 activates Id3 and promotes colorectal tumorigenesis. Int J Cancer. 2015;137:2803-14
30. Dang W, Qin Z, Fan S, Wen QY, Lu YJ, Wang J. et al. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7:32421-32
31. Li YC, Zhao CH, Yu Z, Chen JR, She XL, Li PY. et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016;7:68585-96
32. Ou CL, Sun ZQ, Zhang H, Xiong W, Ma J, Zhou M. et al. SPLUNC1 reduces the inflammatory response of nasopharyngeal carcinoma cells infected with the EB virus by inhibiting the TLR9/NF-kappaB pathway. Oncol Rep. 2015;33:2779-88
33. Ou CL, Sun ZQ, Li X, Li X, Ren W, Qin Z. et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53-63
34. Tang ZF, Li CW, Kang BX, Gao G, Li C, Zhang ZM. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-102
35. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
36. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-463
37. Yang L, Tang Y, Xiong F, He Y, Wei F, Zhang S. et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2018;9:1426-1443
38. Sun ZQ, Ou CL, Liu JB, Chen C, Zhou QB, Yang SX. et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627-2644
39. Jiang X, Zhu QY, Wu PX, Zhou FS, Chen J. Upregulated Long Noncoding RNA LINC01234 Predicts Unfavorable Prognosis for Colorectal Cancer and Negatively Correlates With KLF6 Expression. Ann Lab Med. 2020;40:155-163
40. Bo H, Fan LQ, Li JJ, Liu ZZ, Zhang SS, Shi L. et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer. 2018;9:4677-4683
41. Yin DD, He XZ, Zhang E, Kong R, De W, Zhang ZH. et al. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:253
42. Ou CL, Sun ZQ, He XY, Li XL, Fan SQ, Zheng X. et al. Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal. Cancer.Adv Sci (Weinh). 2020;7:1901380
43. Liu F, Ai FY, Zhang DC, Tian L, Yan ZY, Liu SJ. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2020;9:1079-91
44. Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25
45. Wang X, Mo FM, Bo H, Xiao L, Chen GY, Zeng PW. et al. Upregulated Expression of Long Non-Coding RNA, LINC00460, Suppresses Proliferation of Colorectal Cancer. J Cancer. 2018;9:2834-43
46. Rondeau G, Moreau I, Bezieau S, Petit JL, Heilig R, Fernandez S. et al. Comprehensive analysis of a large genomic sequence at the putative B-cell chronic lymphocytic leukaemia (B-CLL) tumour suppresser gene locus. Mutat Res. 2001;458:55-70
47. Wu WM, Zhao YH, Gao EJ, Li Y, Guo X, Zhao TC. et al. LncRNA DLEU2 accelerates the tumorigenesis and invasion of non-small cell lung cancer by sponging miR-30a-5p. J Cell Mol Med. 2020;24:441-450
48. Ma W, Zhang CQ, Dang CX, Cai HY, Li HL, Miao GY. et al. Upregulated long-non-coding RNA DLEU2 exon 9 expression was an independent indicator of unfavorable overall survival in patients with esophageal adenocarcinoma. Biomed Pharmacother. 2019;113:108655
49. Leng J, Song QX, Zhao YG, Wang ZY. miR15a represses cancer cell migration and invasion under conditions of hypoxia by targeting and downregulating Bcl2 expression in human osteosarcoma cells. Int J Onco. 2018;52:1095-104
50. Xu BL Gong XF, Zi L, Li G, Dong SX, Chen XR. et al. Silencing of DLEU2 suppresses pancreatic cancer cell proliferation and invasion by upregulating microRNA-455. Cancer Sci. 2019;110:1676-1685
51. Xie ZC, Li XJ, Chen H, Zeng AL, Shi Y, Tang Y. The lncRNA-DLEU2/miR-186-5p/PDK3 axis promotes the progress of glioma cells. Am J Transl Res. 2019;11:4922-34
52. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761-72
Author contact
![]() Corresponding author: Chunlin Ou. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. E-mail: ouchunlinedu.cn.
Corresponding author: Chunlin Ou. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. E-mail: ouchunlinedu.cn.

 Global reach, higher impact
Global reach, higher impact