Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(4):1240-1248. doi:10.7150/jca.51179 This issue Cite
Research Paper
Prognostic significance of KRT19 in Lung Squamous Cancer
1. Department of Oncology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, 1095 Jiefang Avenue, Wuhan 430030, P.R. China.
2. Department of Molecular Pathology, The Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou 450008, China.
Received 2020-7-27; Accepted 2020-12-4; Published 2021-1-1
Abstract
Backgroud: Keratin 19 (KRT19) is the intermediate filament that constitutes the cytoskeleton and regulates cell-cycle and cell death.
Objective: We aimed to assess whether KRT19 was involved in lung cancer development.
Methods: The expression of KRT19 in lung cancer was evaluated from mRNA expression on open databse and protein abundance on tumor tissue array.
Results: Using open microarray gene expression datasets and differential expression analysis, we found that KRT19 was upregulated in lung cancer compared with normal tissue. Further analysis suggested that KRT19 mRNA expression was correlated with tumor progression and overall survival in lung cancer patients. As KRT19 was overexpressed in adenocarcinoma (AC) and squamous cell carcinoma (SCC), we examined the prognostic value of KRT19 protein abundance by tissue microarray (TMA). The results suggested that protein expression of KRT19 was significantly associated with overall survival of SCC.
Conclusions: Giving the prognostic role of KRT19 in lung cancer, KRT19 could be considered as an potential molecular marker in lung cancer, especially in SCC.
Keywords: KRT19, lung cancer, squamous cell carcinoma, prognosis
Introduction
Lung cancer, a heterogeneous group of malignancies, remains the leading cause of cancer-related deaths in the world. The different histological types of lung cancer are divided into two main groups, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), the latter includes adenocarcinoma (AC), squamous cell carcinoma (SCC) and large cell lung cancer (LCC). AC and SCC are the most common histological types of lung cancer and account for approximately 70% of lung cancers [1]. Small cell lung cancer (SCLC) is usually reported to comprise about 15% of all lung cancer cases recorded [2]. Improved understanding of the biology of lung cancer has resulted in the development of new biomarker-directed therapies targeting molecular alterations (eg, EGFR mutations, ALK rearrangements, ROS1 rearrangements, and BRAF V600E mutations), which have prolonged survival of lung cancer patients [3]. However, the progress is slow, with about 5% improvements in 5-year survival rates for the past 20 years [4]. Therefore, it is crucial to identify additional prognostic biomarkers to generate individualized treatment and follow-up schedules [5, 6].
There is an urgent need for a highly reproducible, inexpensive biomarker, which might help to identify patients at high risk for disease-progression, recurrence, metastasis and death [7]. Hence, we analyzed gene expression pattern in publicly available cohorts of lung cancer patients and identified a signature of genes which might be involved in lung cancer development and prognosis of patients. Among the identified genes, KRT19 is a member of the keratin family, which is responsible for the structural integrity of epithelial cells, including bronchial epithelial cells [8, 9]. KRT19 expresses in normal lung tissue, as well as lung cancer tissues [10]. However, when necrosis occurs within the lung tumor, KRT19 degrades and fragments are released into the blood. This leads to an increase of the blood CYFRA 21-1 level [11, 12]. Hence, the serum level of KRT19 fragments could be applied in the clinic as a tumor marker.
Many studies have investigated the prognostic value of serum tumor biomarkers, including CYFRA 21-1 in lung cancer [13-16]. However, the value of KRT19 in lung cancer is still unknown. In the present study, we evaluated the significance of KRT19 in lung cancer from mRNA and protein level.
Materials and methods
Analysis of Gene Expression data
In order to determine whether genes were differentially expressed between primary lung tumor and normal lung tissue, differential gene expression analysis was performed by the SAM method using histological type as the variable [17]. Gene lists were created using a cut-off of Q <0.05, >1.5-fold change. The raw data for differential expression analysis are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-GEOD-18842 and E-GEOD-27262.
Six other independent datasets, available through the Gene Expression Omnibus (GEO) database in public repositories and containing gene expression and clinical data, were analyzed to evaluate the expression and prognosis of the biomarkers (GSE31210, GSE32863, GSE19188, GSE3141, GSE14814, GSE30219) [18].
Lung cancer tissue microarray and immunohistochemical staining
To evaluate the clinical significance of KRT19 in patients with lung cancer, commercially available tissue microarray (TMA) slide (LC811, Alenabio, Inc., Xian, China) with pathological characteristic information was purchased for immunohistochemistry (IHC) analysis [19]. Other tissue microarrays (HLug-Ade150Sur-02 and HLug-Squ150Sur-01, Shanghai Biochip Co., Ltd., Shanghai, China) both with 75 matched pairs of primary lung cancer samples and adjacent lung tissues were applied to evaluate the prognostic value of KRT19 based on its detailed survival data. Specific primary antibody against KRT19 (polyclonal rabbit antibody, 1:100; ProteinTech Group, Chicago, USA) was used for IHC with a 2-step protocol. Immunohistochemical staining was performed using a standard technique, as previously described [20, 21]. Level of KRT19 expression was measured using Image-Pro Plus® software version 6.0.0.260.
Statistical analysis
The Student's t-test and one-way ANOVA were applied to evaluate the differences in groups as appropriate and the significance level was set at 0.05. For the univariate analysis, patients' clinical end points were calculated using the Kaplan-Meier method and compared by the log-rank test. Forward stepwise multivariate Cox proportion hazard analysis was performed to determine the influence of age, gender, histologic subtype and TNM stage on overall survival (OS). Hazard ratios (HRs) with corresponding 95% confidence intervals (CIs) were estimated from the Cox analysis. We also employed Harrell's concordance index (C-index) to assess the model's prognostic accuracy in the multivariate analysis. A two-sided P<0.05 was considered statistically significant. All statistical analyses were performed with R, version 3.6.2 (http://www.r-project.org/).
Results
Transcriptional levels of KRT19 characterize lung cancer
In an effort to characterize histological type-specific gene signature of lung cancer, we analyzed gene expression patterns in microarrays through a method, Significance Analysis of Microarrays (SAM). Using a public dataset GSE18842 with 50 primary lung tumor and 50 paired adjacent lung tissues, SAM identified differential expression genes that changed at least 1.5-fold. Among them, KRT19 were significantly overexpressed in lung cancer (Table 1). It was also validated in another dataset GSE27262 with 25 primary lung tumor and 25 paired adjacent lung tissue.
Differential expression analysis identified genes KRT19 from GSE18842 and GSE27262
| Gene Symbol | Gene Description | Dataset | Normal | Tumor | Fold Change | p-value |
|---|---|---|---|---|---|---|
| KRT19 | Keratin 19 | GSE18842 | 10.264 | 12.054 | 3.458 | 0.008 |
| GSE27262 | 10.663 | 11.648 | 1.979 | 0.003 |
Then, we evaluated KRT19 transcript levels in microarrays downloaded from ArrayExpress dataset. In accordance with previous conclusion, the relative abundance of KRT19 mRNA increased 2- to 3-fold when normal tissue transformed into tumor (t-test, p<0.001) (Figure 1A-C).
The mRNA expression of KRT19 in lung cancer and normal tissue. (A-C) KRT19 mRNA were overexpressed in tumor compared with normal lung tissue. (D-E) KRT19 mRNA was overexpressed in SCC. (F) KRT19 mRNA level was higher in wt lung cancer than in EGFR mutated lung cancer. (G-H) KRT19 expression increased with the presence of higher stage. (I) KRT19 expression was associated with OS in lung cancer patients from GSE30219. (J-K) KRT19 expression was associated with RFS and OS in lung cancer patients from GSE31210.
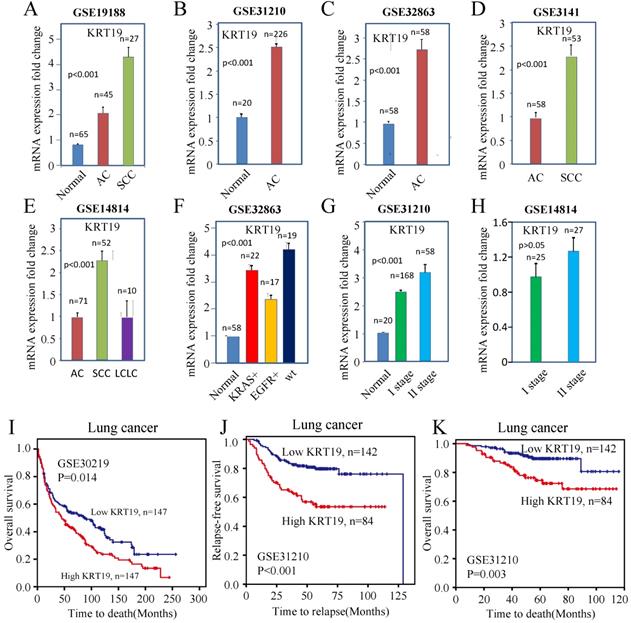
To investigate the association between the KRT19 with distinct lung cancer subtypes, we compared KRT19 expression with subtypes of lung cancer in microarrays. The expression of KRT19 was higher in SCC than in AC and LCLC (Figure 1D-E). In addition, KRT19 mRNA was increased about 2-fold in wild type (wt) lung cancer compared with EGFR mutated lung cancer, suggesting that EGFR pathway might affect KRT19 gene expression (Figure 1F).
In order to test whether KRT19 is associated with lung cancer progression, we examined the association of KRT19 expression with TNM staging. An increased expression of KRT19 was correlated with higher TNM stage (Figure 1G-H). These analyses indicated that KRT19 could be a tumor marker for diagnosis of lung cancer, especially of SCC.
To understand the value of KRT19 in the prediction of lung cancer's prognosis, we examined expression of KRT19 in microarrays which provided survival data. The results indicated that KRT19 was statistically associated with the relapse-free survival (RFS) and OS rate of lung cancer (Figure 1I-K). Combined, these results revealed that KRT19 could be a diagnostic and prognostic biomarker of lung cancer.
Tissue expression of KRT19 protein characterize lung cancer
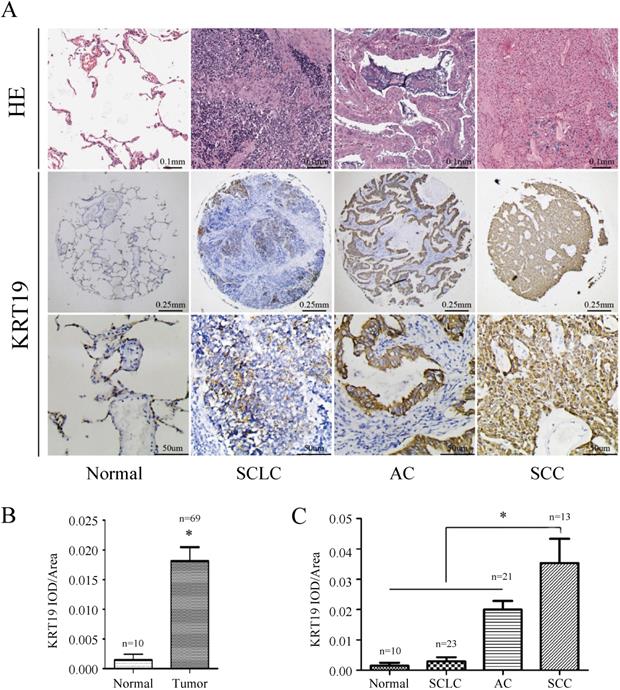
In order to further explore the role of KRT19 in lung cancer, we analyzed KRT19 protein levels in a TMA containing 69 informative patients with distinct histological subtypes and additional normal lung tissues. KRT19 protein was predominantly detected in the cytoplasm, and representative images of immunohistochemical staining for noncancerous and cancerous tissue are shown in Figure 2A. We examined the potential association of KRT19 expression with the clinicopathological parameters by using quantitative criteria (Table 2). Statistical significance was found for KRT19 expression in AC, SCC and SCLC (f-test, p<0.001). Importantly, KRT19 was significantly overexpressed in SCC compared with AC (t-test, p=0.039). Immunohistochemical staining confirmed that KRT19 expression was high in lung tumor, especially in SCC (Figure 2B-C).
Association between biomarker expression and clinicopathological characteristics in TMA (LC811)
| Characteristics | Total | High expression | |
|---|---|---|---|
| KRT19 | P-value | ||
| Number n (%) | 69 | 35 (50.7) | - |
| Age (years) (n [%]) | 0.022 | ||
| < 53 | 29 (42.0) | 10 (34.5) | |
| ≥ 53 | 40 (58.0) | 25(62.5) | |
| Gender (n [%]) | 0.155 | ||
| Female | 19 (27.5) | 7 (36.8) | |
| Male | 50 (72.5) | 28 (56.0) | |
| Histology (n [%]) | <0.001 | ||
| AC | 33 (47.8) | 21 (63.6) | |
| SCC | 13 (18.8) | 12 (92.3) | |
| SCLC | 23 (33.4) | 2 (8.7) | |
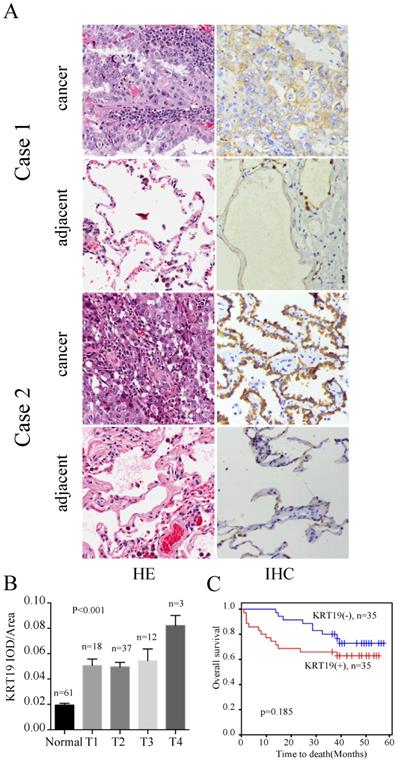
To understand the value of KRT19 in the prediction of ACs' prognosis, we examined expression of KRT19 in the other tissue microarray, which provided survival data. The profiles of KRT19 of two representative cases were shown in Figure 3A. Case1 had the longest survival and case2 had shortest survival in this cohort. Intriguinly, expression of KRT19 was strongly correlated with the tumor T stage (Figure 3B). However, segregation of the patients into KRT19-high and low expression groups did not reveal significant relationship with clinicopathological parameters of sex, age, tumor size, location, grade (Table 3). We also investigated the association between cumulative overall survival-rates and clinicopathological factors by univariate and multivariate Cox regression analysis. As shown in Table 4, tumor size was prognostic factor for OS, whereas other clinicopathological factors were not directly related to the clinical outcome of AC. Besides, KRT19 expression was not correlated with OS in AC (Figure 3C).
Immunohistochemistry analysis of KRT19 in lung cancer tissues. Representative images of KRT19 expression in normal lung tissue and different historical type of lung cancer were shown (A) with quantitative result displayed as mean ± SE (B-C).
Immunohistochemistry analysis of KRT19 in AC patients. In a five years cohort, representative images of the cancerous and adjacent tissue from the longest survival patient and the shortest one were shown by using HE and IHC staining (A). The quantification of KRT19 expression in different stages was displayed as mean ± SE (B). Kaplan-Meier survival curve of KRT19 (C) was also analyzed.
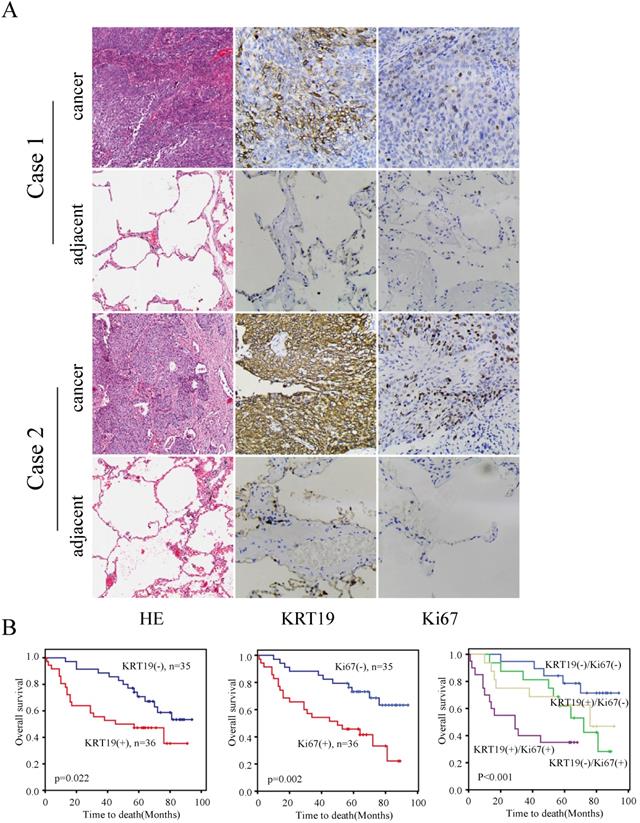
To further explore the prognostic role of KRT19 in SCC, we employed another tissue microarray containing 70 SCC patients with complete survival data and detailed information about Ki-67 expression. The profiles of KRT19 and Ki-67 of three representative cases were shown in Figure 4A, which showed the positive correlation between KRT19 and Ki-67. In comparison of early-stage lung cancer tissue, the majorty of late-stage lung cancer samples showed increased expression of KRT19 and Ki-67 in this five years cohort (Figure 4B-C). Segregation of these patients into KRT19-high and low expression groups did not reveal significant relationship with clinicopathological parameters of sex, age, tumor size, location, but noticeably associated with tumor grading, TNM staging (Table 5). Univariate and multivariate Cox regression analysis were perfomed to analyze the association between overall survival-rates and clinicopathological factors. Age, TNM stage, Ki-67 and KRT19 were prognostic factors for OS, whereas other clinicopathological factors were not directly related to the clinical outcomes of SCC (Table 6). Two representative cases of the longest survival patient and the shortest survival patient were shown in Figure 5A. As expected, high expression of KRT19 and Ki-67 tended to predict worse clinical outcome (Kaplan Meier log-rank test, p=0.022; p=0.002). Combined examination of KRT19 with Ki-67 would provide more precise information for the clinical outcome of SCC patients. We performed the forward variable-selection procedure using the two factors and patients characterized by low levels of KRT19 and Ki-67 had the longest OS in this study (Kaplan Meier log-rank test, p<0.001).
Immunohistochemistry analysis of KRT19 in SCC tissues. In a five years cohort, representative images of KRT19 expression in normal lung tissue and SCC were shown (A) with quantitative result displayed as mean ± SE (B-C).
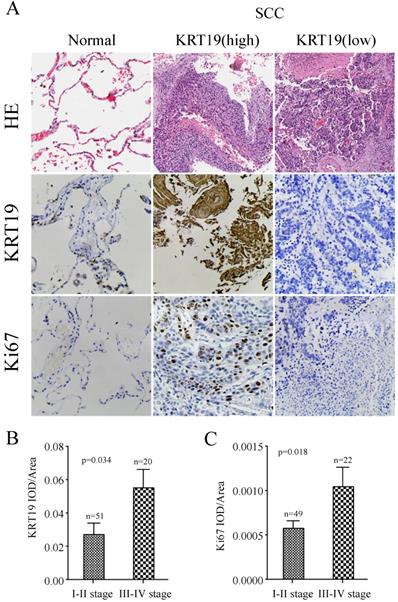
KRT19 is an independent prognostic factor of overall survival in SCC patients. In a five years cohort, representative images of the cancerous and adjacent tissue from the longest survival patient and the shortest one were shown by using HE and IHC staining (A). Kaplan-Meier survival curve of KRT19 alone or in combination with Ki-67was also analyzed (B).
Discussion
KRTs are intermediate filament proteins that constitute the cytoskeleton and regulate cell-cycle, apoptosis, cellular response to stress and death through dynamic interactions with a range of cellular proteins, including kinases, receptors, adaptors, and effectors [22]. There are 20 types of KRTs, as classified by the molecular weight and isoelectric point. These KRTs are categorized to 2 subtypes: low-molecular weight acidic type I and high-molecular weight basic or neutral type II [23]. KRT19 is the smallest (40 kDa) known type I KRTs and has been related to some diseases [24]. In a mouse model, knockdown of KRT19 caused skeletal myopathy via mitochondrial and sarcolemmal reorganization [25]. Subsequently, KRT19 was identified as tumor-associated biomarker in various cancers, including esophageal squamous cell carcinoma, cervical cancer, breast cancer and HCC [26-29]. Gene amplification of KRT19 was found in HER2-positive breast tumor, indicating a relationship between KRT19 and HER2 [30]. Then, it was found that KRT19 could interact with HER2 on the cell membrane and stabilize HER2. However, inhibition of KRT19 downregulated HER2 by increased ubiquitination and destabilization of HER2, and reduced cell viability [31]. MicroRNA-492, originated from KRT19, showed significant association with the liver enzyme BAAT and was overexpressed in metastatic hepatoblastoma (HB) [32]. In addition, it was found that KRT19 correlated with tumor size, tumor differentiation, metastasis and micovascular invasion of HCC. Mechanically, KRT19 increased invasiveness and induced chemoresistance in HCC cells [33]. Furthermore, expression of KRT19 was associated with survival of HCC patients [34].
Association between biomarker expression and clinicopathological characteristics in AC TMA (HLug-Ade150Sur-02)
| Characteristics | Total | High expression | |
|---|---|---|---|
| KRT19 | P-value | ||
| Number n (%) | 70 | 37 (52.9) | |
| Age (years) (n [%]) | 0.808 | ||
| < 60 | 29 (41.4) | 14 (48.3) | |
| ≥ 60 | 41 (58.6) | 21 (51.2) | |
| Gender (n [%]) | 1.000 | ||
| Female | 34 | 17 (50.0) | |
| Male | 36 | 18 (50.0) | |
| Maximal tumor size (n [%]) | 0.051 | ||
| ≤ 5 cm | 53 | 23 (43.4) | |
| > 5 cm | 17 | 12 (70.6) | |
| Tumor location (n [%]) | 0.626 | ||
| Left | 28 | 15 (53.6) | |
| Right | 42 | 20 (47.6) | |
| Grade (n [%]) | 1.000 | ||
| 1+2 | 56 | 28 (50.0) | |
| 3+4 | 14 | 7 (50.0) | |
Univariate and multivariate Cox regression analyses of multiple variables for OS (HLug-Ade150Sur-02)
| Characteristics | OS | |||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (years) (n [%]) | ||||
| < 60 | 1 (Reference) | 1 (Reference) | ||
| ≥ 60 | 1.233 (0.517,2.940) | 0.637 | 0.701 (0.269,1.824) | 0.466 |
| Gender (n [%]) | ||||
| Female | 1 (Reference) | 1 (Reference) | ||
| Male | 1.903 (0.798,4.540) | 0.147 | 1.434 (0.556,3.700) | 0.456 |
| Maximal tumor size (n [%]) | ||||
| ≤ 5 cm | 1 (Reference) | 1 (Reference) | ||
| > 5 cm | 3.686 (1.584,8.575) | 0.002 | 5.147 (1.834,14.443) | 0.002 |
| Tumor location (n [%]) | ||||
| Left | 1 (Reference) | 1 (Reference) | ||
| Right | 1.337 (0.540,3.313) | 0.531 | 0.949 (0.365,2.468) | 0.915 |
| Grade (n [%]) | ||||
| 1+2 | 1 (Reference) | 1 (Reference) | ||
| 3+4 | 0.971 (0.329,2.869) | 0.957 | 0.359 (0.095,1.352) | 0.130 |
| KRT19 | ||||
| Negative | 1 (Reference) | 1 (Reference) | ||
| Positive | 1.760 (0.751,4.123) | 0.185 | 1.184 (0.477,2.943) | 0.716 |
Association between biomarker expression and clinicopathological characteristics in SCC TMA (HLug-Squ150Sur-01)
| Characteristics | Total | High expression | |
|---|---|---|---|
| KRT19 | P-value | ||
| Number n (%) | 71 | 36 (50.7) | |
| Age (years) (n [%]) | 0.727 | ||
| < 64 | 33 (46.5) | 16 (48.5) | |
| ≥ 64 | 38 (53.5) | 20 (52.6) | |
| Gender (n [%]) | 0.290 | ||
| Female | 4 (5.6) | 1 (0.3) | |
| Male | 67 (94.4) | 35 (52.2) | |
| Maximal tumor size (n [%]) | 0.188 | ||
| ≤ 7 cm | 61 (85.9) | 29 (47.5) | |
| > 7 cm | 10 (14.1) | 7 (0.7) | |
| Tumor location (n [%]) | 0.734 | ||
| Left | 29 (40.8) | 14 (48.3) | |
| Right | 42 (59.2) | 22 (52.4) | |
| Grade (n [%]) | 0.007 | ||
| 1+2 | 58 (81.7) | 25 (43.1) | |
| 3+4 | 13 (18.3) | 11 (84.6) | |
| TNM (n [%]) | 0.048 | ||
| I+II | 49 (69.0) | 21 (42.9) | |
| III+IV | 22 (31.0) | 15 (68.2) | |
Univariate and multivariate Cox regression analyses of multiple variables for OS (HLug-Squ150Sur-01)
| Characteristics | OS | |||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (years) (n [%]) | ||||
| < 64 | 1 (Reference) | 1 (Reference) | ||
| ≥ 64 | 3.017 (1.405,6.479) | 0.005 | 2.753 (1.190,6.369) | 0.018 |
| Gender (n [%]) | ||||
| Female | 1 (Reference) | 1 (Reference) | ||
| Male | 0.411 (0.144,1.176) | 0.097 | 0.244 (0.071,0.840) | 0.025 |
| Maximal tumor size (n [%]) | ||||
| ≤ 7 cm | 1 (Reference) | 1 (Reference) | ||
| > 7 cm | 1.224 (0.472,3.172) | 0.677 | 0.787 (0.285,2.176) | 0.645 |
| Tumor location (n [%]) | ||||
| Left | 1 (Reference) | 1 (Reference) | ||
| Right | 0.751 (0.380,1.481) | 0.408 | 0.578 (0.279,1.196) | 0.139 |
| Grade (n [%]) | ||||
| 1+2 | 1 (Reference) | 1 (Reference) | ||
| 3+4 | 1.568 (0.679,3.619) | 0.292 | 0.632 (0.237,1.682) | 0.358 |
| TNM (n [%]) | ||||
| I+II | 1 (Reference) | 1 (Reference) | ||
| III+IV | 2.232 (1.106,4.504) | 0.025 | 2.119 (0.920,4.882) | 0.078 |
| Ki-67 | ||||
| Negative | 1 (Reference) | 1 (Reference) | ||
| Positive | 2.575 (1.263,5.248) | 0.009 | 2.686 (1.191,6.059) | 0.017 |
| KRT19 | ||||
| Negative | 1 (Reference) | 1 (Reference) | ||
| Positive | 2.206 (1.097,4.437) | 0.027 | 2.904 (1.269,6.643) | 0.012 |
Importantly, KRT19 can be used as a molecular marker for reverse transcriptase PCR (RT-PCR) and ELISA-mediated detection of disseminated tumor cells in regional lymph nodes (LNs), peripheral blood (PB) and bone marrow from susceptible cancerous patients [12, 35-36]. Recently, KRT19 was also used as a novel marker by circulated tumor cells (CTC) detection to identify tumor cells in breast cancer and colorectal cancer [37-39], and its positivity was demonstrated to be a prognostic indicator [40]. However, KRT19 has not been well characterized in lung cancer, specifically with regard to the prognostic value. Our study has found that KRT19 is associated with clinical progression and is predictive of clinical outcomes in lung cancer as a whole in gene transcription and protein expression level.
Using microarray gene expression datasets and differential expression analysis, KRT19 was found to overexpress in lung cancer compared with normal tissue. Moreover, KRT19 mRNA was significantly associated with the presence of SCC, EGFR wt lung cancer, higher stage and shorter survival time of lung cancer patients. It was reported that serum level of KRT19 was associated with the efficacy of EGFR-TKI treatment in NSCLC harboring EGFR mutation, and that patients with high KRT19 responded poorly to EGFR-TKI, leading to a shorter survival time [41]. As KRT19 represents the extent of the EGFR wt component in lung tumor, EGFR-TKI's efficacy in NSCLC is poor in high serum KRT19 group compared with normal serum KRT19 group. Our study confirmed the difference of KRT19 expression in different molecular phenotype of lung cancer. As KRT19 correlated with EGFR wt lung cancer, KRT19 may be a valuable predictive marker for EGFR-TKI use. Further clinical trials are needed to validate the significance of KRT19 in EGFR mutated patients.
We also employed tissue microarray to validate the value of KRT19 in protein level. Distinguishing SCC from AC could provide better treatment decisions. However, the histological classification mainly depended on biopsy. In addition, biopsy occasionally leads to controversial diagnosis in lung cancer for the inherent histologic heterogeneity that exists in a subset of lung cancer [42]. Our data indicated that KRT19 levels were much higher in SCC than in AC, and KRT19 showed the strongest correlation with stage and grade in SCC. Liquid bead array hybridization assay has been applied as a predictive and prognostic index of cancer patients and guiding clinical treatment, making KRT19 based CTC a promising diagnostic method for SCC. We speculated that interaction between KRT19 and other proteins including cell proliferatin marker Ki-67, initiated signaling cascades that promoted tumorigenesis, tumor invasion and metastasis. Rencently, KRT19 was found to interact with β-catenin/RAC1 complex and regulated NUMB and Notch1 expression, leading to the enhancement of the cancer stem-like cell properties in breast cancer [43]. Furthemore, KRT19 expression was identified as an independent prognostic factor for the overall survival for SCC patients. Hence, the function of KRT19 in SCC is remarkable, and it would be interesting to explore the underlying mechanisms in detail.
In summary, KRT19 is a prognostic marker in lung cancer. High KRT19 expression in lung cancer was associated with clinical progression and could be used as a clinically relevant marker in lung cancer patients, especially in SCC patients. Recently, liquid biopsy has been used to identify cancer patients based on KRT19 expression. Further studies are needed to compare the predictive value of CTC detection and tumor tissue IHC in SCC.
Acknowledgements
Availability of data and materials
The datasets generated for this study can be found in the GEO dataset: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?, using code GSE18842, GSE19188, GSE31210, GSE32863, GSE3141, GSE14814, GSE30219 and GSE27262.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82003310 and 81874120), Wuhan Science and Technology Bureau (No. 2017060201010170), and also supported in part by National Cancer Center Climbing Foundation Key project (NCC201816B046).
Authors' contributions
XY participated in the conception of the study, analysis of data and preparation of the manuscript. YM and BD help to the revision for important intellectual content. QC and KW designed the project and participated in the supervison of the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ono A, Takahashi T, Mori K. et al. Prognostic impact of serum CYFRA 21-1 in patients with advanced lung adenocarcinoma: a retrospective study. BMC Cancer. 2013;13:354
2. Ding X, Cao H, Chen X. et al. Cellular immunotherapy as maintenance therapy prolongs the survival of the patients with small cell lung cancer. J Transl Med. 2015;13:158
3. Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA. 2019;322:764-774
4. Johnson DH, Schiller JH, Bunn PA Jr. Recent clinical advances in lung cancer management. J Clin Oncol. 2014;32:973-982
5. Yuan X, Wu H, Han N. et al. Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J Hematol Oncol. 2014;7:87
6. Ma YZ, Yuwen DL, Chen JW. et al. Exosomal Transfer Of Cisplatin-Induced miR-425-3p Confers Cisplatin Resistance in NSCLC through activating Autophagy. Int J Nanomedicine. 2019;14:8121-8132
7. Chaudhary K, Poirion OB, Lu L. et al. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259
8. Moll R, Franke WW, Schiller DL. et al. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11-24
9. Saha SK, Kim K, Yang GM. et al. Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-like Cells to less aggressive and more drug-sensitive cells. Int J Mol Sci. 2018;19:1423
10. Wang W, He J, Lu H. et al. KRT8 and KRT19, associated with EMT, are hypomethylated and overexpressed in lung adenocarcinoma and link to unfavorable prognosis. Biosci Rep. 2020: BSR20193468.
11. Dohmoto K, Hojo S, Fujita J. et al. The role of caspase 3 in producing cytokeratin 19 fragment (CYFRA21-1) in human lung cancer cell lines. Int J Cancer. 2001;91:468-473
12. Nordgard O, Singh G, Solberg S. et al. Novel molecular tumor cell markers in regional lymph nodes and blood samples from patients undergoing surgery for non-small cell lung cancer. PLoS One. 2013;8:e62153
13. Edelman MJ, Hodgson L, Rosenblatt PY. et al. CYFRA 21-1 as a prognostic and predictive marker in advanced non-small-cell lung cancer in a prospective trial: CALGB 150304. J Thorac Oncol. 2012;7:649-654
14. Satoh H, Ishikawa H, Fujiwara M. et al. Production of cytokeratin 19 fragment by human squamous lung cancer cell lines. Am J Respir Cell Mol Biol. 1997;16:597-604
15. Pujol JL, Grenier J, Daures JP. et al. Serum fragment of cytokeratin subunit 19 measured by CYFRA 21-1 immunoradiometric assay as a marker of lung cancer. Cancer Res. 1993;53:61-66
16. Park SY, Lee JG, Kim J. et al. Preoperative serum CYFRA 21-1 level as a prognostic factor in surgically treated adenocarcinoma of lung. Lung Cancer. 2013;79:156-160
17. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116-5121
18. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207-210
19. Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26:5630-5637
20. Liu Q, Li A, Yu S. et al. DACH1 antagonizes CXCL8 to repress tumorigenesis of lung adenocarcinoma and improve prognosis. J Hematol Oncol. 2018;11:53
21. Kong D, Li A, Liu Y. et al. SIX1 activates STAT3 signaling to promote the proliferation of thyroid carcinoma via EYA1. Front Oncol. 2019;9:1450
22. Coulombe PA, Wong P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol. 2004;6:699-706
23. Rhee H, Kim HY, Choi JH. et al. Keratin 19 Expression in Hepatocellular Carcinoma is regulated by Fibroblast-derived HGF via a MET-ERK1/2-AP1 and SP1 Axis. Cancer Res. 2018;78:1619-1631
24. Wu YJ, Rheinwald JG. A new small (40 kd) keratin filament protein made by some cultured human squamous cell carcinomas. Cell. 1981;25:627-635
25. Stone MR, O'Neill A, Lovering RM. et al. Absence of keratin 19 in mice causes skeletal myopathy with mitochondrial and sarcolemmal reorganization. J Cell Sci. 2007;120:3999-4008
26. Zhang J, Wang K, Zhang J. et al. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res. 2011;10:2863-2872
27. Okamoto S, Niikura H, Nakabayashi K. et al. Detection of sentinel lymph node metastases in cervical cancer: assessment of KRT19 mRNA in the one-step nucleic acid amplification (OSNA) method. Gynecol Oncol. 2013;130:530-536
28. Ju JH, Yang W, Lee KM. et al. Regulation of cell proliferation and migration by keratin19-induced nuclear import of early growth response-1 in breast cancer cells. Clin Cancer Res. 2013;19:4335-4346
29. Zeng SS, Yamashita T, Kondo M. et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127-134
30. Lamy PJ, Fina F, Bascoul-Mollevi C. et al. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast Cancer Res. 2011;13:R15
31. Ju JH, Oh S, Lee KM. et al. Cytokeratin19 induced by HER2/ERK binds and stabilizes HER2 on cell membranes. Cell Death Differ. 2015;22:665-676
32. von Frowein J, Pagel P, Kappler R. et al. MicroRNA-492 is processed from the keratin 19 gene and up-regulated in metastatic hepatoblastoma. Hepatology. 2011;53:833-842
33. Govaere O, Komuta M, Berkers J. et al. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut. 2014;63:674-685
34. Nault JC, De Reynies A, Villanueva A. et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176-187
35. Gilje B, Nordgard O, Tjensvoll K. et al. Comparison of molecular and immunocytochemical methods for detection of disseminated tumor cells in bone marrow from early breast cancer patients. BMC Cancer. 2014;14:514
36. Guzvic M, Braun B, Ganzer R. et al. Combined genome and transcriptome analysis of single disseminated cancer cells from bone marrow of prostate cancer patients reveals unexpected transcriptomes. Cancer Res. 2014;74:7383-7394
37. Wang XM, Zhang Z, Pan LH. et al. KRT19 and CEACAM5 mRNA-marked circulated tumor cells indicate unfavorable prognosis of breast cancer patients. Breast Cancer Res Treat. 2019;174:375-385
38. Mostert B, Sieuwerts AM, Bolt-de Vries J. et al. mRNA expression profiles in circulating tumor cells of metastatic colorectal cancer patients. Mol Oncol. 2015;9:920-932
39. Guo M, Li X, Zhang S. et al. Real-time quantitative RT-PCR detection of circulating tumor cells from breast cancer patients. Int J Oncol. 2015;46:281-289
40. Ignatiadis M, Xenidis N, Perraki M. et al. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J Clin Oncol. 2007;25:5194-5202
41. Tanaka K, Hata A, Kaji R. et al. Cytokeratin 19 fragment predicts the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitor in non-small-cell lung cancer harboring EGFR mutation. J Thorac Oncol. 2013;8:892-898
42. Travis WD, Rekhtman N, Riley GJ. et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: a paradigm shift. J Thorac Oncol. 2010;5:411-414
43. Saha SK, Choi HY, Kim BW. et al. KRT19 directly interacts with β-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene. 2017;36:332-349
Author contact
![]() Corresponding author: Kongming Wu, Department of Oncology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology. 1095 Jiefang Avenue, Wuhan 430030, P.R. China. Tel.: 0086-135-1719-6182 Fax: 0086-027-8366-2834, E-mail: kmwutjmu.edu.cn.
Corresponding author: Kongming Wu, Department of Oncology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology. 1095 Jiefang Avenue, Wuhan 430030, P.R. China. Tel.: 0086-135-1719-6182 Fax: 0086-027-8366-2834, E-mail: kmwutjmu.edu.cn.

 Global reach, higher impact
Global reach, higher impact