Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(5):1270-1283. doi:10.7150/jca.51621 This issue Cite
Research Paper
A pharmacogenetics study of platinum-based chemotherapy in lung cancer: ABCG2 polymorphism and its genetic interaction with SLC31A1 are associated with response and survival
1. Center for Medical Research and Innovation, Shanghai Pudong Hospital and Pudong Medical Center, Shanghai Medical College, Fudan University, Shanghai, China.
2. Ministry of Education Key Laboratory of Contemporary Anthropology and Department of Anthropology and Human Genetics, School of Life Sciences, Fudan University, Shanghai, China.
3. Department of Respiratory and Critical Care Medicine, Changhai Hospital, the Second Military Medical University, Shanghai, China.
4. Department of Pulmonary Medicine, Zhongshan Hospital of Fudan University, Shanghai, China.
5. Department of Pneumology, Chest Hospital, Shanghai Jiao Tong University, Shanghai, China.
6. Department of Cardiothoracic Surgery, Changzheng Hospital of the Second Military Medical University, Shanghai, China.
7. Department of Clinical Pharmacology, Xiangya Hospital; Hunan Key Laboratory of Pharmacogenomics, Institute of Clinical Pharmacology, Central South University, Changsha, China.
Received 2020-8-6; Accepted 2020-12-3; Published 2021-1-1
Abstract
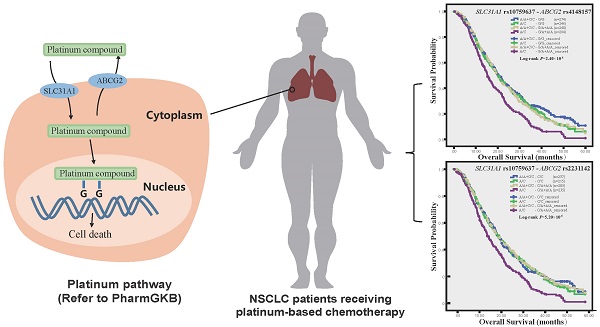
Objective: The expression and function of platinum transporters affect drug tissue concentration and therapeutic effects. We had previously characterized functional variant of platinum intake transporter SLC31A1 gene. We aimed to investigate the association of platinum efflux transporter gene ABCG2 polymorphism and combined ABCG2 and SLC31A1 polymorphisms with clinical outcomes of NSCLC patients receiving platinum-based chemotherapy.
Methods: We genotyped thirteen tagging and functional SNPs of ABCG2 in 1004 patients, and assessed their association with response, toxicity and survival using unconditional logistic regression and Cox proportional hazards regression analyses respectively.
Results: Nonsynonymous rs2231142 (odds ratio [OR] 2.07; 95 % confidence interval [CI] 1.26-3.63), rs1871744 (OR 0.60; 95 % CI 0.42-0.87) and their haplotype and diplotype were associated with objective response. Rs4148157 was associated with shorter overall survival (Log-rank P = 0.002; hazard ratio [HR] 1.22; 95 % CI 1.05-1.42). Furthermore, the combined SLC31A1 rs2233914 and ABCG2 rs1871744 genotype was significantly associated with poor response (OR 0.31; 95 % CI 0.17-0.56; Pinteraction = 0.003). And the combined genotypes of the functional rs10759637 of SLC31A1 and the nonsynonymous rs2231142 (Log-rank P = 5.20×10-5; HR 1.47; 95 % CI 1.19-1.81; Pinteraction = 0.007) or linked rs4148157 of ABCG2 were significantly associated with poor survival.
Conclusion: This study reveals divergent association of ABCG2 polymorphism with response and survival of NSCLC patients receiving platinum-based chemotherapy, demonstrates the combined effects of functional variants of ABCG2 and SLC31A1 on clinical outcomes, and highlights pharmacogenetic relevance of platinum transporter genes interaction.
Keywords: NSCLC, pharmacogenetics, platinum transporter, ABCG2, SLC31A1, SNP
Introduction
Lung cancer is one of the most common cancers in both man and woman with high mortality worldwide [1]. Non-small cell lung cancer (NSCLC) is the main type (80%) primary lung cancer, and often presents with advanced stage (Ⅲ/Ⅳ) upon first diagnosis [2]. The standard treatment for advanced NSCLC is platinum-based combination chemotherapy using cytotoxic compounds such as paclitaxel, navelbine and gemcitabine. However, the response rate is only 20% to 30% and the five-year survival rate is less than 15% [3]. Therapeutic response and efficacy are linked to pharmacokinetics and pharmacodynamics pathways including alteration in intracellular drug accumulation mediated by transporters, genetic polymorphisms in these pathways may influence interindividual variability as predictive markers for tailoring chemotherapy with better efficacy and minimal toxicity [3].
Accumulating evidences from studies in cell lines and in clinical setting support reduced drug accumulation as a significant mechanism of platinum resistance [4]. The intracellular accumulation of platinum drugs is determined by the plasma-membrane transporters that are responsible for their intake and efflux. Their uptake into cells is mainly mediated by SLC31A1 (solute carrier family 31 member 1), also known as CTR1 (copper transporter 1). Their efflux out of cells is largely mediated by the ATP-binding cassette (ABC) transporters including ABCG2, also known as the breast cancer resistance protein (BCRP), one of most important ABC transporters involved in multidrug resistance of cancer cells. ABCG2 is also expressed in normal tissues including liver, small intestine, colon, kidney and lung, notably in the bronchial epithelium and seromucinous glands, and thus affects the bioavailability and tissue distribution of its substrates [5]. In clinical setting of platinum-based chemotherapy for NSCLC, patients with undetectable SLC31A1 in tumors have reduced platinum concentration, decreased tumor response and shorter survival [4, 6], while ABCG2 expression in biopsy specimen predicts shorter survival [7, 8]. The aberrant expression and dysfunction of platinum transporters, which are largely ascribed to the functional polymorphisms of their coding genes, may influence interindividual variability in drug tissue concentration and therapeutic effects. We have recently reported that a functional SNP, rs10759637, at the 3′ untranslated region (3′UTR) of SLC31A1 gene could affect the microRNA-3′UTR interaction, modulate gene expression, and thereby is associated with toxicity and survival of NSCLC patients receiving platinum-based chemotherapy [9]. Although the association of ABCG2 polymorphism with drug resistance has been addressed in a range of solid tumors including lung cancer [5, 10-13], its relevance in outcome prediction for platinum-based chemotherapy of NSCLC is still elusive, and particularly, pharmacogenetic interaction between platinum uptake and efflux transporter genes is largely unknown.
In the present study of NSCLC patients receiving platinum-based treatment (n = 1004), we assessed the association of tagging and functional ABCG2 SNPs with objective response, survival and toxicities, and also tested the joint effects of ABCG2 and SLC31A1 polymorphisms on these clinical outcomes.
Materials and methods
Patient recruitment and follow-up
This NSCLC pharmacogenetics study involved two cohorts of patients, the discovery (A) panel (n = 237) and the replication (B) panel (n = 767). All of the 1004 patients are Chinese Han and were histologically diagnosed with stage Ⅲ-Ⅳ NSCLC between March 2005 and January 2010 from five hospitals in the East of China: Shanghai Chest Hospital, Shanghai Zhongshan Hospital, Shanghai Changhai Hospital, Shanghai Changzheng Hospital, and Cancer Hospital of Jiangsu Province. The recruitment criteria, the demographic and baseline characteristics including gender, age at diagnosis, smoking status, ECOG performance status, TNM stage, and histological type, were described in detail in our previous reports [9, 14-17]. The chemotherapeutic regimens were as follows: either cisplatin (75 mg/m2) or carboplatin (at an area under the curve 5), both administered on day 1 every 3 weeks, in combination with navelbine (25 mg/m2) on days 1 and 8 every 3 weeks, or gemcitabine (1250 mg/m2) on days 1 and 8 every 3 weeks, or paclitaxel (175 mg/m2) on day 1 every 3 weeks, or docetaxel (75 mg/m2) on day 1 every 3 weeks. A few patients received other platinum-based treatment (n = 49). Drugs were administered intravenously and treatments lasted for 2 to 6 cycles.
Clinical outcomes including responses, toxicities and survival were assessed. Tumor responses were evaluated after the first two cycles of the course according to the Response Evaluation Criteria in Solid Tumors (RECIST) guidelines version 1.0 [18] , which are classified into four categories: complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). Toxicity was assessed from the end of the first two cycles of treatment according to the Common Terminology Criteria for Adverse Events version 3.0 (CTCAE v3.0) [19]. Progression-free survival (PFS) was calculated from the date of chemotherapy beginning to the date of disease progression or death (whichever occurred first) or the last progression-free follow-up. Overall survival (OS) was calculated from the date of chemotherapy beginning to the date of death. The study was approved by the Ethical Review Committees of Fudan University School of Life Sciences and the participating hospitals, and was conducted in accordance with the Declaration of Helsinki. The written informed consent was obtained from each subject.
SNPs selection and genotyping
Thirteen tagging and functional SNPs of ABCG2 were selected. The tagging SNPs were screened from the Han Chinese in Beijing (CHB) population dataset of HapMap phase II database by Haploview 4.1 (http://www.broadinstitute.org/haploview) using a minor allele frequency (MAF) cutoff of 0.05 and a correlation coefficient (r2) threshold of 0.8. In the setting of common variations and candidate gene based strategy for genetic association study, the linkage disequilibrium (LD)-selected tagSNPs at a relatively stringent r2 threshold (r2 > 0.8) resolve >80% of all haplotypes diversity, regardless of recombination, and tag specific haplotypes and clades of related haplotypes in nonrecombinant regions, and analysis of the tagSNP set can comprehensively interrogate for main effects from common functional variation [20]. Genomic DNA was extracted from whole blood using the QIAamp DNA Maxi Kit (Qiagen GmbH, Hilden, Germany). Genotyping was performed using iSelect HD BeadChip (Illumina, San Diego, CA, USA).
Statistical analysis
We used Pearson χ2 tests to examine Hardy-Weinberg equilibrium (HWE) for genotype data and test their distribution between groups with various clinical outcomes. We used PHASE version 2.1 to estimate haplotypes [21], used Haploview 4.1 to plot linkage disequilibrium (LD, D' and r2). We measured the association between SNPs and dichotomous clinical phenotypes by calculating odds ratios (OR) and their 95% confidence intervals (CIs) in multivariate unconditional logistic regression analysis, with adjustment of gender, age, smoking status, ECOG performance status, TNM status, histological types and treatment regimen. Haplotype-based association analysis was performed using Haplo.stats package in R-plus (Version: 1.6.8). We tested the null hypotheses of multiplicative gene-gene interactions by evaluating departures from multiplicative joint effect model. Departure from the multiplicative model was assessed by including main effect variables and their product terms in the logistic regression model [22]. We analyzed the genetic association with survival by log-rank test with adjustment for covariates. We calculated the hazard ratios (HR) and 95%CI with multivariate Cox proportional hazards regression by adjustment for covariates, and plotted survival curve with Kaplan-Meier method. All statistical analysis was performed by SPSS (version 22). We use the two-side test for all P values. A P value < 0.05 was considered statistically significant. To account for the issue of multiple testing of SNPs in the Pearson χ2 tests of statistical significance of genotypic frequencies of ABCG2 SNP between response and non-response patients, or between toxicologically graded G0-2 and G3-4 patients, we used SNPSpD to correct the significance threshold taking into account LD between polymorphisms [23]. The popular Bonferroni method for multiple test correction may reduce statistical power when the analyzed SNPs show genetic association or in strong LD. Instead, on the basis of the spectral decomposition (SpD) of matrics of pairwise LD between SNPs, the SNPSpD method generates the experiment-wide significance threshold required to keep the Type I error rate at <5%. To account for the issue of multiple testing for log rank comparisons of more than two survival curves in the survival analysis, we used the Bonferroni method for adjustment [24].
Results
Patient Characteristics and Clinical Outcomes
The main characteristics for the 1004 eligible patients of the two panel cohorts in total and their clinical outcomes including objective response, toxicity and survival are summarized in Table 1 were documented in detail in our recent report [9]. Briefly, 177 (18.1%) of the 975 patients evaluated were responders (one was complete response, CR, and 176 were partial response, PR), and 798 (81.9%) were non-responders (610 showed stable disease, SD, and 188 showed progressive disease, PD). 29.9% of patients evaluated (n = 952) showed severe overall toxicity. 8.3% of patients evaluated (n = 964) showed severe gastrointestinal toxicity (nausea/vomiting). 23.9% of patients evaluated (n = 969) manifested severe hematological toxicity, among which severe thrombocytopenia, neutropenia, leukopenia, anemia were observed in 34 (3.6%), 115 (12.3%), 149 (15.2%) and 29 (3.1%) patients respectively. In the survival analysis, the mortality rate of the cohort patients was 74.9% during a median follow-up of 46.5 months until July 2012 for the final data collection, with median PFS and median OS being 6.5 and 16.0 months respectively. There was no significant difference in the distributions of the demographic and clinical characterizations between the two panels of patients cohorts. In addition, there were comparable rates of objective response and toxicities between our study population and those previously reported in large randomized clinical trials [25].
We selected thirteen tagging and functional SNPs of ABCG2, including eleven intronic (rs6857600, rs3109823, rs2725252, rs17731538, rs2231138, rs11931123, rs1871744, rs2231146, rs12505410, rs4148157 and rs2231164) and two nonsynonymous (rs2231137, Val12Met; rs2231142, Gln141Lys) variations, among which the nonsynonymous rs2231142 reportedly leads to reduced ABCG2 protein level and efflux transporter functionality [26]. In order to address the consistency of genetic association for the multi-center NSCLC cohorts, we genotyped all of these polymorphisms in both the discovery panel A and the replication panel B. All SNPs are common (MAF > 0.05) in this study population. Their genotypic distributions in panel A (n = 237), panel B (n = 767) and the combined cohort (n = 1004) were all in HWE, and were also comparable with those in the general healthy Chinese Han population in the 1000genome dataset (n = 208) (Supplemental Table 1). The LD analysis illustrated relatively strong association between the ABCG2 polymorphisms in the NSCLC Chinese Han population (Supplemental Figure 1), which is largely consistent with the haplotype architecture of ABCG2 in Chinese population we previously described by resequencing its exons and regulatory regions [27].
Association of ABCG2 polymorphism with response
We compared genotypic distributions of ABCG2 polymorphisms between responders (CR+PR) and non-responders (SD + PD) (Supplemental Table 2). Rs1871744 consistently showed differential genotypic distribution between responders and non-responders in either panel A (P = 0.023), panel B (P = 0.042) or the combined cohort (P = 0.022). Particularly, the imbalanced distribution of its genotypes, in under-dominant model, that is A/G vs A/A+G/G, remained statistically significant after multiple test correction in the combined cohort (P = 0.007). Significantly differential genotypic distribution was also observed for rs2231142 in either the total cohort (P = 0.005), or panel B (P = 0.002), but was not observed in panel A that was much less statistically powered due to its small sample size as compared with panel B. By using logistic regression analysis (Table 2), we found that the A/G genotype of rs1871744, in under-dominant model, was significantly associated with poor response in the total cohort (OR 0.60; 95% CI 0.42-0.87; P = 0.006), and this association was of significantly marginal significance in either panel A or panel B. The variant A/A genotype of rs2231142, in recessive model, was significantly associated with favorite response in either the total cohort (OR 2.07; 95% CI 1.26-3.63; P = 0.004) or panel B (OR 2.73; 95% CI 1.53-4.85; P = 0.001) but not panel A. We also estimated the haplotype and diplotype frequencies for rs1871744 and rs2231142 in the total cohort (Table 3). We predicted three haplotypes in the 975 patients evaluated for response with differential frequencies between responders and non-responders (P = 0.046), which was due to overrepresentation in responders of Hap2_AA (composed with their response prone alleles) (P = 0.019). Agreeing with the genotype-based association results as above, we consistently observed significant association of favorite response with the Hap2 haplotype (OR 1.45; 95% CI 1.10-1.90; P = 0.008) and the Hap2/Hap2 diplotype (OR 2.13; 95% CI 1.28-3.55; P = 0.004). Interestingly, in further stratification analysis by demographic and clinical characteristics, the associations of rs1871744 and rs2231142 with response were consistently pronounced in specific subgroups of patients such as men, older than 58, with ECOG PS 0-1, or with squamous cell carcinoma (Supplemental Table 3).
Patient characteristics and clinical outcomes (n = 1004)
| Characteristic | Panel A | Panel B | P value d | All | |||
|---|---|---|---|---|---|---|---|
| Total Number | n (%) | Total Number | n (%) | Total Number | n (%) | ||
| All patients | 237 | 767 | 1004 | ||||
| Sex | 237 | 767 | 1004 | ||||
| Male | 175 (73.8) | 531 (69.2) | 0.202 | 706 (70.3) | |||
| Female | 62 (26.2) | 236 (30.8) | 298 (29.7) | ||||
| Median age (yrs) | 237 | 59 | 767 | 58 | 1004 | 58 | |
| ≤ Median age | 123 (51.9) | 404 (52.7) | 0.893 | 518 (51.6) | |||
| > Median age | 114 (48.1) | 363 (47.3) | 486 (48.4) | ||||
| Smoking Status | 237 | 763 | 1000 | ||||
| Ever Smoker | 140 (59.1) | 435 (56.6) | 0.628 | 575 (57.5) | |||
| Nonsmokera | 97 (40.9) | 328 (43.4) | 425 (42.5) | ||||
| ECOG performance statusb | 234 | 756 | 990 | ||||
| 0-1 | 217 (92.7) | 687 (90.9) | 0.453 | 904 (91.3) | |||
| 2 | 17 (7.3) | 69 (9.1) | 86 (8.7) | ||||
| TNM stage | 226 | 773 | 999 | ||||
| IIIA | 19 (8.0) | 62 (8.1) | 0.073 | 81 (8.1) | |||
| IIIB | 83 (35.2) | 210 (27.5) | 293 (29.3) | ||||
| IV | 134 (56.8) | 491 (64.4) | 625 (62.6) | ||||
| Histological type | 237 | 767 | 1004 | ||||
| Adenocarcinoma (AC) | 143 (60.3) | 489 (63.8) | 0.533 | 632 (62.9) | |||
| Squamous cell carcinoma (SCC) | 53 (22.4) | 168 (21.9) | 221 (22.1) | ||||
| Adenosquamocarcinoma | 7 (2.9) | 13 (1.7) | 20 (2.0) | ||||
| Othersc | 34 (14.4) | 97 (12.6) | 131 (13.0) | ||||
| Chemotherapy regimens | 237 | 767 | 1004 | ||||
| Platinum (cisplatin)-navelbine | 84 (35.4) | 232 (30.2) | 0.110 | 316 (31.5) | |||
| Platinum (cisplatin)-gemcitabine | 52 (21.9) | 187 (24.4) | 239 (23.8) | ||||
| Platinum (carboplatin)-paclitaxel | 61 (25.7) | 252 (32.9) | 313 (31.2) | ||||
| Platinum-docetaxel | 25 (10.5) | 62 (8.1) | 87 (8.7) | ||||
| Other platinum combinations | 15 (6.5) | 34 (4.4) | 49 (4.9) | ||||
| Objective response | 234 | 741 | 975 | ||||
| Complete response (CR) | 1 (0.4) | 0 (0.0) | 0.106 | 1 (0.1) | |||
| Partial response (PR) | 43 (18.4) | 133 (17.9) | 176 (18.0) | ||||
| Stable disease (SD) | 154 (65.8) | 456 (61.5) | 610 (62.6) | ||||
| Progressive disease (PD) | 36 (15.4) | 152 (20.6) | 188 (19.3) | ||||
| Toxicity outcome | |||||||
| Grade 3 or 4 gastrointestinal toxicity | |||||||
| Nausea/vomiting | 225 | 16 (7.1) | 739 | 64 (8.7) | 0.587 | 964 | 80 (8.3) |
| Grade 3 or 4 hematologic toxicity | 223 | 52 (23.3) | 746 | 180 (24.1) | 0.914 | 969 | 232 (23.9) |
| Anemia | 223 | 12 (5.4) | 721 | 17 (2.4) | 0.048 | 944 | 29 (3.1) |
| Leukopenia | 227 | 34 (15.0) | 753 | 115 (15.3) | 1.000 | 980 | 149 (15.2) |
| Neutropenia | 217 | 19 (8.8) | 718 | 96 (13.4) | 0.133 | 935 | 115 (12.3) |
| Thrombocytopenia | 226 | 9 (4.0) | 724 | 25 (3.5) | 0.876 | 950 | 34 (3.6) |
| Grade 3 or 4 overall toxicity | 222 | 60 (27.0) | 730 | 225 (30.8) | 0.472 | 952 | 285 (29.9) |
| Median time to outcomes (months) | 228 | 744 | 972 | ||||
| Progression-free survival (PFS) | 6.6 | 6.5 | 6.5 | ||||
| Overall survival (OS) | 17.5 | 15.7 | 16.0 | ||||
a Nonsmokers were defined as those who had smoked <1 cigarette per day and for <1 year in their lifetime.
b ECOG PS, Eastern Cooperative Oncology Group performance status.
c Other carcinomas included mixed cell or undifferentiated carcinoma.
d P values of Pearson χ2 tests for differences between panel A and B.
Association between ABCG2 SNPs and objective response
| Genotype | Panel A | Panel B | All | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Response (CR+PR/SD+PD) | P valuea | OR (95%CI)b | Response (CR+PR/SD+PD) | P value a | OR (95%CI)b | Response (CR+PR/SD+PD) | P valuea | OR (95%CI)b | |
| rs2231164 | |||||||||
| A/A | 16/51 | 0.274 | 1.00 (ref) | 33/173 | 0.028 | 1.00 (ref) | 49/224 | 0.035 | 1.00 (ref) |
| A/G | 16/92 | 0.54 (0.23-1.26) | 60/316 | 1.09(0.68-1.77) | 76/408 | 0.89 (0.59-1.34) | |||
| G/G | 12/47 | 0.77 (0.31-1.95) | 40/119 | 1.94 (1.13-3.32) | 52/166 | 1.47 (0.93-2.32) | |||
| G/G vs A/A+A/G | 32/143 | 0.528 | 1.12 (0.51-2.46) | 93/489 | 0.008 | 1.83 (1.17-2.84) | 125/632 | 0.013 | 1.58 (1.08-2.32) |
| rs4148157 | |||||||||
| G/G | 23/100 | 0.433 | 1.00 (ref) | 63/335 | 0.036 | 1.00 (ref) | 86/435 | 0.043 | 1.00 (ref) |
| G/A | 16/75 | 0.69 (0.32-1.53) | 57/245 | 1.35 (0.90-2.04) | 73/320 | 1.17 (0.82-1.68) | |||
| A/A | 5/15 | 1.86 (0.57-6.05) | 13/28 | 2.65(1.23-5.70) | 18/43 | 2.12 (1.13-4.00) | |||
| A/A vs G/G+G/A | 39/175 | 0.458 | 2.19 (0.71-6.75) | 120/580 | 0.018 | 2.31 (1.11-4.84) | 159/755 | 0.017 | 2.00 (1.08-3.71) |
| rs1871744 | |||||||||
| A/A | 25/98 | 0.023 | 1.00 (ref) | 77/279 | 0.042 | 1.00 (ref) | 102/377 | 0.022 | 1.00 (ref) |
| A/G | 9/72 | 0.51 (0.21-1.23) | 44/254 | 0.59 (0.39-0.91) | 53/326 | 0.59 (0.41-0.86) | |||
| G/G | 10/20 | 2.35 (0.87-6.31) | 12/75 | 0.59 (0.30-1.18) | 22/95 | 0.92 (0.54-1.58) | |||
| A/G vs A/A+G/G | 35/118 | 0.022 | 0.43 (0.18-0.99) | 91/362 | 0.064 | 0.65 (0.43-0.98) | 124/472 | 0.007 c | 0.60 (0.42-0.87) |
| rs2231142 | |||||||||
| C/C | 21/89 | 0.618 | 1.00 (ref) | 55/292 | 0.002 | 1.00 (ref) | 76/381 | 0.005 c | 1.00 (ref) |
| C/A | 17/80 | 0.83 (0.39-1.78) | 55/270 | 1.17 (0.76-1.79) | 72/350 | 1.07 (0.74-1.54) | |||
| A/A | 6/21 | 1.13 (0.37-3.43) | 23/46 | 2.96 (1.59-5.45) | 29/67 | 2.14 (1.26-3.63) | |||
| A/A vs C/C+C/A | 38/169 | 0.348 | 1.24 (0.44-3.51) | 110/562 | 4.71×10-4 | 2.73 (1.53-4.85) | 148/731 | 0.001 c | 2.07 (1.26-3.63) |
a P values of Pearson χ2 tests.
b Odds ratios (OR) and their 95% confidence intervals (CIs) and P values were calculated with unconditional logistic regression analysis, with adjustment of gender, age, smoking status, ECOG performance status, TNM status, histological types, and treatment regimen.
c Statistical significance remained after multiple tests adjustment taking into account linkage disequilibrium between polymorphisms.
Association between ABCG2 haplotype and diplotype and objective response
| Haplotype or Diplotypea | Response (CR+PR/SD+PD) | P-valueb | OR (95% CI)c | P-valuec |
|---|---|---|---|---|
| Haplotype frequency | ||||
| Hap1_AC | 127/596 | 0.605 | 1.00 | |
| Hap2_AA | 130/484 | 0.019 | 1.45 (1.10-1.90) | 0.008 |
| Hap3_GC | 97/516 | 0.071 | 1.08 (0.62-1.88) | 0.781 |
| Diplotype frequency | ||||
| Non-Hap2 carriers | 104/530 | 0.005 | 1.00 | |
| Hap2-Others carriers | 44/201 | 1.10 (0.73-1.65) | 0.643 | |
| Hap2-Hap2 carriers | 29/67 | 2.13 (1.28-3.55) | 0.004 | |
a Haplotypes were predicted with PHASE basing on rs1871744(A/G) and rs2231142 (C/A) that were associated with ORR outcomes
b P-values of Pearson χ2 test for the difference in haplotype and diplotype frequencies.
c Odds ratios (OR) and their 95% confidence intervals (CIs) and P values were calculated with unconditional logistic regression analysis, with adjustment of gender, age, smoking status, ECOG performance status, TNM status, histological types, and treatment regimen.
Joint association of ABCG2 and SLC31A1 polymorphisms with response
We have recently reported that SLC31A1 polymorphisms overall were not associated with response [9], except rs2233914 (G/A), a common variant at 5' flanking region upstream the transcription start site that was associated with poor response in dominant model (OR 0.67; 95% CI 0.48-0.95; P = 0.024) (Supplemental Table 4). In an effort to interrogate pharmacogenetically relevant genetic interaction between platinum drug intake and export pathways, we investigated the joint effect of ABCG2 (rs1871744 and rs2231142) and SLC31A1 (rs2233914) genotypes on response in the total cohort (Table 4). The combined rs1871744 A/G and rs2233914 (G/A+A/A) group was significantly underrepresented in responders than in non-responders (9.04% versus 23.68%; P = 2.404×10-5, Pearson's Chi-squared test with Yates' continuity correction). Compared to the reference, the presence of only one non-responsive genotype, either of rs1871744 A/G or rs2233914 G/A+A/A were not associated with response. However, their combined group was significantly associated with reduced response opportunity (OR 0.31; 95% CI 0.17-0.56; Pinteraction = 0.003). We did not observe significant joint effect of SLC31A1 rs2233914 and ABCG2 and rs2231142 genotypes on response (Supplemental Table 5). In further stratification analysis for the joint effect, the association of combined genotype of rs1871744 A/G and rs2233914 G/A+A/A with poor response was highly significant in male patients (OR 0.20; 95% CI 0.10-0.42), patients older than 58 (OR 0.16; 95% CI 0.06-0.41), patients with ECOG PS 0-1 (OR 0.30; 95% CI 0.16-0.57), smoker patients (OR 0.25; 95% CI 0.11-0.54) and patients with squamous cell carcinoma (OR 0.14; 95% CI 0.04-0.53), which is largely consistent with the stratification spectrum of rs1871744. These results suggest genetic interaction between SLC31A1 (rs2233914) and ABCG2 (rs1871744) associated with tumor response to platinum drug chemotherapy.
Association of ABCG2 polymorphism with survival
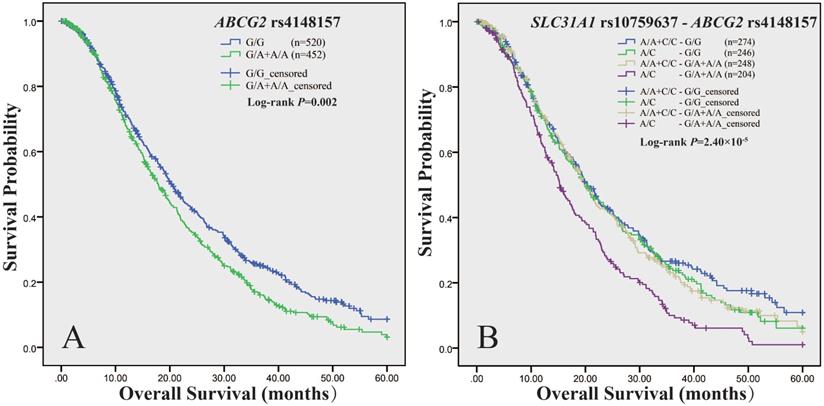
We measured association of ABCG2 polymorphisms with survival in panel A, panel B and the total cohort respectively (Supplemental Table 6). As to PFS, we did not observe any association signal. As to OS, the log-rank test showed that rs4148157, in dominant model, was consistently associated with survival in either panel A (P = 0.006), or panel B (P = 0.04), or the combined cohort (P = 0.002) (Table 5). As shown in Figure 1A, in the total cohort, the median OS time of patients with rs4148157 G/A+A/A was significantly shorter than those with G/G (17.9 vs 20.4, P = 0.002). Cox proportional hazards regression analysis showed that rs4148157 was associated with disease progression (HR 1.22; 95% CI 1.05-1.42). Notably, in further stratification analysis (Supplemental Table 7), the association of rs4148157 with OS were pronounced in patients with ECOG PS 0-1, patients with IIIB TNM stage, patients with adenocarcinoma, and patients treated with platinum-gemcitabine, respectively. We also observed that rs2231142, which was in strong LD with rs4148157 (D′ = 0.94 r2 = 0.69), was associated with survival in either panel A (log-rank P = 0.007) or the total cohort (log-rank P = 0.028) but not panel B. These data show that the two linked SNPs of ABCG2 were associated with survival.
Joint association of ABCG2 and SLC31A1 polymorphisms with survival
We have recently reported that the functional rs10759637 of SLC31A1 was associated with shorter OS [9]. We here further analyzed the joint effect of SLC31A1 (rs10759637) and ABCG2 (rs4148157 and rs2231142) on survival in the total cohort (Table 6). The combined genotypes of SLC31A1 rs10759637 and ABCG2 rs4148157 were significantly associated with survival (log-rank P = 2.50×10-5) (Figure 1B). The median OS times of patients with different genotypic combination were 20.4, 20.0, 19.9, and 15.3 months, respectively. The combined group of rs10759637 A/C and rs4148157 G/A+A/A was significantly associated with disease progression (HR 1.58; 95% CI 1.28-1.96). Furthermore, stratification analysis showed that their joint association with shorter OS were significant in male patients (HR 1.54; 95% CI 1.20-1.98), patients older than 58 (HR 1.61; 95% CI 1.19-2.17), patients with ECOG PS 0-1 (HR 1.62; 95% CI 1.30-2.02), smoker patients (HR 1.67; 95% CI 1.27-2.21), patients with IIIB TNM stage (HR 1.75; 95% CI 1.19-2.57), patients with adenocarcinoma (HR 1.49; 95% CI 1.14-1.96), and patients treated with platinum-gemcitabine (HR 2.30; 95% CI 1.42-3.75), respectively. Similarly, joint effect on OS and accordant stratification spectrum with pronounced association signal were also observed for SLC31A1 (rs10759637) and ABCG2 (rs2231142) combination (Supplemental Table 8, Supplemental Figure 2). Of note, we also observed significant interactions between SLC31A1 (rs10759637) and ABCG2 (rs4148157, Pinteraction = 0.03; rs2231142, Pinteraction = 0.007) associated with OS. These data suggest that genetic interaction between SLC31A1 and ABCG2 polymorphisms may be linked with survival outcome.
Kaplan-Meier curve of estimated overall survival for the NSCLC cohort according to ABCG2 and SLC31A1 polymorphisms. The curves were plotted with SPSS software according to the genotypes of ABCG2 rs4148157 (A), and the combined genotypes of ABCG2 rs4148157 and SLC31A1 rs10759637 (B). For ABCG2 rs4148157 G/A, the G/A+A/A genotypes group was compared to the wild G/G as reference in dominant model. For SLC31A1rs10759637 A/C, which had been genotyped in our previous report (Ref 9), the A/C heterozygote was compared to the A/A+C/C homozygotes group as reference in under-dominant model.
Joint association of SLC31A1 rs2233914 (G/A) and ABCG2 rs1871744 (A/G) with objective response
| Stratification subgroup | Genotype (SLC31A1—ABCG2)a | Response (CR+PR/SD+PD) | OR (95% CI)b | P valueb |
|---|---|---|---|---|
| All | [G/G] - [A/A+G/G] | 58/215 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 66/257 | 0.93 (0.62-1.41) | 0.740 | |
| [G/G] - [A/G] | 37/137 | 0.96 (0.59-1.57) | 0.875 | |
| [A/G+A/A] - [A/G] | 16/189 | 0.31 (0.17-0.56)c | 1.23×10-4 | |
| Gender | ||||
| Male | [G/G] - [A/A+G/G] | 49/141 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 50/189 | 0.73 (0.45-1.17) | 0.190 | |
| [G/G] - [A/G] | 22/94 | 0.56 (0.31-1.04) | 0.066 | |
| [A/G+A/A] - [A/G] | 10/134 | 0.20 (0.10-0.42) | 2.50×10-5 | |
| Female | [G/G] - [A/A+G/G] | 9/74 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 16/68 | 1.65 (0.64-4.24) | 0.299 | |
| [G/G] - [A/G] | 15/43 | 3.19 (1.22-8.33) | 0.018 | |
| [A/G+A/A] - [A/G] | 6/55 | 0.85 (0.27-2.67) | 0.778 | |
| Age | ||||
| ≤58 | [G/G] - [A/A+G/G] | 23/126 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 28/127 | 1.12 (0.60-2.11) | 0.719 | |
| [G/G] - [A/G] | 22/70 | 1.51 (0.76-3.03) | 0.243 | |
| [A/G+A/A] - [A/G] | 10/96 | 0.54 (0.24-1.23) | 0.141 | |
| >58 | [G/G] - [A/A+G/G] | 35/89 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 38/130 | 0.69 (0.39-1.23) | 0.211 | |
| [G/G] - [A/G] | 15/67 | 0.53 (0.26-1.11) | 0.091 | |
| [A/G+A/A] - [A/G] | 6/93 | 0.16 (0.06-0.41) | 1.46×10-4 | |
| ECOG PS | ||||
| 0-1 | [G/G] - [A/A+G/G] | 53/199 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 64/223 | 1.02 (0.66-1.58) | 0.922 | |
| [G/G] - [A/G] | 31/123 | 0.95 (0.57-1.60) | 0.849 | |
| [A/G+A/A] - [A/G] | 14/171 | 0.30 (0.16-0.57) | 2.44×10-4 | |
| 2 | [G/G] - [A/A+G/G] | 5/12 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 2/29 | 0.04 (0.00-0.53) | 0.014 | |
| [G/G] - [A/G] | 5/12 | 0.47 (0.04-5.36) | 0.546 | |
| [A/G+A/A] - [A/G] | 2/17 | 0.18 (0.01-2.98) | 0.230 | |
| Smoking status | ||||
| Nonsmoker | [G/G] - [A/A+G/G] | 20/103 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 22/100 | 1.15 (0.57-2.32) | 0.703 | |
| [G/G] - [A/G] | 21/57 | 2.06 (1.00-4.28) | 0.052 | |
| [A/G+A/A] - [A/G] | 6/82 | 0.40 (0.15-1.08) | 0.070 | |
| Smoker | [G/G] - [A/A+G/G] | 38/112 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 44/156 | 0.80 (0.47-1.36) | 0.409 | |
| [G/G] - [A/G] | 16/80 | 0.52 (0.26-1.06) | 0.072 | |
| [A/G+A/A] - [A/G] | 10/107 | 0.25 (0.11-0.54) | 4.27×10-4 | |
| TNM stage | ||||
| IIIA | [G/G] - [A/A+G/G] | 9/12 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 9/17 | 0.47 (0.13-1.80) | 0.272 | |
| [G/G] - [A/G] | 6/10 | 0.53 (0.11-2.62) | 0.436 | |
| [A/G+A/A] - [A/G] | 3/13 | 0.24 (0.05-1.24) | 0.088 | |
| IIIB | [G/G] - [A/A+G/G] | 19/72 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 19/63 | 1.20 (0.56-2.60) | 0.637 | |
| [G/G] - [A/G] | 15/32 | 1.66 (0.70-3.93) | 0.247 | |
| [A/G+A/A] - [A/G] | 3/61 | 0.18 (0.05-0.66) | 0.010 | |
| IV | [G/G] - [A/A+G/G] | 30/130 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 38/175 | 0.86 (0.50-1.50) | 0.604 | |
| [G/G] - [A/G] | 16/93 | 0.76 (0.38-1.50) | 0.423 | |
| [A/G+A/A] - [A/G] | 10/115 | 0.38 (0.17-0.82) | 0.013 | |
| Histological type | ||||
| AC | [G/G] - [A/A+G/G] | 24/146 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 24/173 | 0.84 (0.45-1.56) | 0.577 | |
| [G/G] - [A/G] | 19/84 | 1.34 (0.68-2.64) | 0.391 | |
| [A/G+A/A] - [A/G] | 11/131 | 0.50 (0.23-1.07) | 0.072 | |
| SCC | [G/G] - [A/A+G/G] | 20/37 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 24/46 | 0.99 (0.46-2.12) | 0.969 | |
| [G/G] - [A/G] | 15/38 | 0.72 (0.30-1.70) | 0.451 | |
| [A/G+A/A] - [A/G] | 3/34 | 0.14 (0.04-0.53) | 0.004 | |
| Therapy regimens | ||||
| Pt-navelbine | [G/G] - [A/A+G/G] | 21/62 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 28/78 | 0.97 (0.49-1.94) | 0.936 | |
| [G/G] - [A/G] | 17/36 | 1.41 (0.63-3.15) | 0.406 | |
| [A/G+A/A] - [A/G] | 7/53 | 0.33 (0.13-0.87) | 0.025 | |
| Pt-gemcitabine | [G/G] - [A/A+G/G] | 13/54 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 15/61 | 0.90 (0.38-2.17) | 0.820 | |
| [G/G] - [A/G] | 4/29 | 0.52 (0.14-1.89) | 0.323 | |
| [A/G+A/A] - [A/G] | 5/50 | 0.47 (0.15-1.47) | 0.195 | |
| Pt-paclitaxe | [G/G] - [A/A+G/G] | 20/72 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 13/77 | 0.59 (0.25-1.38) | 0.223 | |
| [G/G] - [A/G] | 13/54 | 0.89 (0.38-2.08) | 0.786 | |
| [A/G+A/A] - [A/G] | 3/58 | 0.19 (0.05-0.70) | 0.012 | |
| Pt-docetaxel | [G/G] - [A/A+G/G] | 3/15 | 1.00 (ref) | |
| [A/G+A/A] - [A/A+G/G] | 4/29 | 1.54 (0.16-15.34) | 0.711 | |
| [G/G] - [A/G] | 3/10 | 0.55 (0.06-5.16) | 0.597 | |
| [A/G+A/A] - [A/G] | 1/20 | 0.25 (0.01-4.70) | 0.352 |
a SLC31A1 rs2233914 (G/A) for the study subjects had been genotyped in our previous report (Ref 9).
b Odds ratios (OR) and their 95% confidence intervals (CIs) and P values were calculated with unconditional logistic regression analysis, with adjustment of gender, age, smoking status, ECOG performance status, TNM status, histological types, and treatment regimen.
c Test of interaction for the cohort of all patients with P value being 0.003.
ABCG2 polymorphism and toxicity
We also investigated the effects of ABCG2 polymorphism on toxicological outcomes. Genotypic distributions between groups with respective mild or severe toxicities are shown in Supplemental Table 9. We found that rs12505410, rs1871744, rs2231142 and rs2231138 displayed differential genotypic distribution, with marginal significance, between groups with mild and severe neutropenia or anemia, and rs12505410 C/C genotype was significantly associated, in recessive model, with neutropenia risk (OR 2.09; 95% CI 1.23-3.57) (Supplemental Table 10).
Association between ABCG2 SNPs and overall survival
| Genotype | Panel A | Panel B | All | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/Na | MST (m)b | Log-rank P | HR (95% CI)c | n/Na | MST (m)b | Log-rank P | HR (95% CI)c | n/Na | MST (m)b | Log-rank P | HR (95% CI)c | |
| rs2231164 | ||||||||||||
| A/A | 47/65 | 22.4 | 0.025 | 1.00 (ref) | 152/205 | 19.1 | 0.383 | 1.00 (ref) | 199/270 | 20.7 | 0.039 | 1.00 (ref) |
| A/G | 79/104 | 19.3 | 1.36 (0.92-2.00) | 276/379 | 18.8 | 1.01 (0.83-1.24) | 355/483 | 19.0 | 1.08 (0.91-1.29) | |||
| G/G | 47/59 | 15.8 | 1.86 (1.18-2.94) | 127/160 | 18.2 | 1.15 (0.91-1.47) | 174/219 | 18.0 | 1.26 (1.02-1.55) | |||
| rs4148157 | ||||||||||||
| G/G | 84/118 | 21.8 | 0.018 | 1.00 (ref) | 287/402 | 19.0 | 0.120 | 1.00 (ref) | 371/520 | 20.4 | 0.008d | 1.00 (ref) |
| G/A | 72/90 | 15.6 | 1.35 (0.95-1.92) | 236/300 | 18.9 | 1.20 (1.00-1.43) | 308/390 | 17.9 | 1.21 (1.04-1.42) | |||
| A/A | 17/0 | 15.1 | 1.67 (0.95-2.93) | 32/42 | 20.2 | 1.14 (0.78-1.66) | 49/62 | 17.0 | 1.25 (0.92-1.70) | |||
| G/A+A/A vs G/G | 89/110 | 15.4 | 0.006 | 1.40 (1.01-1.96) | 268/342 | 18.9 | 0.040 | 1.19 (1.01-1.41) | 357/452 | 17.9 | 0.002 | 1.22 (1.05-1.42) |
| rs12505410 | ||||||||||||
| A/A | 92/114 | 18.7 | 0.083 | 1.00 (ref) | 245/327 | 17.0 | 0.350 | 1.00 (ref) | 337/441 | 17.1 | 0.064 | 1.00 (ref) |
| A/C | 60/85 | 21.4 | 0.73 (0.51-1.04) | 241/327 | 19.7 | 0.85 (0.71-1.03) | 301/412 | 20.7 | 0.84 (0.72-0.99) | |||
| C/C | 21/29 | 21.5 | 0.82 (0.49-1.36) | 69/90 | 22.2 | 0.78 (0.59-1.03) | 90/119 | 21.6 | 0.77 (0.61-0.96) | |||
| rs2231142 | ||||||||||||
| C/C | 76/106 | 21.7 | 0.025 | 1.00 (ref) | 248/327 | 19.0 | 0.326 | 1.00 (ref) | 327/452 | 20.4 | 0.043 | 1.00 (ref) |
| C/A | 73/95 | 15.4 | 1.49 (1.05-2.10) | 248/329 | 18.9 | 1.06 (0.88-1.27) | 321/422 | 17.9 | 1.14 (0.97-1.33) | |||
| A/A | 24/27 | 21.2 | 1.46 (0.88-2.42) | 60/76 | 16.5 | 1.21 (0.90-1.63) | 80/98 | 19.6 | 1.27 (0.99-1.64) | |||
| C/A+A/A vs C/C | 97/122 | 16.6 | 0.007 | 1.48 (1.07-2.06) | 308/405 | 18.9 | 0.336 | 1.08 (0.91-1.29) | 401/520 | 18.0 | 0.028 | 1.16 (1.00-1.35) |
a Numbers indicate the death event for NSCLC patients during the following-up time among all individuals in the same genotype group.
b MST: median survival time.
c Hazard ratios (HR) and their 95% confidence intervals (CIs) and P values were calculated with by multivariate Cox proportional hazards regression with adjustment for covariates.
d Statistical significance remained after Bonferroni multiple tests.
Joint association of SLC31A1 rs10759637 (A/C) and ABCG2 rs4148157 (G/A) with overall survival
| Stratification subgroup | Genotype (SLC31A1—ABCG2)a | n/Nb | MST (m)c | Log-rank P | HR (95% CI)d | P valued |
|---|---|---|---|---|---|---|
| All | [A/A+C/C] - [G/G] | 188/274 | 20.4 | 2.50×10-5 f | 1.00 (ref) | |
| [A/C] - [G/G] | 183/246 | 20.0 | 1.08 (0.87-1.33) | 0.482 | ||
| [A/A+C/C] - [G/A+A/A] | 184/248 | 19.9 | 1.07 (0.86-1.31) | 0.557 | ||
| [A/C] - [G/A+A/A] | 173/204 | 15.3 | 1.58 (1.28-1.96)e | 2.40×10-5 | ||
| Gender | ||||||
| Male | [A/A+C/C] - [G/G] | 143/196 | 19.5 | 0.002 f | 1.00 (ref) | |
| [A/C] - [G/G] | 123/168 | 19.1 | 1.04 (0.81-1.33) | 0.752 | ||
| [A/A+C/C] - [G/A+A/A] | 138/184 | 19.2 | 1.02 (0.80-1.30) | 0.888 | ||
| [A/C] - [G/A+A/A] | 124/141 | 14.9 | 1.54 (1.20-1.98) | 0.001 | ||
| Female | [A/A+C/C] - [G/G] | 45/78 | 28.8 | 0.013 | 1.00 (ref) | |
| [A/C] - [G/G] | 60/78 | 23.8 | 1.18 (0.78-1.79) | 0.428 | ||
| [A/A+C/C] - [G/A+A/A] | 46/64 | 21.4 | 1.30 (0.84-2.03) | 0.243 | ||
| [A/C] - [G/A+A/A] | 49/63 | 16.8 | 1.73 (1.12-2.66) | 0.013 | ||
| Age | ||||||
| ≤58 | [A/A+C/C] - [G/G] | 103/157 | 22.3 | 0.010 | 1.00 (ref) | |
| [A/C] - [G/G] | 86/126 | 22.7 | 0.98 (0.73-1.32) | 0.896 | ||
| [A/A+C/C] - [G/A+A/A] | 94/128 | 21.2 | 1.09 (0.81-1.46) | 0.572 | ||
| [A/C] - [G/A+A/A] | 76/94 | 17.9 | 1.51 (1.11-2.06) | 0.009 | ||
| >58 | [A/A+C/C] - [G/G] | 85/117 | 18.8 | 0.004f | 1.00 (ref) | |
| [A/C] - [G/G] | 97/120 | 17.7 | 1.11 (0.82-1.51) | 0.485 | ||
| [A/A+C/C] - [G/A+A/A] | 90/120 | 19.3 | 0.96 (0.70-1.31) | 0.788 | ||
| [A/C] - [G/A+A/A] | 97/110 | 14.3 | 1.61 (1.19-2.17) | 0.002 | ||
| ECOG PS | ||||||
| 0-1 | [A/A+C/C] - [G/G] | 172/253 | 21.0 | 3.00×10-6f | 1.00 (ref) | |
| [A/C] - [G/G] | 156/215 | 20.9 | 1.06 (0.85-1.32) | 0.604 | ||
| [A/A+C/C] - [G/A+A/A] | 168/223 | 20.0 | 1.08 (0.87-1.34) | 0.491 | ||
| [A/C] - [G/A+A/A] | 160/188 | 15.2 | 1.62 (1.30-2.02) | 1.60×10-5 | ||
| 2 | [A/A+C/C] - [G/G] | 12/16 | 17.8 | 0.800 | 1.00 (ref) | |
| [A/C] - [G/G] | 23/27 | 12.4 | 0.82 (0.34-1.97) | 0.658 | ||
| [A/A+C/C] - [G/A+A/A] | 15/24 | 19.1 | 0.43 (0.16-1.14) | 0.089 | ||
| [A/C] - [G/A+A/A] | 11/13 | 21.4 | 0.67 (0.24-1.88) | 0.444 | ||
| Smoking status | ||||||
| Nonsmoker | [A/A+C/C] - [G/G] | 72/116 | 22.4 | 0.042 | 1.00 (ref) | |
| [A/C] - [G/G] | 76/101 | 23.8 | 1.03 (0.73-1.44) | 0.870 | ||
| [A/A+C/C] - [G/A+A/A] | 75/101 | 20.3 | 1.04(0.73-1.46) | 0.843 | ||
| [A/C] - [G/A+A/A] | 68/87 | 17.2 | 1.42 (1.00-2.00) | 0.048 | ||
| Smoker | [A/A+C/C] - [G/G] | 116/158 | 20.2 | 0.001 f | 1.00 (ref) | |
| [A/C] - [G/G] | 106/144 | 18.1 | 1.10 (0.84-1.45) | 0.483 | ||
| [A/A+C/C] - [G/A+A/A] | 108/146 | 19.2 | 1.07 (0.82-1.40) | 0.624 | ||
| [A/C] - [G/A+A/A] | 103/115 | 14.3 | 1.67(1.27-2.21) | 2.99×10-4 | ||
| TNM stage | ||||||
| IIIA | [A/A+C/C] - [G/G] | 14/20 | 31.3 | 0.192 | 1.00 (ref) | |
| [A/C] - [G/G] | 15/21 | 15.3 | 2.54 (1.04-6.20) | 0.041 | ||
| [A/A+C/C] - [G/A+A/A] | 13/21 | 25.5 | 3.20 (1.28-8.00) | 0.013 | ||
| [A/C] - [G/A+A/A] | 14/14 | 16.5 | 2.81 (1.06-7.47) | 0.038 | ||
| IIIB | [A/A+C/C] - [G/G] | 64/89 | 21.0 | 0.004 f | 1.00 (ref) | |
| [A/C] - [G/G] | 43/57 | 22.5 | 0.99 (0.66-1.50) | 0.976 | ||
| [A/A+C/C] - [G/A+A/A] | 55/75 | 17.0 | 1.37 (0.95-1.99) | 0.095 | ||
| [A/C] - [G/A+A/A] | 53/62 | 15.1 | 1.75 (1.19-2.57) | 0.004 | ||
| IV | [A/A+C/C] - [G/G] | 110/165 | 18.3 | 0.019 | 1.00 (ref) | |
| [A/C] - [G/G] | 124/167 | 19.6 | 1.02 (0.79-1.33) | 0.875 | ||
| [A/A+C/C] - [G/A+A/A] | 115/150 | 21.3 | 0.93 (0.71-1.22) | 0.604 | ||
| [A/C] - [G/A+A/A] | 104/126 | 15.6 | 1.42 (1.08-1.87) | 0.013 | ||
| Histological type | ||||||
| AC | [A/A+C/C] - [G/G] | 117/175 | 21.8 | 0.004 f | 1.00 (ref) | |
| [A/C] - [G/G] | 119/158 | 22.5 | 1.02 (0.78-1.32) | 0.900 | ||
| [A/A+C/C] - [G/A+A/A] | 113/148 | 19.6 | 1.18 (0.91-1.54) | 0.214 | ||
| [A/C] - [G/A+A/A] | 105/131 | 16.5 | 1.49 (1.14-1.96) | 0.004 | ||
| SCC | [A/A+C/C] - [G/G] | 48/66 | 19.3 | 0.044 | 1.00 (ref) | |
| [A/C] - [G/G] | 34/50 | 13.1 | 1.26 (0.79-2.01) | 0.334 | ||
| [A/A+C/C] - [G/A+A/A] | 37/54 | 21.9 | 0.75 (0.47-1.19) | 0.218 | ||
| [A/C] - [G/A+A/A] | 42/43 | 14.3 | 1.48 (0.93-2.33) | 0.096 | ||
| Therapy regimens | ||||||
| Pt-navelbine | [A/A+C/C] - [G/G] | 61/90 | 21.8 | 0.178 | 1.00 (ref) | |
| [A/C] - [G/G] | 55/72 | 23.3 | 1.04 (0.71-1.52) | 0.837 | ||
| [A/A+C/C] - [G/A+A/A] | 63/84 | 21.3 | 0.89 (0.62-1.28) | 0.532 | ||
| [A/C] - [G/A+A/A] | 50/60 | 16.1 | 1.48 (1.01-2.16) | 0.045 | ||
| Pt-gemcitabine | [A/A+C/C] - [G/G] | 37/62 | 22.5 | 0.002f | 1.00 (ref) | |
| [A/C] - [G/G] | 52/68 | 20.9 | 1.24 (0.80-1.92) | 0.343 | ||
| [A/A+C/C] - [G/A+A/A] | 43/57 | 19.1 | 1.50 (0.95-2.39) | 0.084 | ||
| [A/C] - [G/A+A/A] | 42/49 | 13.3 | 2.30 (1.42-3.75) | 0.001 | ||
| Pt-paclitaxe | [A/A+C/C] - [G/G] | 62/79 | 18.2 | 0.112 | 1.00 (ref) | |
| [A/C] - [G/G] | 53/76 | 19.1 | 0.88 (0.60-1.29) | 0.511 | ||
| [A/A+C/C] - [G/A+A/A] | 58/80 | 20.0 | 0.80 (0.55-1.15) | 0.232 | ||
| [A/C] - [G/A+A/A] | 55/65 | 15.3 | 1.14 (0.78-1.67) | 0.494 | ||
| Pt-docetaxel | [A/A+C/C] - [G/G] | 20/29 | 20.4 | 0.527 | 1.00 (ref) | |
| [A/C] - [G/G] | 17/22 | 15.2 | 2.14 (0.88-5.22) | 0.093 | ||
| [A/A+C/C] - [G/A+A/A] | 10/14 | 17.0 | 0.91 (0.41-2.06) | 0.826 | ||
| [A/C] - [G/A+A/A] | 17/18 | 17.4 | 1.78 (0.86-3.71) | 0.121 |
a SLC31A1 rs2233914 (G/A) for the study subjects had been genotyped in our previous report (Ref 9).
b Numbers indicate the death event for NSCLC patients during the following-up time among all individuals in the same genotype group.
c MST: median survival time.
d Hazard ratios (HR) and their 95% confidence intervals (CIs) and P values were calculated with by multivariate Cox proportional hazards regression with adjustment for covariates.
e Test of interaction for the cohort of all patients with P value being 0.030.
f Statistical significance remained after Bonferroni multiple tests.
Discussion
As the standard treatment for advanced NSCLC, platinum-based chemotherapy achieved varying response and efficacy in patients. The in situ expression of plasma-membrane transporters that are responsible for the intake and efflux of platinum drug has been shown to directly affect tissue concentration of drug and thereby be correlated with chemotherapy outcomes [4]. In this multi-institutions-based study of NSCLC patients with platinum-based chemotherapy, we characterized divergent association of platinum efflux transporter gene ABCG2 polymorphism with response and survival, and identified interaction between ABCG2 and the platinum uptake transporter gene SLC31A1 associated with clinical outcomes, furthering the pharmacogenetics understanding of platinum-based chemotherapy.
Our result that ABCG2 nonsynonymous rs2231142 was associated with tumor response to platinum-based chemotherapy is parallel in several lines to both laboratory findings and population observations. Rs2231142 encodes a change from glutamine to lysine at the 141 amino acid in the nucleotide-binding domain of ABCG2. Cell-based studies showed that this missense codon leads to reduced ABCG2 protein level and functionality not only by disrupting protein folding and increasing ubiquitin-mediated lysosomal degradation, but also via enhancing the 3′UTR and microRNA-dependent repression [28-31]. Rs2231142 variant reportedly results in decreased substrates efflux in cell lines and affects the pharmacokinetics of, response to, and toxicity of a variety of substrate in clinical settings. For examples, in embryonic kidney cells transfected with ABCG2, the variant construct had an 80% higher intracellular concentration of the carcinogenic heterocyclic anime as compared with the wild construct [32]. In myelogenous leukemia cell lines, the variant correlated with reduced transport activity, increased cytotoxicity and efficacy after treatment with tyrosine kinase inhibitors [33]. Rs2231142 variant genotype has been reported to correlate with reduced in vivo intestinal transport capacity [34], high drug plasma in HIV-infected patients treated with dolutegravir [35], much greater plasma area under the concentration-time curve (AUC) values of sulfasalazine, a drug used as ABCG2 probe in vivo [36], better low-density lipoprotein cholesterol-lowering efficacy of rosuvastatin in patients with hypercholesterolemia [37], and more favorite response rate of metastatic colorectal cancer to first-line chemotherapy [38]. The functional rs2231142 and the intronic rs1871744 are, in genome wide association studies, correlated with serum level of urate, one of endogenous substrates of ABCG2, the susceptibility of gout, which is caused by hyperuricemia, and response to allopurinol in patients with gout [39-41]. Consistent with these reports, we observed that the rs2231142 variant genotypes, and its combined diplotype with rs1871744, were associated with favorite response of NSCLC patients to platinum-based chemotherapy. The biological plausibility may be that rs2231142 could decrease ABCG2 expression in overall tissues especially in intestine and lung, increase the bioavailability and the intracellular accumulation of substrate drugs, and thus reinforce their pharmacokinetics properties and pharmacodynamics efficacy.
The functional variant of rs2231142, which is expected to have reduced ABCG2 expression level and capacity, not only correlated with favorite tumor response, as discussed above, but also was reported to be associated with longer survival in the settings of chemotherapy for a few types of cancer [5, 42, 43]. However, in contrast to their association with favorite response, we unexpectedly observed that the variant genotypes of rs2231142 and linked rs4148157 of ABCG2 were associated with shorter survival and disease progression. There are possible explanations for this discrepancy. First, although improved survival is the “gold standard” for evaluating the efficacy of oncologic therapy, and tumor response is a logical surrogate endpoint, since advancing tumor burden is the predominant mechanism by which the disease causes morbidity and mortality, the extent to which tumor response correlates with survival varies [44]. Response to chemotherapy could be affected by both a patient's genetic background in genomic DNA and somatic mutation in tumors. During the course of chemotherapy, the selection of increasingly mutated tumor cells will progressively change the tumors genetic makeup from its germ line origin and reduce the impact of original host-specific relevant polymorphism, which could explain why there are some effects on the early tumor response that are not observed again later on survival [10]. In accordance with this assumption, we in this study (Supplemental Table 11 and 12) and Muller et al. consistently observed that in NSCLC patients receiving first-line chemotherapy, favorable response after the second cycle of the course was significantly correlated with PFS but not with OS [10]. Second, the wide ranges of substrates of ABCG2 include not only chemotherapeutic agents such as platinum but also endogenous cell-growth-promoting metabolite molecules such as folates [45, 46]. The de-functional ABCG2 rs2231142, on one hand, could maintain accumulation of chemotherapeutic compounds, which would achieve reduced tumor burden. On the other hand, however, it could retain more folic acid, which would lead to high rates of proliferation and worse survival. Indeed, it was recently reported that, for prostate cancer patients with docetaxel treatment, ABCG2 rs2231142 correlates with improved drug response, but also correlates with poorer outcomes possibly through increasing intratumoral folate levels and thereby enhancing tumor cell proliferation [47]. Third, because in this study the association of rs4148157 (P = 0.008) with shorter survival was much more significant than the marginal signal of the functional rs2231142 (P = 0.043), and rs4148157 reportedly has implications for the pharmacokinetics of xenobiotic and endogenous substrates [48, 49], it is plausible that potential cis-regulatory function of rs4148157, or its associated causative SNP, may overwhelm the well-established role of rs2231142, and the observed signal for rs2231142 may be due to genetic hitchhiking. Consistent with these scenarios for herein observed divergent associations of ABCG2 polymorphism with response and survival, similar counter-intuitive results were also reported. In platinum-treated lung cancer patients, carriers with ABCG2 variant genotype, which are expected to have reduced ABCG2 level, show a shorter OS [10]. In patients with head and neck squamous cell carcinoma, high expressions of ABCB1 and ABCC1 were associated with favorable survival [50]. In patients with childhood neuroblastoma, low (rather than high) ABCC3 expression were predictive of poor survival [51].
We have previously reported the association of platinum uptake transporter gene SLC31A1 with clinical outcomes of platinum-based chemotherapy in NSCLC patients, rs10759637 at 3′UTR correlated with shorter OS through reducing SLC31A1 expression, and rs2233914 at 5' flanking region correlated with poor response [9]. With an effort to systemically investigate the pharmacogenetic relevance of variations in platinum uptake and efflux transport pathways, we here genotyped the functional and tagging SNPs of ABCG2 and other five transporter genes (SLC31A2, ATP7A, ATP7B, ABCB1 and ABCC2), but did not observe any significant association signal other than ABCG2 (data not shown). Consistent with these negative results, it was reported that the expression of ABCB1 in NSCLC cell lines does not correlate with sensitivity to cisplatin or intracellular platinum accumulation, its expression in NSCLC tissues does not correlate with response to cisplatin [4]. Abcc2 knockout in mice does not affect cisplatin disposition and toxicity, ABCC2 polymorphisms do not correlate with ABCC2 expression and cisplatin-induced cytotoxicity in NCI60 panel and are not associated with cisplatin pharmacokinetics and efficacy in cancer patients [52]. In clinical NSCLC specimens, only SLC31A1, but not ATP7A or ATP7B, predicts clinical outcome after platinum-based chemotherapy [53]. Of interest, we found that SLC31A1 rs2233914 and ABCG2 rs1871744 were jointly associated with response, SLC31A1 rs10759637 and ABCG2 rs4148157 were jointly associated with survival. Particularly, the genetic interactions were concomitantly pronounced in subgroups of patients demographically stratified as males, older than 58, ECOG PS 0-1, and smokers. It was reported that reduced ABCG2 and increased SLC22A1 mRNA expression are associated with imatinib response in chronic myeloid leukemia [54], and simultaneous high expressions of SLC31A1 and ABCG2 are associated with poor survival of HNSCC patients [50]. Cui et al. also recently reported the combined effect of ABCG2 rs2231142 and carboxylesterase 5A gene SNP is associated with platinum-based chemotherapy-induced toxicity in NSCLC patients [55]. In a genome-wide association study, additive genetic risk score of nonsynonymous variants of ABCG2 (rs2231142) and SLC2A9 (rs16890979) showed graded associations with uric acid and gout [56]. It was also recently reported that the association of ABCG2 rs2231142 with hyperuricemia is modified by SLC2A9 polymorphism in an elderly Chinese population [57]. Although the causative mechanism for the synergistic effects of polymorphisms in platinum intake and export pathways, such as SLC31A1 and ABCG2, is still largely unknown, the association of their interaction with clinical outcomes suggests the combined relevant genotypes of SLC31A1 and ABCG2 as potential pertinent and actionable pharmacogenetic biomarkers for platinum-based chemotherapy of NSCLC, especially for some demographically stratified subgroups patients.
Notably, the functional polymorphisms of SLC31A1 and ABCG2 genes have highly variable frequencies depending on ethnicity. In public SNP database, the low-expression-related ancestral allele (C) of SLC31A1 rs10759637 is common in African populations, while the high-expression-related derived allele (A) is dominant in Caucasian and Chinese Han population. The low-function-related derived allele (A) of ABCG2 rs2231142 is prevalent in Eastern Asian populations (25-35%), common in Caucasian (8-15%) but rare in Sub-Saharan (0.9%) and African American (0-5%). The pronounced overrepresentation of the high-expression allele of uptake transporter SLC31A1 and the low-function allele of efflux transporter ABCG2 in non-Africans, as compared with in Africans, strongly suggest divergent pharmacokinetics of platinating agents between them. Indeed, in vitro study showed that B-lymphoblastoid cell lines derived from Caucasians of the HapMap project are more susceptible to cytotoxicity induced by carboplatin than those cells from Africans [58]. In a cohort of NSCLC patients with neoadjuvant platinum-based chemotherapy, Kim et al. reported that the African American had significantly reduced SLC31A1 expression in tumor, lowered tissue platinum concentration and decreased tumor shrinkage as compared to Caucasians [6]. Thus, our findings that the genetic interaction between SLC31A1 and ABCG2 polymorphisms was associated with clinical outcomes of NSCLC patients receiving platinum-based chemotherapy may have important implications for the pharmacoethnicity of platinating agents in cancer chemotherapy.
We should acknowledge that this work may have some limitations. All of the 1004 patients are Chinese Han and were recruited from five hospitals in East China. In order to address the replicability of genetic association, we grouped this cohort into the discovery (A) panel (n = 237) and the replication (B) panel (n = 767) according to their institutional and regional sources, and genotyped all of the selected SNPs of ABCG2 in the total cohort. There was no significant difference in the characterized demographic and clinical features for the two panels. Furthermore, the observed genotypic frequencies of ABCG2 polymorphisms in either panel A, panel B and the total cohort all fit well with the Hardy-Weinberg law, and are also much comparable to those in the general healthy Chinese Han population in the 1000 genome dataset. Our previous genomic dissection of population substructure of Chinese Han also indicates that there is no apparent population differentiation in East Chinese Han, the general natural population in this study, from the one-dimensional “north-south” structure [59]. It is thus at least possible that the association of ABCG2 polymorphisms with clinical outcomes of NSCLC patients receiving platinum-based chemotherapy could be due to sampling and ascertainment biases and population stratification. We here emphasized rs1871744 and rs4148157 because only the two SNPs were consistently associated with response and survival respectively in both panel A, panel B and the total cohort. However, we still found differential and even divergent association signals among the three populations for other ABCG2 SNPs including the nonsynonymous rs2231142, which was associated with response in panel B and with survival in panel A respectively. Therefore, further validations of the findings of this work in cohorts of different ethnic populations with larger sample size are highly warranted.
In summary, this pharmacogenetic study demonstrates that platinum efflux transporter gene ABCG2 polymorphism was divergently associated with objective response and overall survival of NSCLC patients receiving platinum-based chemotherapy, and its interaction with the platinum import transporter gene SLC31A1 polymorphism was also associated with clinical outcomes. These findings may provide potential predictive markers for clinical outcomes of platinum-based chemotherapy of NSCLC and have implications for pharmacogenetics of platinating agents based cancer chemotherapy.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank all the enrolled patients who made this study possible, and acknowledge greatly the collaboration from the participating hospitals and staff. This work was supported by the National Key Research Development Programs of China (2018YFC0910700, 2016YFC0901903, 2016YFC0905000), the International S&T Cooperation Program of China (2015DFE32790), National Natural Science Foundation of China (81172093, 81372526, 81572404, 81372236) and Shanghai Pujiang Program (11PJD005).
Author Contributions
H.W., D.L., J.C.W., B.H., C.B. and Q.L. designed this study. H.W., D.L., J.C.W. and J.Q. generated the genotyping data. J.J.W., Z.G. and J.X. collected the samples and clinical information. L.W, C.S. and X.L. performed data analysis and produced figures and tables. H.W. wrote the paper. C.M., J.Y. and Z.L. actively participated in writing the paper. L.J. supervised the study. All authors reviewed and approved the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Spiro SG, Silvestri GA. The treatment of advanced non-small cell lung cancer. Curr Opin Pulm Med. 2005;11:287-91
3. Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9-23
4. Kim ES, Lee JJ, He G, Chow CW, Fujimoto J, Kalhor N. et al. Tissue platinum concentration and tumor response in non-small-cell lung cancer. J Clin Oncol. 2012;30:3345-52
5. Heyes N, Kapoor P, Kerr ID. Polymorphisms of the Multidrug Pump ABCG2: A Systematic Review of Their Effect on Protein Expression, Function, and Drug Pharmacokinetics. Drug Metab Dispos. 2018;46:1886-99
6. Kim ES, Tang X, Peterson DR, Kilari D, Chow CW, Fujimoto J. et al. Copper transporter CTR1 expression and tissue platinum concentration in non-small cell lung cancer. Lung Cancer. 2014;85:88-93
7. Yoh K, Ishii G, Yokose T, Minegishi Y, Tsuta K, Goto K. et al. Breast cancer resistance protein impacts clinical outcome in platinum-based chemotherapy for advanced non-small cell lung cancer. Clin Cancer Res. 2004;10:1691-7
8. Ota S, Ishii G, Goto K, Kubota K, Kim YH, Kojika M. et al. Immunohistochemical expression of BCRP and ERCC1 in biopsy specimen predicts survival in advanced non-small-cell lung cancer treated with cisplatin-based chemotherapy. Lung Cancer. 2009;64:98-104
9. Sun C, Zhang Z, Qie J, Wang Y, Qian J, Wang J. et al. Genetic polymorphism of SLC31A1 is associated with clinical outcomes of platinum-based chemotherapy in non-small-cell lung cancer patients through modulating microRNA-mediated regulation. Oncotarget. 2018;9:23860-77
10. Muller PJ, Dally H, Klappenecker CN, Edler L, Jager B, Gerst M. et al. Polymorphisms in ABCG2, ABCC3 and CNT1 genes and their possible impact on chemotherapy outcome of lung cancer patients. Int J Cancer. 2009;124:1669-74
11. Kim SH, Kim MJ, Cho YJ, Jeong YY, Kim HC, Lee JD. et al. Clinical Significance of ABCG2 Haplotype-tagging Single Nucleotide Polymorphisms in Patients With Unresectable Non-Small Cell Lung Cancer Treated With First-line Platinum-based Chemotherapy. Am J Clin Oncol. 2015;38:294-9
12. Han JY, Lim HS, Yoo YK, Shin ES, Park YH, Lee SY. et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110:138-47
13. Chen L, Manautou JE, Rasmussen TP, Zhong XB. Development of precision medicine approaches based on inter-individual variability of BCRP/ABCG2. Acta Pharm Sin B. 2019;9:659-74
14. Song X, Wang S, Hong X, Li X, Zhao X, Huai C. et al. Single nucleotide polymorphisms of nucleotide excision repair pathway are significantly associated with outcomes of platinum-based chemotherapy in lung cancer. Sci Rep. 2017;7:11785
15. Li X, Shao M, Wang S, Zhao X, Chen H, Qian J. et al. Heterozygote advantage of methylenetetrahydrofolate reductase polymorphisms on clinical outcomes in advanced non-small cell lung cancer (NSCLC) patients treated with platinum-based chemotherapy. Tumour Biol. 2014;35:11159-70
16. Zhao X, Wang S, Wu J, Li X, Wang X, Gao Z. et al. Association of TERT Polymorphisms with Clinical Outcome of Non-Small Cell Lung Cancer Patients. PLoS One. 2015;10:e0129232
17. Wang S, Song X, Li X, Zhao X, Chen H, Wang J. et al. RICTOR polymorphisms affect efficiency of platinum-based chemotherapy in Chinese non-small-cell lung cancer patients. Pharmacogenomics. 2016;17:1637-47
18. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L. et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205-16
19. Carbone DP, Minna JD. Chemotherapy for non-small cell lung cancer. BMJ. 1995;311:889-90
20. Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J hum Genet. 2004;74:106-20
21. Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162-9
22. Brennan P. Gene-environment interaction and aetiology of cancer: what does it mean and how can we measure it? Carcinogenesis. 2002;23:381-7
23. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765-9
24. Tripathi A, Pandey A. Post-Hoc Comparison in Survival Analysis: An Easy Approach. J Biosci Med. 2017;5:112-9
25. Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J. et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92-8
26. Sarkadi B, Homolya L, Hegedus T. The ABCG2/BCRP transporter and its variants - from structure to pathology. FEBS Lett. 2020 doi: 10.1002/1873-3468.13947. Online ahead of print
27. Wang H, Hao B, Zhou K, Chen X, Wu S, Zhou G. et al. Linkage disequilibrium and haplotype architecture for two ABC transporter genes (ABCC1 and ABCG2) in Chinese population: implications for pharmacogenomic association studies. Ann Hum Genet. 2004;68:563-73
28. Tamura A, Wakabayashi K, Onishi Y, Takeda M, Ikegami Y, Sawada S. et al. Re-evaluation and functional classification of non-synonymous single nucleotide polymorphisms of the human ATP-binding cassette transporter ABCG2. Cancer Sci. 2007;98:231-9
29. Furukawa T, Wakabayashi K, Tamura A, Nakagawa H, Morishima Y, Osawa Y. et al. Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm Res. 2009;26:469-79
30. Ripperger A, Benndorf RA. The C421A (Q141K) polymorphism enhances the 3'-untranslated region (3'-UTR)-dependent regulation of ATP-binding cassette transporter ABCG2. Biochem Pharmacol. 2016;104:139-47
31. Mizuarai S, Aozasa N, Kotani H. Single nucleotide polymorphisms result in impaired membrane localization and reduced atpase activity in multidrug transporter ABCG2. Int J Cancer. 2004;109:238-46
32. Gardner ER, Ahlers CM, Shukla S, Sissung TM, Ockers SB, Price DK. et al. Association of the ABCG2 C421A polymorphism with prostate cancer risk and survival. BJU Int. 2008;102:1694-9
33. Skoglund K, Boiso Moreno S, Jonsson JI, Vikingsson S, Carlsson B, Green H. Single-nucleotide polymorphisms of ABCG2 increase the efficacy of tyrosine kinase inhibitors in the K562 chronic myeloid leukemia cell line. Pharmacogenet Genomics. 2014;24:52-61
34. Tanaka Y, Kitamura Y, Maeda K, Sugiyama Y. Quantitative Analysis of the ABCG2 c.421C>A Polymorphism Effect on In vivo Transport Activity of Breast Cancer Resistance Protein (BCRP) Using an Intestinal Absorption Model. J Pharm Sci. 2015;104:3039-48
35. Tsuchiya K, Hayashida T, Hamada A, Oki S, Oka S, Gatanaga H. High plasma concentrations of dolutegravir in patients with ABCG2 genetic variants. Pharmacogenet Genomics. 2017;27:416-9
36. Yamasaki Y, Ieiri I, Kusuhara H, Sasaki T, Kimura M, Tabuchi H. et al. Pharmacogenetic characterization of sulfasalazine disposition based on NAT2 and ABCG2 (BCRP) gene polymorphisms in humans. Clin Pharmacol Ther. 2008;84:95-103
37. Tomlinson B, Hu M, Lee VW, Lui SS, Chu TT, Poon EW. et al. ABCG2 polymorphism is associated with the low-density lipoprotein cholesterol response to rosuvastatin. Clin Pharmacol Ther. 2010;87:558-62
38. Zhao J, Li W, Zhu D, Yu Q, Zhang Z, Sun M. et al. Association of single nucleotide polymorphisms in MTHFR and ABCG2 with the different efficacy of first-line chemotherapy in metastatic colorectal cancer. Med Oncol. 2014;31:802
39. Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A. 2009;106:10338-42
40. Nakayama A, Nakaoka H, Yamamoto K, Sakiyama M, Shaukat A, Toyoda Y. et al. GWAS of clinically defined gout and subtypes identifies multiple susceptibility loci that include urate transporter genes. Ann Rheum Dis. 2017;76:869-77
41. Wen CC, Yee SW, Liang X, Hoffmann TJ, Kvale MN, Banda Y. et al. Genome-wide association study identifies ABCG2 (BCRP) as an allopurinol transporter and a determinant of drug response. Clin Pharmacol Ther. 2015;97:518-25
42. Limviphuvadh V, Tan CS, Konishi F, Jenjaroenpun P, Xiang JS, Kremenska Y. et al. Discovering novel SNPs that are correlated with patient outcome in a Singaporean cancer patient cohort treated with gemcitabine-based chemotherapy. BMC Cancer. 2018;18:555
43. Tian C, Ambrosone CB, Darcy KM, Krivak TC, Armstrong DK, Bookman MA. et al. Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a Gynecologic Oncology Group study. Gynecol Oncol. 2012;124:575-81
44. Gollob JA, Bonomi P. Historic evidence and future directions in clinical trial therapy of solid tumors. Oncology (Williston Park). 2006;20:10-8
45. Breedveld P, Pluim D, Cipriani G, Dahlhaus F, van Eijndhoven MA, de Wolf CJ. et al. The effect of low pH on breast cancer resistance protein (ABCG2)-mediated transport of methotrexate, 7-hydroxymethotrexate, methotrexate diglutamate, folic acid, mitoxantrone, topotecan, and resveratrol in in vitro drug transport models. Mol Pharmacol. 2007;71:240-9
46. Bistulfi G, Diegelman P, Foster BA, Kramer DL, Porter CW, Smiraglia DJ. Polyamine biosynthesis impacts cellular folate requirements necessary to maintain S-adenosylmethionine and nucleotide pools. FASEB J. 2009;23:2888-97
47. Sobek KM, Cummings JL, Bacich DJ, O'Keefe DS. Contrasting roles of the ABCG2 Q141K variant in prostate cancer. Exp Cell Res. 2017;354:40-7
48. Roberts JK, Birg AV, Lin T, Daryani VM, Panetta JC, Broniscer A. et al. Population Pharmacokinetics of Oral Topotecan in Infants and Very Young Children with Brain Tumors Demonstrates a Role of ABCG2 rs4148157 on the Absorption Rate Constant. Drug Metab Dispos. 2016;44:1116-22
49. Bhatnagar V, Richard EL, Wu W, Nievergelt CM, Lipkowitz MS, Jeff J. et al. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin Kidney J. 2016;9:444-53
50. Warta R, Theile D, Mogler C, Herpel E, Grabe N, Lahrmann B. et al. Association of drug transporter expression with mortality and progression-free survival in stage IV head and neck squamous cell carcinoma. PLoS One. 2014;9:e108908
51. Henderson MJ, Haber M, Porro A, Munoz MA, Iraci N, Xue C. et al. ABCC multidrug transporters in childhood neuroblastoma: clinical and biological effects independent of cytotoxic drug efflux. J Natl Cancer Inst. 2011;103:1236-51
52. Sprowl JA, Gregorc V, Lazzari C, Mathijssen RH, Loos WJ, Sparreboom A. Associations between ABCC2 polymorphisms and cisplatin disposition and efficacy. Clin Pharmacol Ther. 2012;91:1022-6
53. Chen HH, Yan JJ, Chen WC, Kuo MT, Lai YH, Lai WW. et al. Predictive and prognostic value of human copper transporter 1 (hCtr1) in patients with stage III non-small-cell lung cancer receiving first-line platinum-based doublet chemotherapy. Lung Cancer. 2012;75:228-34
54. de Lima LT, Vivona D, Bueno CT, Hirata RD, Hirata MH, Luchessi AD. et al. Reduced ABCG2 and increased SLC22A1 mRNA expression are associated with imatinib response in chronic myeloid leukemia. Med Oncol. 2014;31:851
55. Cui JJ, Wang LY, Zhu T, Gong WJ, Zhou HH, Liu ZQ. et al. Gene-gene and gene-environment interactions influence platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Sci Rep. 2017;7:5082
56. Dehghan A, Kottgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F. et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953-61
57. Liu J, Yang W, Li Y, Wei Z, Dan X. ABCG2 rs2231142 variant in hyperuricemia is modified by SLC2A9 and SLC22A12 polymorphisms and cardiovascular risk factors in an elderly community-dwelling population. BMC Med Genet. 2020;21:54
58. O'Donnell PH, Dolan ME. Cancer pharmacoethnicity: ethnic differences in susceptibility to the effects of chemotherapy. Clin Cancer Res. 2009;15:4806-14
59. Xu S, Yin X, Li S, Jin W, Lou H, Yang L. et al. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 2009;85:762-74
Author contact
![]() Corresponding author: Haijian Wang, Shanghai Pudong Hospital and School of Life Sciences, Fudan University, Shanghai, China, E-mail: haijianwangedu.cn.
Corresponding author: Haijian Wang, Shanghai Pudong Hospital and School of Life Sciences, Fudan University, Shanghai, China, E-mail: haijianwangedu.cn.

 Global reach, higher impact
Global reach, higher impact