3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(5):1307-1317. doi:10.7150/jca.49702 This issue Cite
Research Paper
Identification of novel biomarkers and candidate small-molecule drugs in cutaneous melanoma by comprehensive gene microarrays analysis
1. Department of Clinical Laboratory, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, 450052, China.
2. Department of Biotechnology and Food Engineering, Anyang Institute of Technology, Anyang, Henan 455000, PR China
3. Department of Human Anatomy and Histoembryology, Henan Vocational College of Nursing, Anyang, Henan, 400500, China.
4. Department of Orthopaedics, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China.
Abstract
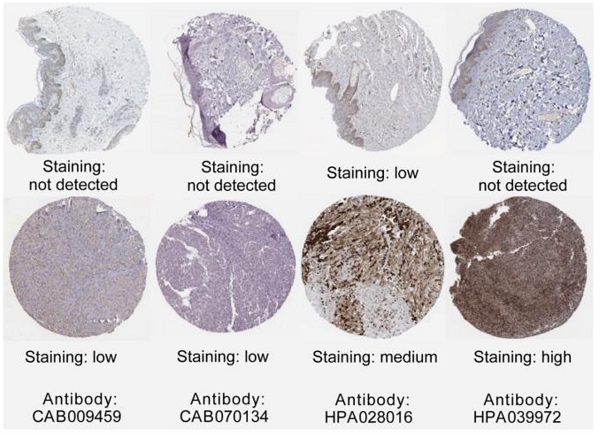
Background: Melanoma is a pernicious skin cancer with high aggressiveness. This study aimed to identify potential novel biomarkers associated with the prognosis and pathogenesis of cutaneous melanoma and to explore new targeted drugs for melanoma. Methods: Two Gene Expression Omnibus (GEO) microarray datasets, GSE3189 and GSE7553 were combined to analyze the differentially expressed genes (DEGs). To better understand the DEGs in the melanoma pathogenesis, we performed gene enrichment analyses and established a protein-protein interaction network (PPI). The survival analyses for key genes were conducted based on the GEPIA platform. Finally, we mined the CMap database to explore potential small-molecule drugs to target the obtained DEGs. Results: In short, we identified 500 DEGs between cutaneous melanoma samples and normal samples. The PPI network was established with 349 nodes and 1251 edges. Signaling pathway analysis showed that these genes play a vital role in ECM-receptor interactions, the PPAR signaling pathway and pathways in cancer. Eight DEGs with a relatively high degree of connectivity (CDC45, CENPF, DTL, FANCI, GINS2, HJURP, TPX2 and TRIP13) were selected as hub-genes that remarkably correlated to a poor survival rate. Based on 500 DEGs, 20 small-molecule drugs that potentially target genes with abnormal expression in cutaneous melanoma were obtained from the CMap database. Among these compounds, we found that menadione has the greatest therapeutic value for melanoma. Conclusions: In conclusion, we identified the 8 candidate biomarkers and potential key signaling pathways in cutaneous melanoma through comprehensive microarray analyses. The identified candidate drugs have provided several directive significances for the synthesis medicine for melanoma.
Keywords: cutaneous melanoma, novel biomarkers, candidate small molecules, prognostic factor

 Global reach, higher impact
Global reach, higher impact