Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(5):1483-1492. doi:10.7150/jca.52395 This issue Cite
Research Paper
miR-361-3p Regulates Liver Tumor-initiating Cells Expansion and Chemo-resistance
1. Department of Hepatic Surgery, Third Affiliated Hospital of Second Military Medical University, Shanghai, 200438, China.
2. Department of General surgery, First Affiliated Hospital of Second Military Medical University, Shanghai, 200433, China.
#These authors contributed equally to this work.
Received 2020-8-25; Accepted 2020-12-15; Published 2021-1-1
Abstract
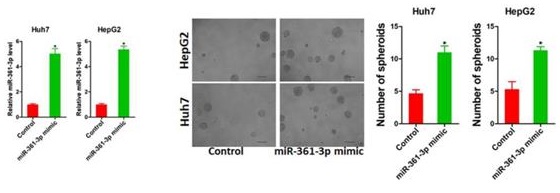
Increasing evidence shows that liver tumor-initiating cells (T-ICs) closely associated with the progression, metastasis, recurrence and chemo-resistance of hepatocellular carcinoma (HCC). However, the underlying mechanism for the propagation of liver T-ICs remains unclear. Here we show that miR-361-3p is upregulated in liver T-ICs. Knockdown of miR-361-3p impairs the self-renewal and tumorigenicity liver T-ICs. Conversely, forced miR-361-3p expression enhances the self-renewal and tumorigenicity liver T-ICs. Mechanistically, miR-361-3p directly targets SOX1 via binding its 3'-UTR in liver T-ICs. Moreover, miR-361-3p knockdown hepatoma cells are more sensitive to cisplatin or sorafenib treatment. Clinical cohort analysis demonstrates that miR-361-3p low HCC patients are benefited from TACE (transcatheter arterial chemoembolization) or sorafenib treatment. In conclusion, our findings revealed the crucial role of the miR-361-3p in liver T-IC expansion and TACE or sorafenib response, rendering miR-361-3p an optimal target for the prevention and intervention in HCC.
Keywords: Tumor-initiating cells, hepatocellular carcinoma, miR-361-3p, SOX1, cisplatin, sorafenib
Background
With nearly 700,000 new cases per year, hepatocellular carcinoma (HCC) ranks as the sixth most common cancer in the world [1]. However, the early symptoms of HCC are not obvious. Most HCC patients are diagnosed at advanced stage [2]. Due to high grade invasive HCC can develop to life threatening metastases and less than 20% patients with Portal vein thrombus-only have 2-year overall survival [3]. Currently, most of drugs which used for clinical HCC patients were proved to be disappointed. So, the underlying mechanism of HCC initiation and progression needs to be deeply explored.
Tumor-initiating cells (T-ICs) or cancer stem cells (CSCs) are a subgroup of cancer cells, which have the self-renewal ability and tumorigenesis capacity [4, 5]. T-ICs or CSCs were first discovered in blood system diseases [6]. Later studies also confirmed the existence of T-ICs or CSCs in solid tumors, including breast cancer, liver cancer and glioma [7-9]. Currently researches suggest that T-ICs or CSCs are responsible for the progression, metastasis, recurrence and chemo-resistance of cancers [10-12]. Therefore, identification of the underlying mechanisms governing T-ICs or CSCs propagation may lead to the discovery of promising therapeutic strategies for cancer patients.
MicroRNAs (miRNAs) are a class of small noncoding RNA molecules that contain approximately 22 nucleotides [13]. It was reported to be essential for many physiological processes, such as cellular homeostasis, development, differentiation, cell survival and death, tumor initiation and progression [14, 15]. miR-361-3p is a newly discovered miRNA, and its function and mechanism of action in biological processes and diseases are not completely understood. Previous studies found that miR-361-3p promotes human breast cancer cell viability by inhibiting the E2F1/P73 signaling pathway [16]. Moreover, miR-361-3p regulates ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal adenocarcinoma [17]. However, the function of miR-361-3p in liver T-ICs is unclear.
In this study, we for first find that miR-361-3p is highly expressed in liver T-ICs. Next, by using loss-of-function analysis and gain-of-function analysis in liver T-ICs, we demonstrate that miR-361-3p promotes the self-renewal capacity and tumorigenicity of liver T-ICs. Further mechanism study reveals that miR-361-3p directly targets SOX1 in liver T-ICs. Interestingly, miR-361-3p knockdown hepatoma cells are more sensitive to cisplatin or sorafenib treatment. Clinical cohort analysis demonstrates that miR-361-3p low HCC patients are benefited from TACE or sorafenib treatment. Altogether, we discover that miR-361-3p promotes the expansion of liver T-ICs via interacting with SOX1.
Materials and Methods
Cell Culture
Huh7 and HepG2 cells were maintained at 37°C in 5% CO2 incubator with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 25 µg/ml of gentamicin. FBS was purchased from Nova-Tech (Grand Island, NE). miR-361-3p mimic virus or miR-361-3p sponge virus was purchased from RIBOBIO (Guangzhou, China). siRNA SOX1 was purchased from Genechem (Shanghai, China).
Patients and specimens
HCC tissues were obtained from patients who underwent liver resection in the Eastern Hepatobiliary Surgery Hospital (EHBH). Samples were frozen in liquid nitrogen immediately after surgical resection for further RNA extraction. A total of 70 patients received TACE therapy after surgery for primary HCC at EHBH from 2010-2015 were included in Cohort 1. Detailed clinicopathological features of these patients are described in supplementary Table 1. A total of 68 patients received adjuvant sorafenib therapy after surgery for primary HCC at EHBH from 2011-2015 were included in Cohort 2. Detailed clinicopathological features of these patients are described in supplementary Table 2. Another group of 50 HCC specimens were used for analyzing the correlation between SOX1 and miR-361-3p expression. Patient informed consent was obtained and the procedure of human sample collection was approved by the Ethics Committee of EHBH.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from the cells using Trizol reagent (Invitrogen, 15596-018). Total cDNAs were synthesized by ThermoScript TM RT-PCR system (Invitrogen, 11146-057). The mRNA amount presented in the cells was measured by semi quantitative RT-PCR. PCR conditions included 1 cycle at 94 °C for 5 minutes, followed by up to 40 cycles of 94 °C for 15 seconds (denaturation), 60 °C for 30 seconds (annealing) and 72 °C for 30 seconds (extension). The sequences of primers used was listed in Supplementary Table 3.
Luciferase reporter assay
A 300-bp fragment of the SOX1 3'UTR containing the conserved miR-361-3p-binding sites was inserted into a luciferase reporter plasmid. The SOX1 3'UTR mutant luciferase plasmid contained changes in potential miR-361-3p-binding base sequence “CUGGGGG” to “GACUCUA”. HCC cells were transfected with SOX1 3'UTR luciferase reporter in combination with the pRL-TK vector (Promega, E2241) as an internal control. The dual luciferase assay kit was purchased from Promega (0000060417). The luciferase activities were determined using a luminometer (Wallac 1420 Victor 2 multilabel counter system) as described in previous studies [18].
Western Blotting Assay
The cells collected by cell lysis buffer, then disposed like we described before [19]. Equal aliquots of cell extracts were separated on SDS-polyacrylamide gels. The proteins were then transferred to PVDF membranes (Bio-Rad), blocked, and probed with the antibodies. Primary antibody-bound proteins were detected by using an alkaline phosphatase-linked secondary antibody and an ECF Western Blotting system (Amersham). The densitometric analyses of the protein bands vs. the individual loading controls were performed using the ImageQuant 5.2 software (GE Healthcare). The primary antibodies used were listed in supplementary Table 4. The results shown were representative one of three independent experiments.
Spheroid formation assay
HCC cells were digested and then cultured in a 96-well ultra-low attachment (300 cells per well) and cultured in DMEM/F12 (Gibco) media, supplemented with 1% FBS, 20 ng/mL bFGF and 20 ng/mL EGF for 7 days. The total number of spheres was counted under the microscope (Olympus).
In vitro limiting dilution assay
Various numbers of Huh7/HepG2 miR-361-3p mimic or miR-361-3p sponge and their control cells were digested and seeded into 96-well ultra-low attachment (2, 4, 8, 16, 32, 64 cells per well, n=8) and cultured in DMEM/F12 (Gibco) supplemented with 1% FBS, 20 ng/mL bFGF and 20 ng/mL EGF for seven days. The CSC proportions were analyzed using Poisson distribution statistics and the L-Calc Version 1.1 software program (Stem Cell Technologies, Inc., Vancouver, Canada) as previously described [20].
In vivo limiting dilution
For the in vivo limiting dilution assay, Huh7 miR-361-3p mimic or Huh7 miR-361-3p sponge and their control HCC cells digested and then mixed with Matrigel (BD) at a ratio of 1:1 and injected subcutaneously at indicated cell doses (1X103, 5X103, 1X104, 5X104) per NOD-SICD mouse (n=6). After two months, tumors formation was evaluated.
Apoptosis Assay
Huh7 miR-361-3p sponge and control cells were treated with cisplatin (10 μg/ml) or sorafenib (10 μM) for 48 hours, followed by staining with Annexin V and 7-AAD for 15 minutes at room temperature in the dark. Apoptotic cells were determined by an Annexin VFITC Apoptosis Detection Kit I (BD Pharmingen, San Diego, CA) and detected by flow cytometry according to the manufacturer's instructions.
Statistical Analysis
GraphPad Prism (GraphPad Software, Inc. La Jolla, USA) was used for all statistical analyses. Statistical analysis was carried out using t test or Bonferroni Multiple Comparisons Test: *p<0.05. A p value of less than 0.05 was considered significant.
Results
miR-361-3p is upregulated in liver T-ICs
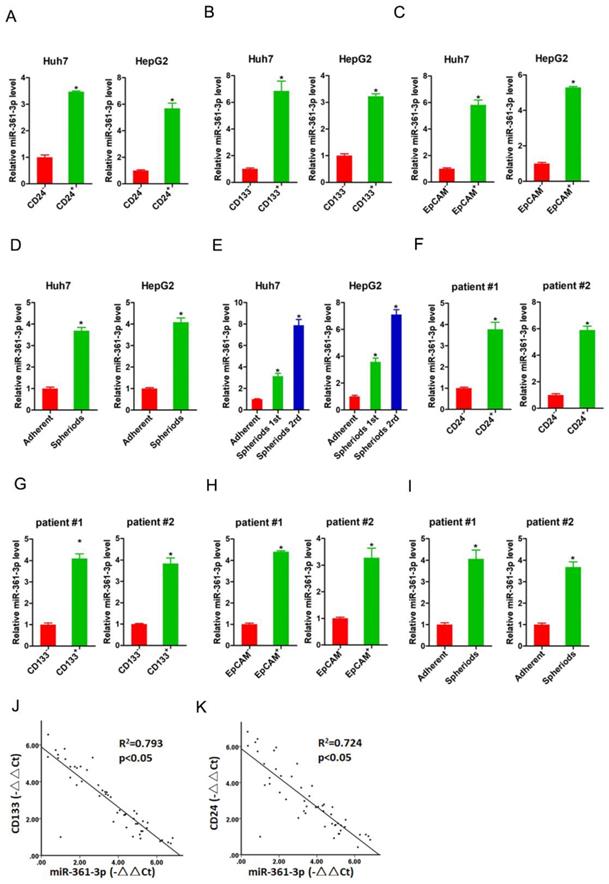
It was well accepted that CD24, CD133 and EpCAM were liver T-ICs markers [21-23]. Therefore, we isolated CD24, CD133 and EpCAM positive HCC cells by flow cytometry. As expected, the expression of miR-361-3p was dramatically increased in CD24, CD133 and EpCAM positive HCC cells compared with their negative HCC cells (Figure 1A-C). Spheroid culture of cancer cells is a routine approach to enrich T-ICs. We observed that expression of miR-361-3p was significantly upregulated in the self-renewing spheroids compared with the attached cells (Figure 1D). In serial passages of Huh7 or HepG2 spheroids, miR-361-3p expression was gradually increased (Figure 1E). Consistently, miR-361-3p expression was also upregulated in CD24, CD133 and EpCAM positive primary HCC cells compared with negative primary HCC cells (Figure 1F-H). The level of miR-361-3p was increased in primary HCC spheroids compared with the attached cells (Figure 1I). More importantly, in HCC tissues, pearson correlation analysis demonstrated that miR-361-3p levels were negatively correlated with the expression of CD24 and CD133 (Figure 1J&K). Taken together, our data demonstrated that miR-361-3p was upregulated in liver T-ICs.
miR-361-3p promotes liver T-ICs self-renew and tumorigenesis
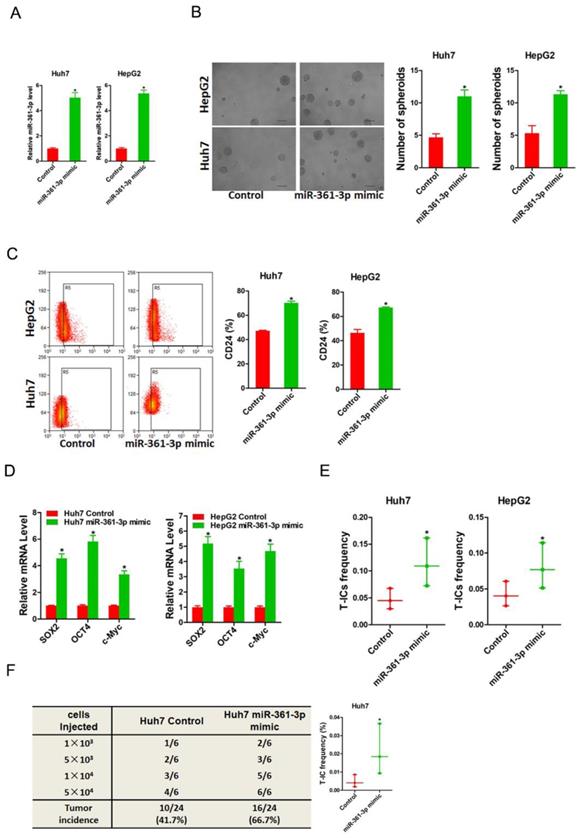
To evaluate the potential role of miR-361-3p in liver T-ICs, Huh7 and HepG2 cells were infected with miR-361-3p overexpression virus. The overexpression effect of miR-361-3p was determined by real-time PCR (Figure 2A). Spheroids formation is a routine approach to assess self-renewal ability. We found that Huh7/HepG2 miR-361-3p mimic cells formed much more spheres than control cells (Figure 2B). Moreover, the expression of liver T-ICs markers was upregulated in miR-361-3p mimic HCC cells compared with control cells (Figure 2C). Consistently, the expression of stemness associated transcription factors was increased in miR-361-3p mimic HCC cells compared with control cells (Figure 2D). Furthermore, the in vitro and in vivo limiting dilution assay indicated that the proportion of liver T-ICs and tumorigenic ability were enhanced in miR-361-3p mimic HCC cells (Figure 2E&F).
miR-361-3p knockdown inhibits liver T-ICs self-renew and tumorigenesis
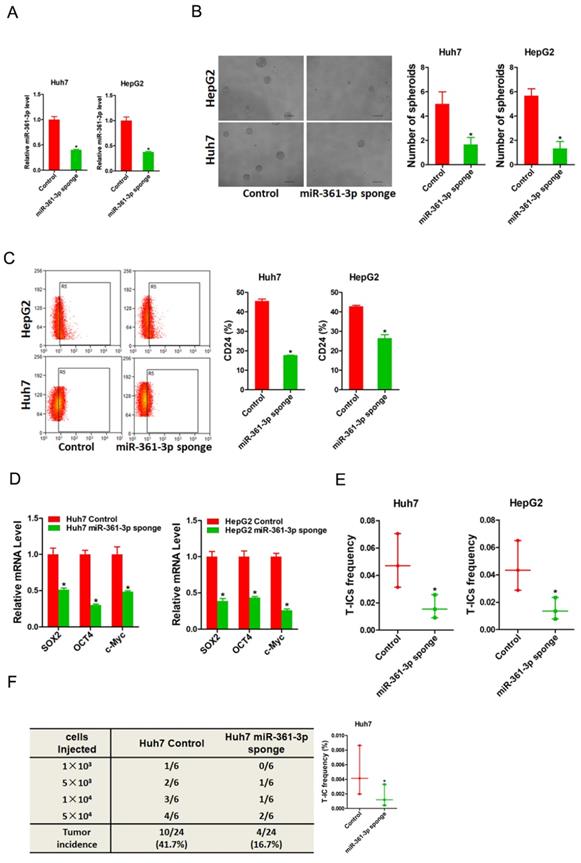
To further explore the role of miR-361-3p in liver T-ICs, Huh7 and HepG2 cells were infected with miR-361-3p interference virus. The knockdown effect of miR-361-3p was determined by real-time PCR (Figure 3A). Spheroids formation is a routine approach to assess self-renewal ability. We found that Huh7/HepG2 miR-361-3p knockdown cells formed much less spheres than control cells (Figure 3B). Moreover, the expression of liver T-ICs markers was downregulated in miR-361-3p knockdown HCC cells compared with control cells (Figure 3C). Consistently, the expression of stemness associated transcription factors was decreased in miR-361-3p knockdown HCC cells compared with control cells (Figure 3D). Furthermore, the in vitro and in vivo limiting dilution assay indicated that the proportion of liver T-ICs and tumorigenic ability were impaired in miR-361-3p knockdown HCC cells (Figure 3E&F). Collectively, our results demonstrated that miR-361-3p promoted liver T-ICs expansion.
miR-361-3p was upregulated in liver T-ICs. A. The expression of miR-361-3p in CD24+ and CD24- HCC cells was assessed by real-time PCR assay. B. The expression of miR-361-3p in CD133+ and CD133- HCC cells was detected by real-time PCR assay. C. The expression of miR-361-3p in EpCAM+ and EpCAM- HCC cells was checked by real-time PCR assay. D. The expression of miR-361-3p in hepatoma spheroids was examined by real-time PCR assay. E. miR-361-3p expression in serial passages of hepatoma spheroids was analyzed by real-time PCR. F. The expression of miR-361-3p in CD24+ and CD24- primary HCC cells was assessed by real-time PCR assay. G. The expression of miR-361-3p in CD133+ and CD133- primary HCC cells was detected by real-time PCR assay. H. The expression of miR-361-3p in EpCAM+ and EpCAM- primary HCC cells was checked by real-time PCR assay. I. The expression of miR-361-3p in primary hepatoma spheroids was examined by real-time PCR assay. J&K. The correlation between the transcription level of miR-361-3p and CD24 or CD133 in fifty HCC tissues was determined by real-time PCR analysis. Data were normalized to U6 or β-actin as △Ct and analyzed by Spearman's correlation analysis.
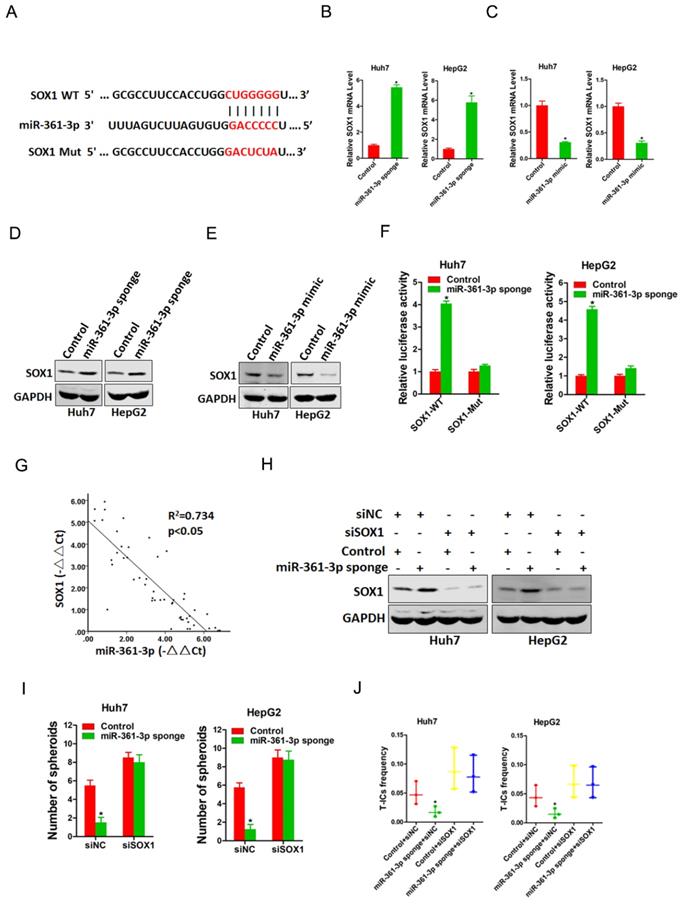
SOX1 is required in miR-361-3p-mediated liver T-ICs expansion
To elucidate mechanism underlying miR-361-3p-mediated liver T-ICs expansion, TargetScan and miRbase were used to predicted the potential targets of miR-361-3p in liver T-ICs. Bioinformatics analysis found that miR-361-3p has a putative binding site in SOX1 mRNA 3'-UTR (Figure 4A). As expected, both the mRNA and protein expression of SOX1 were upregulated in miR-361-3p knockdown HCC cells and downregulated in miR-361-3p overexpression HCC cells (Figure 4B-E). Luciferase reporter assays with a vector that included the wild-type (WT) 3'-UTR or mutant-type (MUT) 3'-UTR of SOX1 were then performed to determine whether miR-361-3p could directly regulate SOX1. The results showed that miR-361-3p knockdown significantly enhanced the relative luciferase activity compared with control group. However, such effects were diminished when the predicted binding site was mutated (Figure 4F). These results indicated that miR-361-3p directly suppressed SOX1 expression by binding to its 3'-UTR. In addition, a significant negative correlation was identified between SOX1 and miR-361-3p expression in clinical samples of HCC (R2 = 0.734, P < 0.05, n=50) (Figure 4G).
miR-361-3p promoted liver T-ICs self-renew and tumorigenesis. A. Huh7 and HepG2 cells were transfected with miR-361-3p mimic virus. The RNA was extracted and identified by real-time PCR. B. Representative images of hepatoma spheroids generated from Huh7/HepG2 miR-361-3p mimic and control cells. The number of spheroids was counted and compared. C. Flow cytometry analysis of CD24+ populations in spheroids derived from Huh7/HepG2 miR-361-3p mimic and control cells. Representative results from three independent experiments were shown. D. The expression of transcription factors (SOX2, OCT4 and c-Myc) in Huh7/HepG2 miR-361-3p mimic and control cells were detected by real-time PCR. E. The in vitro limiting dilution assay was used to check the population of liver T-ICs in Huh7/HepG2 miR-361-3p mimic and control cells. F. The in vivo limiting dilution assay was used to determine the tumorigenicity and liver T-ICs frequency. The frequency of tumor initiating cells was assessed using ELDA software.
miR-361-3p knockdown inhibited liver T-ICs self-renew and tumorigenesis. A. Huh7 and HepG2 cells were transfected with miR-361-3p sponge virus. The RNA was extracted and identified by real-time PCR. B. Representative images of hepatoma spheroids generated from Huh7/HepG2 miR-361-3p sponge and control cells. The number of spheroids was counted and compared. C. Flow cytometry analysis of CD24+ populations in spheroids derived from Huh7/HepG2 miR-361-3p sponge and control cells. Representative results from three independent experiments were shown. D. The expression of transcription factors (SOX2, OCT4 and c-Myc) in Huh7/HepG2 miR-361-3p sponge and control cells were detected by real-time PCR. E. The in vitro limiting dilution assay was used to check the population of liver T-ICs in Huh7/HepG2 miR-361-3p sponge and control cells. F. The in vivo limiting dilution assay was used to determine the tumorigenicity and liver T-ICs frequency. The frequency of tumor initiating cells was assessed using ELDA software.
Next, we explore the expression of SOX1 in liver T-ICs. As shown in supplementary Figure 1A-C, SOX1 expression was reduced in CD24+, CD133+ and EpCAM+ liver T-ICs that were sorted from HCC cell lines. Compared with the attached cells, SOX1 expression was downregulated in HCC spheres derived from HCC cells (supplementary Figure 1D). Moreover, SOX1 expression was decreased in CD24+, CD133+ and EpCAM+ liver T-ICs that were sorted from primary HCC patients (supplementary Figure 1E-G). Consistently, we also found that SOX1 expression was downregulated in HCC spheres derived from primary HCC patients (supplementary Figure 1H). To further confirm whether SOX1 was required for miR-361-3p mediated liver T-ICs expansion, the special SOX1 siRNA was used (Figure 4H). SOX1 siRNA abrogated the self-renewal ability and liver T-ICs frequency between miR-361-3p knockdown HCC cells and control cells (Figure 4I&J).
miR-361-3p directly targeted SOX1 in liver T-ICs. A. Schematic diagram of luciferase plasmids of SOX1 3'-UTR wild-type (WT) and SXO1 3'-UTR mutant (MUT). B. The SOX1 mRNA expression in Huh7/HepG2 miR-361-3p sponge and control cells were detected by real-time PCR. C. The SOX1 mRNA expression in Huh7/HepG2 miR-361-3p mimic and control cells were detected by real-time PCR. D. The SOX1 protein expression in Huh7/HepG2 miR-361-3p sponge and control cells were detected by western blot. E. The SOX1 protein expression in Huh7/HepG2 miR-361-3p mimic and control cells were detected by western blot. F. HCC cells were stably transfected with SOX1 3'-UTR wild-type (WT) and SXO1 3'-UTR mutant (MUT) luciferase reporter for 24 hours. The luciferase activity was measured as described in “materials and methods”. G. A negative correlation was observed between miR-361-3p and SOX1 expression in 50 clinical samples of HCC (R2 = 0.734, P < 0.05, by Spearman's correlation analysis). H. Huh7/HepG2 miR-361-3p sponge and control cells were transfected with siRNA SOX1 or its vector control. The stable transfectants were identified by Western Blotting. I. Huh7/HepG2 miR-361-3p sponge and control cells transfected with shRNA SOX1 and then subjected to spheroids formation assay. J. Huh7/HepG2 miR-361-3p sponge and control cells transfected with shRNA SOX1 and then subjected to in vitro limiting dilution assay.
miR-361-3p knockdown HCC cells were sensitive to cisplatin and sorafenib treatment. A. The expression of miR-361-3p in cisplatin-resistant or sorafenib-resistant xenograft was measured by real-time PCR. B. The expression of miR-361-3p in cisplatin-resistant HCC cell lines was measured by real-time PCR. C. The expression of miR-361-3p in sorafenib-resistant HCC cell lines was measured by real-time PCR. D. Cell survival curves of Huh7 miR-361-3p sponge and control cells treated with cisplatin or sorafenib. E. Huh7 miR-361-3p sponge and control cells were treated with cisplatin (4 μg/ml) for 48 hours. The apoptotic cells were detected by flow cytometry. F. Huh7 miR-361-3p sponge and control cells were treated with sorafenib (10 μM) for 48 hours. The apoptotic cells were detected by flow cytometry. G. Huh7 miR-361-3p sponge and control cells were treated with cisplatin (4 μg/ml) for 48 hours. The cell extracts were subjected to western blotting with special antibody against PARP. H. Huh7 miR-361-3p sponge and control cells were treated with sorafenib (10 μM) for 48 hours. The cell extracts were subjected to western blotting with special antibody against PARP. I. The total of 70 HCC patients (Cohort 1) were divided into low miR-361-3p group (n = 35) and high miR-361-3p group (n = 35), and the overall survival of the patients in the two groups were compared. J. The total of 68 HCC patients (Cohort 2) were divided into low miR-361-3p group (n = 34) and high miR-361-3p group (n = 34), and the overall survival of the patients in the two groups were compared.
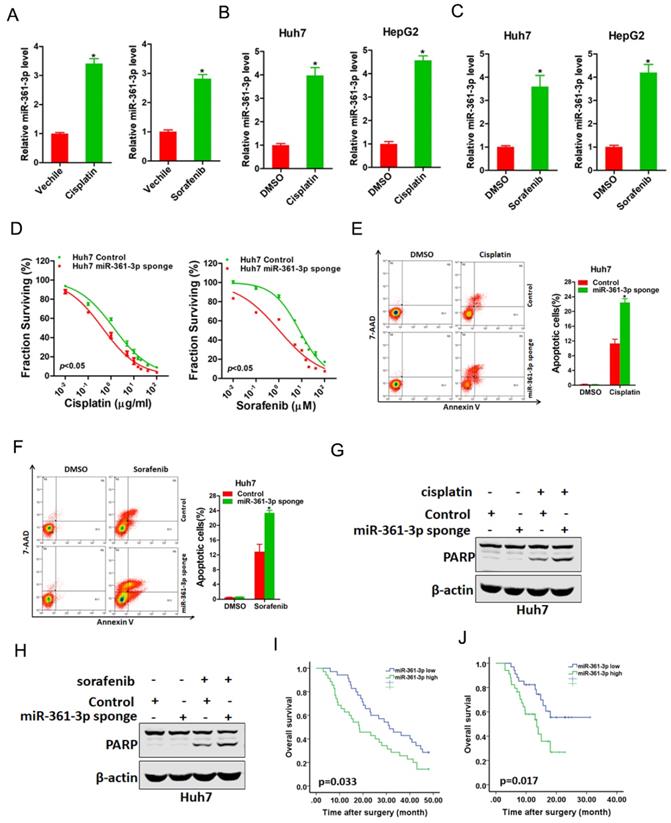
miR-361-3p knockdown HCC cells are sensitive to cisplatin and sorafenib treatment
We next explored whether miR-361-3p was involved in the regulation of chemo-resistance of HCC. As expected, miR-361-3p expression was markedly upregulated in cisplatin-resistance or sorafenib-resistance xenograft (Figure 5A). Consistently, miR-361-3p expression was also significantly increased in cisplatin-resistance or sorafenib-resistance HCC cell lines (Figure 5B&C). Moreover, the sensitivity of cisplatin or sorafenib was increased in miR-361-3p knockdown HCC cells compared with control HCC cells (Figure 5D). We also observed that interference of miR-361-3p sensitized HCC cells to undergo cisplatin-induced or sorafenib-induced cell apoptosis (Figure 5E-H). Furthermore, Kaplan-Meier analysis revealed the survival benefits in adjuvant TACE-treated or sorafenib-treated HCC patients with low miR-361-3p levels (Figure 5I&J).
Discussion
Hepatocellular carcinoma (HCC) is the sixth most common cancer in the world, accounting for 90% of human liver cancer [24]. The incidence of liver cancer is rising due to various factors such as hepatitis, alcoholic fatty liver, nonalcoholic fatty liver and aflatoxin [25, 26]. Hepatectomy and liver transplantation are commonly used in patients with early stage HCC [27]. However, most patients with advanced HCC are contradicting for surgery, and the survival benefit of TACE or targeted drug sorafenib is limited [28]. Therefore, it is necessary to further clarify the development of HCC. In this study, we confirmed for the first time that miR-361-3p was highly expressed in liver T-ICs, which enhanced liver T-ICs self-renewal and tumorigenesis ability. Our clinical data showed that miR-361-3p could be used to predict TACE and sorafenib response in HCC patients.
MiRNAs, a small non-coding RNA molecule (containing about 22 nucleotides), regulates RNA silencing and post-transcriptional of gene expression [29]. miRNAs act as oncogenes or tumor suppressors in tumors dependent on special conditions. miR-361-3p has been reported upregulated in various human tumors, including breast cancer, oral squamous cancer and pancreatic cancer [16, 17, 30]. However, miR-361-3p functions in liver T-ICs have never been investigated before. In current studies, we found that miR-361-3p was upregulated in liver T-ICs. Moreover, we found that miR-361-3p knockdown inhibited liver T-ICs self-renew and tumorigenesis. Conversely, forced miR-361-3p expression promoted liver T-ICs self-renew and tumorigenesis.
SOX1, belonging to SRY (sex determining region Y)-box (SOX) family proteins, is evolutionarily conserved in many species and participated as a key regulator of neural cell fate determination and differentiation [31, 32]. Previous studies have demonstrated that SOX1 plays an essential role in liver cancer progression [33]. In addition, SOX1 was also reported to be involved in the regulation of cancer stemness [34]. We hereby found that miR-361-3p knockdown increased SOX1 mRNA and protein expression in liver T-ICs. Moreover, we found that miR-361-3p directly regulates SOX1 expression via interaction with its 3' UTR. In addition, a significant negative correlation was identified between SOX1 and miR-361-3p expression in clinical samples of HCC. SOX1 siRNA could diminish the self-renewal ability and liver T-ICs frequency between miR-361-3p knockdown HCC cells and control cells.
For advanced HCC patients, TACE (transcatheter arterial chemoembolization) and the targeted drug sorafenib are the major means of treatment for a long time. However, only a small proportion of patients respond to sorafenib or TACE, and the majority of patients not only have no curative effect, but also have serious side effects [35, 36]. Therefore, elucidating the mechanism of TACE and sorafenib resistance has become an important link to prolong the survival time of HCC patients. Due to the different responses to sorafenib and TACE in different HCC patients, how to select biomarkers to predict drug reactivity has become a key issue to improve the clinical efficacy of sorafenib and TACE. In the present study, our finding revealed that miR-361-3p knockdown of HCC cells is more sensitive to cisplatin or sorafenib treatment. The TACE or sorafenib cohort analysis further indicated that a low miR-361-3p level in HCC patients can serve as a reliable predictor for TACE or sorafenib response.
In conclusion, we demonstrate that miR-361-3p is upregulated in liver T-ICs, which in turn promotes the self-renewal and tumorigenicity of liver T-ICs. In addition, miR-361-3p promotes liver T-ICs expansion through directly regulating SOX-1. Moreover, miR-361-3p knockdown HCC cells are more sensitive to cisplatin or sorafenib treatment. These data provide insight into the miR-361-3p as a potential therapeutic target against liver T-ICs and a potential predictor for TACE or sorafenib treatment of HCC patients.
Abbreviations
T-ICs: tumor-initiating cells; CSCs: cancer stem cells; miRNAs: MicroRNAs; 3'-UTR: 3'-untranslational region; DMEM: Dulbecco's modified Eagle's medium; FBS: fetal bovine serum; APC: adenomatous polyposis coli; TACE: transcatheter arterial chemoembolization; HCC: hepatocellular carcinoma; EHBH: Eastern Hepatobiliary Surgery Hospital; RT-PCR: Reverse transcription polymerase chain reaction; SOX: SRY (sex determining region Y)-box; EpCAM: epithelial cell adhesion molecule; CD24: cluster of differentiation 24; CD133: cluster of differentiation 133.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
Funding
This work is supported by the National Natural Science Foundation of China (81672718).
Availability of data and materials
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Craig AJ, von Felden J, Garcia-Lezana T. et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17(3):139-152
2. Vibert E, Schwartz M, Olthoff KM. Advances in resection and transplantation for hepatocellular carcinoma. J Hepatol. 2020;72(2):262-276
3. Wei X, Jiang Y, Zhang X. et al. Neoadjuvant Three-Dimensional Conformal Radiotherapy for Resectable Hepatocellular Carcinoma With Portal Vein Tumor Thrombus: A Randomized, Open-Label, Multicenter Controlled Study. J Clin Oncol. 2019;37(24):2141-2151
4. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283-296
5. Xiang D, Cheng Z, Liu H. et al. Shp2 promotes liver cancer stem cell expansion by augmenting beta-catenin signaling and predicts chemotherapeutic response of patients. Hepatology. 2017;65(5):1566-1580
6. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124-1134
7. Aceto N, Sausgruber N, Brinkhaus H. et al. Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nat Med. 2012;18(4):529-537
8. Xiang DM, Sun W, Zhou T. et al. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut. 2019;68(10):1858-1871
9. Suva ML, Tirosh I. The Glioma Stem Cell Model in the Era of Single-Cell Genomics. Cancer Cell. 2020;37(5):630-636
10. Taniguchi S, Elhance A, Van Duzer A. et al. Tumor-initiating cells establish an IL-33-TGF-beta niche signaling loop to promote cancer progression. Science. 2020 369(6501)
11. Han J, Won M, Kim JH. et al. Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem Soc Rev. 2020 Jul 7
12. Su R, Dong L, Li Y. et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38(1):79-96 e11
13. Vienberg S, Geiger J, Madsen S. et al. MicroRNAs in metabolism. Acta physiologica. 2017;219(2):346-361
14. Tutar Y. miRNA and cancer; computational and experimental approaches. Current pharmaceutical biotechnology. 2014;15(5):429
15. Han T, Zhang Y, Yang X. et al. miR-552 Regulates Liver Tumor-Initiating Cell Expansion and Sorafenib Resistance. Mol Ther Nucleic Acids. 2020;19:1073-1085
16. Hua B, Li Y, Yang X. et al. MicroRNA-361-3p promotes human breast cancer cell viability by inhibiting the E2F1/P73 signalling pathway. Biomed Pharmacother. 2020;125:109994
17. Hu J, Li L, Chen H. et al. MiR-361-3p regulates ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(8):807
18. Wang X, Sun W, Shen W. et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64(6):1283-1294
19. Xiang DM, Sun W, Ning BF. et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 2018Sep;67(9):1704-1715
20. Li XF, Chen C, Xiang DM. et al. Chronic inflammation-elicited liver progenitor cell conversion to liver cancer stem cell with clinical significance. Hepatology. 2017;66(6):1934-1951
21. Lee TK, Castilho A, Cheung VC. et al. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9(1):50-63
22. Yin S, Li J, Hu C. et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120(7):1444-1450
23. Yamashita T, Ji J, Budhu A. et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136(3):1012-1024
24. Han T, Xiang DM, Sun W. et al. PTPN11/Shp2 overexpression enhances liver cancer progression and predicts poor prognosis of patients. J Hepatol. 2015;63(3):651-660
25. Thietart S, Rautou PE. Extracellular Vesicles as Biomarkers in Liver Diseases: A Clinician's Point of View. J Hepatol. 2020
26. Mak LY, Wan-Hin Hui R. et al. Diverse effects of hepatic steatosis on fibrosis progression and functional cure in virologically quiescent chronic hepatitis B. J Hepatol. 2020
27. Chan ACY, Chok KSH, Dai J. et al. Transferability of Liver Transplantation Experience to Complex Liver Resection for Locally Advanced Hepatobiliary Malignancy - Lessons Learnt From 3 Decades of Single Center Experience. Ann Surg. 2020
28. Lencioni R, Llovet JM, Han G. et al. Sorafenib or placebo plus TACE with doxorubicin-eluting beads for intermediate stage HCC: The SPACE trial. J Hepatol. 2016;64(5):1090-1098
29. Campbell K. Do the microRNAs we eat affect gene expression? Nature. 2020;582(7812):S10-S11
30. Ogawa H, Nakashiro KI, Tokuzen N. et al. MicroRNA-361-3p is a potent therapeutic target for oral squamous cell carcinoma. Cancer science. 2020;111(5):1645-1651
31. Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129(18):4193-4203
32. Kan L, Israsena N, Zhang Z. et al. Sox1 acts through multiple independent pathways to promote neurogenesis. Developmental biology. 2004;269(2):580-594
33. Tsao CM, Yan MD, Shih YL. et al. SOX1 functions as a tumor suppressor by antagonizing the WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Hepatology. 2012;56(6):2277-2287
34. Garcia I, Aldaregia J, Marjanovic Vicentic J. et al. Oncogenic activity of SOX1 in glioblastoma. Sci Rep. 2017;7:46575
35. Hindson J. Combined TACE and sorafenib for HCC treatment. Nat Rev Gastroenterol Hepatol. 2020;17(2):66
36. Forner A, Gilabert M, Bruix J. et al. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol. 2014;11(9):525-535
Author contact
![]() Corresponding author: Department of Hepatic Surgery, Third Affiliated Hospital of Second Military Medical University, Shanghai, 200438, China, E-mail: jianzhai1979com (Jian Zhai); WuDongEHBHcom (Dong Wu).
Corresponding author: Department of Hepatic Surgery, Third Affiliated Hospital of Second Military Medical University, Shanghai, 200438, China, E-mail: jianzhai1979com (Jian Zhai); WuDongEHBHcom (Dong Wu).

 Global reach, higher impact
Global reach, higher impact