3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(13):3930-3944. doi:10.7150/jca.52286 This issue Cite
Research Paper
Heterogeneity Analysis of Esophageal Squamous Cell Carcinoma in Cell Lines, Tumor Tissues and Patient-Derived Xenografts
1. Department of Pathophysiology, School of Basic Medical Sciences, College of Medicine, Zhengzhou University, Zhengzhou, China.
2. China-US (Henan) Hormel Cancer Institute, Zhengzhou, Henan, 450008, China.
3. College of Korean Medicine, Dongshin University, Naju-si, Jeollanam-do, 58245, Republic of Korea.
4. Department of Pathology, Henan Cancer Hospital, The Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, Henan, 450008, China.
5. The Hormel Institute, University of Minnesota, Austin, MN, 55912, USA.
* Fayang Ma, Kyle Laster and Wenna Nie equally contributed to this work.
Abstract
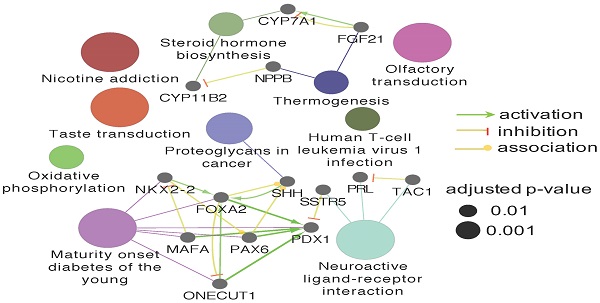
Esophageal Squamous Cell Carcinoma (ESCC) is the predominant type of Esophageal Cancer (EC), accounting for nearly 88% of EC incidents worldwide. Importantly, it is also a life-threatening cancer for patients diagnosed in advanced stages, with only a 20% 5-year survival rate due to a limited number of actionable targets and therapeutic options. Increasing evidence has shown that inter-tumor and intra-tumor heterogeneity are widely distributed across ESCC tumor tissues. In our work, multi-omics data from ESCC cell lines, tumor tissue, normal tissue and Patient-Derived Xenograft (PDX) tissues were analyzed to investigate the heterogeneity among ESCC samples at the DNA, RNA, and protein level. We identified enrichment of ECM-receptor interaction and Focal adhesion pathways from the subset of protein-coding genes with non-silent mutations in ESCC patients. We also found that TP53, TTN, KMT2D, CSMD3, DNAH5, MUC16 and DST are the most frequently mutated genes in ESCC patient samples. Out of the identified genes, TP53 is the most frequently mutated, with 84 distinct non-silent mutation variants. We observed that p.R248Q, p.R175G/H, and p.R273C/H are the most common TP53 mutation variants. The diversity of TP53 mutations reveal its importance in ESCC progression and may also provide promising targets for precision therapeutics. Additionally, we identified the Olfactory transduction as the top signaling pathway, enriched from genes uniquely expressed in The Cancer Genome Atlas (TCGA)-ESCC patient tumor tissues, which may provide implications for the exact roles of the corresponding genes in ESCC. Cyclic nucleotide-gated channel subunit beta 1(CNGB1), a gene belonging to the Olfactory transduction pathway, was found exclusively overexpressed in ESCC. Expression of CNGB1 could serve as a marker, indicating potential diagnostic or therapeutic value. Finally, we investigated heterogeneity in the context of the ESCC PDX model, which is an emerging tool used to predict drug response and recapitulate tumor behavior in vivo. We observed trans-species heterogeneity in as high as 75% of the identified proteins, indicating that the ambiguity of proteins should be addressed by specific strategies to avoid drawing false conclusions. The identification and characterization of gene mutation and expression heterogeneity across different ESCC datasets, including various novel TP53 mutations, ECM-receptor interaction, Focal adhesion, and Olfactory transduction pathways (CNGB1), provide researchers with evidence and implications for accurate research and precision therapeutic development.
Keywords: Esophageal Squamous Cell Carcinoma, Genetic Heterogeneity, Proteomics, Transcriptome, Heterograft, Bioinformatics.

 Global reach, higher impact
Global reach, higher impact