Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(13):3976-3996. doi:10.7150/jca.47695 This issue Cite
Review
Research Progress in Prognostic Factors and Biomarkers of Ovarian Cancer
1. Department of Laboratory Medicine, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China, 210029.
2. National Key Clinical Department of Laboratory Medicine, Nanjing, China, 210029.
Received 2020-5-2; Accepted 2021-4-22; Published 2021-5-13
Abstract
Ovarian cancer is a serious threat to women's health; its early diagnosis rate is low and prone to metastasis and recurrence. The current conventional treatment for ovarian cancer is a combination of platinum and paclitaxel chemotherapy based on surgery. The recurrence and progression of ovarian cancer with poor prognosis is a major challenge in treatment. With rapid advances in technology, understanding of the molecular pathways involved in ovarian cancer recurrence and progression has increased, biomarker-guided treatment options can greatly improve the prognosis of patients. This review systematically discusses and summarizes existing and new information on prognostic factors and biomarkers of ovarian cancer, which is expected to improve the clinical management of patients and lead to effective personalized treatment.
Keywords: ovarian cancer, prognostic factor, biomarker
Introduction
Ovarian cancer is the most fatal gynecological tumor, its incidence is next to cervical cancer and endometrial cancer, but its mortality rate is the first among reproductive system malignancies. According to the data of cancer statistics in 2020, the number of new cases is about 21750 and the number of deaths is 13940 [1]. Ovarian is located in the posterolateral uterine bottom, the onset is insidious, the early symptoms lack specificity, and the screening effect is limited, so the early diagnosis of ovarian cancer is difficult. According to the American congress of obstetricians and gynecologists (ACOG), 70 to 75 percent of ovarian cancers are diagnosed late, and the 5-year survival rate for most women is 20 to 30 percent [2]. Compared with other gynecological tumors, ovarian cancer has complex pathological types, high recurrence rate and poor prognosis. Patients with distant metastasis due to delayed medical treatment and tolerance to chemotherapy have worse prognosis. Therefore, the identification of effective clinical prognostic factors and biomarkers is crucial to improve the prognosis of ovarian cancer patients. With the in-depth study of the molecular changes that drive the transformation of ovarian cancer and tumor progression, many new molecular analysis techniques have been widely used. Recent studies have shown that microRNAs (miRNAs) may play an important role in the pathogenesis of ovarian cancer and serve as potential biomarkers [3].
The main contents of this review are divided into two parts: classic prognostic factors and novel prognostic factors. Classic prognostic factors included clinicopathologic factors (FIGO stage, degree of differentiation, degree of tumor reduction surgery, course of chemotherapy) and serum CA125. New prognostic factors mainly include blood- or tissue-based biomarkers. The ovarian cancer field has lagged in incorporating targeted therapies into standard treatments, these novel biomarkers are expected to provide therapeutic targets for ovarian cancer, thus guiding clinical practice, improving patient prognosis and ultimately reducing the risk of death of ovarian cancer patients.
Search Methods
Based on the topics discussed in this review, we systematically searched the recent medical literatures on novel prognostic biomarkers of ovarian cancer in PubMed and PMC databases by using our search strategy. All the literatures included in the study were published between February 1, 2015 and February 1, 2021. After excluding the duplicated literatures in the two databases, a total of 1,923 literatures met the restriction conditions. Then the retrieved literatures were imported into the literature management software Endnote. Preliminary screening was performed by reading the titles and abstracts of the literatures to exclude irrelevant studies, and then the full text of the included literatures was evaluated. In order to ensure the reliability of the research results, we only selected studies with more than 50 ovarian cancer patients, and the biomarkers studied in the literature were consistent with the clinical results. The inclusion and exclusion criteria and search strategy are provided in the appendix. Finally, a manual search was conducted in major journals and the reference lists of the selected papers to find other relevant citations that were missing by the electronic search.
Search Results
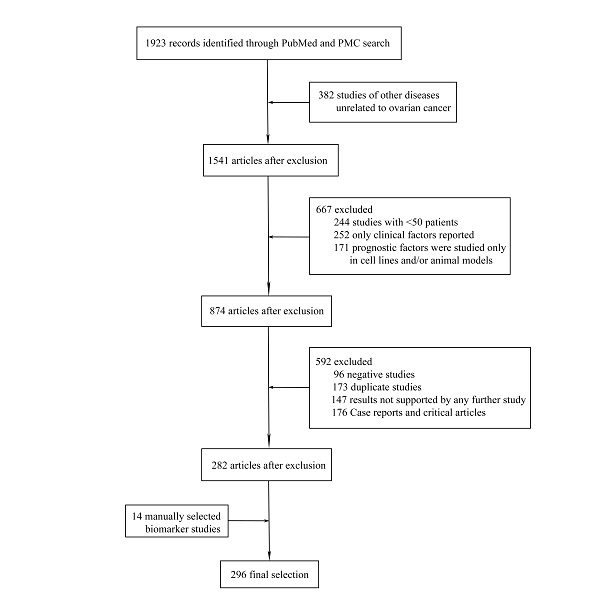
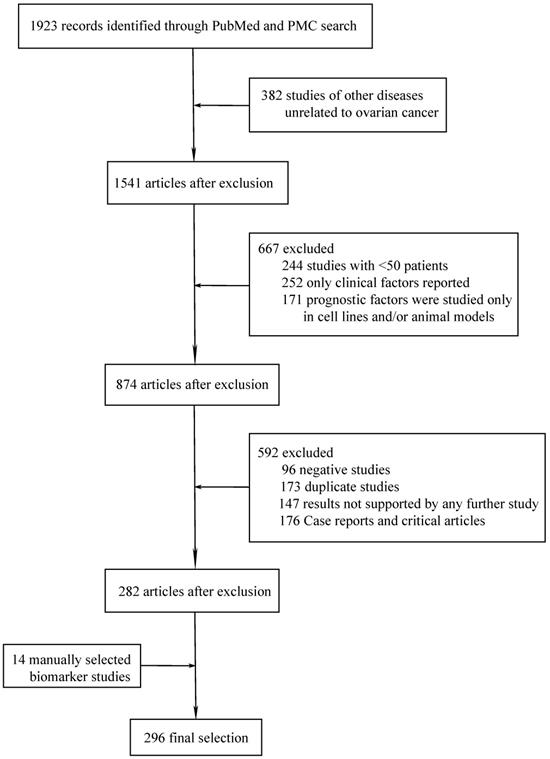
A total of 297 different novel prognostic biomarkers were reported in 296 studies that met the inclusion criteria (Figure 1). These prognostic biomarkers were classified according to the purpose of the study; there were 45 studies on biomarkers in the blood of ovarian cancer patients (Table 1) and 251 studies on biomarkers in tumor tissues (Tables 2-4).
Flowchart of article selection process.
Blood-based biomarkers in ovarian cancer
| Expression or ratio | Potential clinical use | Example study | ||||
|---|---|---|---|---|---|---|
| Study | Studied biomarkers | Subsite | Patients(n) | |||
| Cell proliferation and invasion | ||||||
| Leptin | Increased | Poor prognosis | Kato, S., et al. (2015)16 | Leptin | EOC | 70 |
| miR-429 | Increased | Good prognosis | Meng, X., et al. (2015)17 | miR-429 | EOC | 180 |
| ADAM12 | Increased | Poor prognosis | Cheon, D. J., et al. (2015)18 | ADAM12 | HGSOC | 84 |
| Septin-9, clusterin | Increased | Poor prognosis | Lyu, N., et al. (2018)19 | Septin-9, clusterin | EOC | 137 |
| MMP3, TIMP3 | Increased | Poor prognosis | Cymbaluk-Ploska, A., et al. (2018)20 | MMP3, TIMP3 | OC | 104 |
| MSLN | Increased | Poor prognosis | Karolina Okla et al. (2018)21 | MSLN | EOC | 97 |
| CYFRA21-1 | Increased | Poor prognosis | Jin, C., et al. (2019)22 | CYFRA21-1 | EOC | 203 |
| Inflammation | ||||||
| NLR | Increased | Poor prognosis | Feng, Z., et al. (2016)23 | NLR | HGSOC | 875 |
| NLR | Increased | Poor prognosis | Li, Z., et al. (2017)24 | NLR | EOC | 654 |
| CRP / Alb | Increased | Poor prognosis | Liu, Y., et al. (2017)25 | CRP/Alb | OC | 200 |
| NLR, LDH | Increased | Poor prognosis | Mauricio, P., et al. (2018)26 | NLR, LDH | HGSOC | 128 |
| AFR | Decreased | Poor prognosis | Yu, W., et al. (2019)27 | AFR | EOC | 313 |
| NLR | Increased | Poor prognosis | Ceran, M. U., et al. (2019)28 | NLR | EOC | 244 |
| PLR | Increased | Poor prognosis | Ceran, M. U., et al. (2019)28 | PLR | EOC | 244 |
| NLR | Increased | Poor prognosis | Nomelini, R. S., et al. (2019)29 | NLR | OC | 72 |
| Angiogenesis | ||||||
| Fibulin-4 | Increased | Good prognosis | Chen, J., et al. (2015)30 | Fibulin-4 | OC | 160 |
| VEGF | Increased | Poor prognosis | Dobrzycka, B., et al. (2015)31 | VEGF | SOC | 92 |
| VEGF-A | Increased | Good prognosis | Komatsu, H., et al. (2017)32 | VEGF-A | EOC | 128 |
| LncRNA MALAT1 | Increased | Poor prognosis | Qiu, J. J., et al. (2018)33 | LncRNA MALAT1 | EOC | 60 |
| Antioxidant | ||||||
| 8-OHdG | Increased | Poor prognosis | Pylväs-Eerola, M., et al. (2015)34 | 8-OHdG | EOC | 112 |
| Immune response | ||||||
| TNFa/IL-4 ratio | Increased | Good prognosis | Hao, C. J., et al. (2016)35 | TNFa/IL-4 ratio | OC | 50 |
| sPD-L1 | Increased | Poor prognosis | Chatterjee, J., et al. (2017)36 | sPD-L1 | EOC | 71 |
| s-CD95L | Increased | Good prognosis | De La Motte Rouge, T., et al. (2019)37 | s-CD95L | HGSOC | 51 |
| absolute lymphocyte count | Decreased | Poor prognosis | Lee, Y. J., et al. (2019)38 | absolute lymphocyte count | OC | 537 |
| CD4/CD8 ratio | Decreased | Good prognosis | Waki, K., et al. (2020)39 | CD4/CD8 ratio | OC | 52 |
| Chemotherapeutic sensitivity | ||||||
| CEBPA, C.69.OG>T polymorphism (rs34529039) | Increased | Poor prognosis | Konopka, B., et al. (2016)40 | CEBPA, C.69.OG>T polymorphism (rs34529039) | OC | 118 |
| hyperfibrinogenemia | Increased | Poor prognosis | Feng, Z., et al. (2016)41 | hyperfibrinogenemia | HGSOC | 875 |
| ERCC1 | Expression | Poor prognosis | Chebouti, I., et al. (2017)42 | ERCC1 | OC | 65 |
| miR-135a-3p | Increased | Good prognosis | Fukagawa, S., et al. (2017)43 | miR-135a-3p | OC | 98 |
| Gal-8, Gal-9 | Increased | Poor prognosis | Labrie, M., et al. (2017)44 | Gal-8, Gal-9 | HGSOC | 160 |
| Mitotic process | ||||||
| Aurora A codon 57 SNP | Increased | Good prognosis | Niu, H., et al. (2017)45 | Aurora A codon 57 SNP | OC | 122 |
| EMT and metastasis | ||||||
| miR 200a, miR 200b, miR 200c | Increased | Poor prognosis | Zuberi, M., et al. (2015)46 | miR 200a, miR 200b, miR 200c | EOC | 70 |
| miR-200b, miR-200c | Increased | Poor prognosis | Meng, X., et al. (2016)47 | miR-200b, miR-200c | EOC | 163 |
| Deregulation of the cellular transport | ||||||
| KPNA2 | Increased | Poor prognosis | Huang, L., et al. (2017)48 | KPNA2 | EOC | 162 |
| Apoptosis process | ||||||
| survivin | Increased | Poor prognosis | Dobrzycka, B., et al. (2015)31 | survivin | SOC | 92 |
| Smac/DIABLO | Decreased | Poor prognosis | Dobrzycka, B., et al. (2015)31 | Smac/DIABLO | SOC | 92 |
| Others | ||||||
| miR-200c, miR-141 | Increased | Good prognosis | Gao, Y.C., et al. (2015)49 | miR-200C, miR-141 | EOC | 93 |
| Platelet counts | Increased | Poor prognosis | Chen, Y., et al. (2015)50 | Platelet counts | EOC | 816 |
| SFRA | Increased | Poor prognosis | Kurosaki, A., et al. (2016)51 | SFRA | EOC | 128 |
| OPN | Increased | Poor prognosis | Zivny, J. H., et al. (2016)52 | OPN | SOC | 66 |
| microRNA-125b (miR-125b) | Increased | Poor prognosis | Zuberi, M., et al. (2016)53 | microRNA-125b (miR-125b) | EOC | 70 |
| miR-125b | Increased | Good prognosis | Zhu, T., et al. (2017)54 | miR-125b | EOC | 135 |
| BGA | Expression | Good prognosis | Montavon Sartorius, C., et al. (2018)55 | BGA | OC | 282 |
| RASSF1A rs1989839C > T SNP | Increased | Poor prognosis | He, W., et al. (2018)56 | RASSF1A rs1989839C > T SNP | OC | 1375 |
| MACC1 and S100A4 transcripts | Increased | Poor prognosis | Link, T., et al. (2019)57 | MACC1 and S100A4 transcripts | OC | 79 |
| sP (Hyp-Leu,Glu-Phe-Trp) | Decreased | Good prognosis | Lu, X., et al. (2019)58 | sP (Hyp-Leu,Glu-Phe-Trp) | EOC | 98 |
Abbreviations: miR: MicroRNA; NLR: the ratio of neutrophil count to lymphocyte count; AFR: albumin-to-fibrinogen ratio; PLR: platelet lymphocyte ratio; SNP: single Nucleotide Polymorphism; MSLN: Mesothelin; AAK: Aurora A kinase; Gal: Galectin; VEGF: vascular endothelial growth factor; sPD-L1: soluble PD - L1; OC: ovarian cancer; HGSOC: High grade serous ovarian cancer; EOC: epithelial ovarian cancer.
Tissue-based immunohistochemistry biomarkers in ovarian cancer
| Expression or ratio | Potential clinical use | Example study | ||||
|---|---|---|---|---|---|---|
| Study | Studied biomarkers | Subsite | Patients (n) | |||
| EMT and metastasis | ||||||
| CTHRC1 | Increased | Poor prognosis | Hou, M., et al. (2015)59 | CTHRC1 | EOC | 88 |
| ZEB2 | Increased | Poor prognosis | Prislei, S., et al. (2015)60 | ZEB2 | EOC | 143 |
| CD44v6 | Increased | Poor prognosis | Tjhay, F., et al. (2015)61 | CD44v6 | EOC | 59 |
| miR-506 | Increased | Good prognosis | Sun, Y., et al. (2015)62 | miR-506 | EOC | 204 |
| FILIP1L | Increased | Good prognosis | Kwon, M., et al. (2016)63 | FILIP1L | OC | 369 |
| Par3 | Decreased | Good prognosis | Nakamura, H., et al. (2016)64 | Par3 | OC | 50 |
| MMP-14, CD44 | Double expression | Poor prognosis | Vos, M. C., et al. (2016)65 | MMP-14, CD44 | OC | 97 |
| OTUB1 | Expression | Poor prognosis | Wang, Y., et al. (2016)66 | OTUB1 | OC | 200 |
| ESRP1 | Increased | Good prognosis | Chen, L., et al. (2017)67 | ESRP1 | EOC | 109 |
| MDM2 | Increased | Good prognosis | Chen, Y., et al. (2017)68 | MDM2 | OC | 104 |
| CD24 | Increased | Poor prognosis | Nakamura, K., et al. (2017)69 | CD24 | OC | 174 |
| CCNG1 | Increased | Poor prognosis | Xu, Y., et al. (2019)70 | CCNG1 | HGSOC | 266 |
| DDR2 | Increased | Poor prognosis | Ramalho, S., et al. (2019)71 | DDR2 | HGSOC | 78 |
| Inflammation and immune response | ||||||
| CD8/Treg ratio | Increased | Good prognosis | Knutson, K. L., et al. (2015)72 | CD8/Treg ratio | EOC | 405 |
| PD-1, PD-L1 | Increased | Good prognosis | Darb-Esfahani, S., et al. (2016)73 | PD-1, PD-L1 | HGSOC | 215 |
| Tumour-infiltrating B cell and plasma cell | Increased | Poor prognosis | Lundgren, S., et al. (2016)74 | Tumour-infiltrating B cell and plasma cell | EOC | 154 |
| TIL | Increased | Good prognosis | James, F. R., et al. (2017)75 | TIL | EOC | 707 |
| T-bet+ TILs | Increased | Good prognosis | Xu, Y., et al. (2017)76 | T-bet+ TILs | EOC | 81 |
| PD-L1 | Increased | Poor prognosis | Zhu, J., et al. (2017)77 | PD-L1 | OCCC | 138 |
| Transcription factors WT1 and p53 | Increased | Poor prognosis | Carter, J. H., et al. (2018)78 | Transcription factors WT1 and p53 | OC | 96 |
| SOCS-1 | Increased | Poor prognosis | Nakagawa, S., et al. (2018)79 | SOCS-1 | OC | 83 |
| PD-L1 | Increased | Good prognosis | Kim, K. H., et al. (2019)80 | PD-L1 | EOC | 248 |
| TIL | Increased | Good prognosis | Mauricio, P., et al. (2019)81 | TIL | HGSOC | 128 |
| RCAS1-Ir | Increased | Poor prognosis | Szubert, S., et al. (2019)82 | RCAS1-Ir | EOC | 67 |
| VISTA | Expression | Good prognosis | Zong, L., et al. (2020)83 | VISTA | OC | 146 |
| Co-expression of CD8+ and granzyme B+ | Increased | Good prognosis | Jäntti, T., et al. (2020)84 | Co-expression of CD8+ and granzyme B+ | HGSOC | 67 |
| Antioxidant | ||||||
| Nrf2 | Expression | Poor prognosis | Liew, P. L., et al. (2015)85 | Nrf2 | OC | 108 |
| SOD2 | Increased | Poor prognosis | Amano, T., et al. (2019)86 | SOD2 | EAOC | 61 |
| Angiogenesis | ||||||
| pIKK | Expression | Poor prognosis | Kinose, Y., et al. (2015)87 | pIKK | OC | 94 |
| PDGFβR | Increased | Poor prognosis | Corvigno, S., et al. (2016)88 | PDGFβR | SOC | 186 |
| VEGF-R1, VEGF-R2 | Expression | Good prognosis | Skirnisdottir, I., et al. (2016)89 | VEGF-R1, VEGF-R2 | EOC | 131 |
| Nestin | Increased | Poor prognosis | Onisim, A., et al. (2016)90 | Nestin | SOC | 85 |
| MIG-7 | Increased | Poor prognosis | Huang, B., et al. (2016)91 | MIG-7 | EOC | 121 |
| PTEN | Expression | Good prognosis | Shen, W., et al. (2017)92 | PTEN | OC | 76 |
| HIF-lα and VEGF | Expression | Poor prognosis | Shen, W., et al. (2017)92 | HIF-lα and VEGF | OC | 76 |
| AEG-1 | Increased | Poor prognosis | Yu, X., et al. (2018)93 | AEG-1 | EOC | 170 |
| VEGF, SEMA4D | Expression | Poor prognosis | Chen, Y., et al. (2018)94 | VEGF, SEMA4D | EOC | 124 |
| TBC1D16 | Increased | Good prognosis | Yang, Z., et al. (2018)95 | TBC1D16 | EOC | 156 |
| PGF | Increased | Poor prognosis | Meng, Q., et al. (2018)96 | PGF | EOC | 89 |
| VEGF-A | Decreased | Poor prognosis | Sopo, M., et al. (2019)97 | VEGF-A | OC | 86 |
| vasohibin-1, MACC1 | Increased | Poor prognosis | Yu, L., et al. (2019)98 | vasohibin-1, MACC1 | SOC | 124 |
| Tie-2 | Increased | Poor prognosis | Sopo, M., et al. (2020)99 | Tie-2 | HGSOC | 86 |
| Cell proliferation | ||||||
| FASN | Increased | Poor prognosis | Cai, Y., et al. (2015)100 | FASN | OC | 60 |
| CD73 | Increased | Poor prognosis | Turcotte, M., et al. (2015)101 | CD73 | HGSOC | 208 |
| SPINK1 | Increased | Poor prognosis | Mehner, C., et al. (2015)102 | SPINK1 | EOC | 490 |
| KCNN4, S100A14 | Increased | Poor prognosis | Zhao, H., et al. (2016)103 | KCNN4, S100A14 | SOC | 127 |
| EGFR | Increased | Poor prognosis | Xu, L., et al. (2016)104 | EGFR | EOC | 67 |
| Gab1 | Increased | Poor prognosis | Hu, L. and R. Liu (2016)105 | Gab1 | EOC | 124 |
| IL-36α | Decreased | Poor prognosis | Chang, L., et al. (2017)106 | IL-36α | EOC | 96 |
| DOT1L | Increased | Poor prognosis | Zhang, X., et al. (2017)107 | DOT1L | OC | 250 |
| KRT5, KRT6 | Increased | Poor prognosis | Ricciardelli, C., et al. (2017)108 | KRT5, KRT6 | SOC | 117 |
| hLSR | Increased | Poor prognosis | Hiramatsu, K., et al. (2018)109 | hLSR | EOC | 104 |
| PAUF, TIR4 | TLR4high and PAUFhigh/TLR4high | Poor prognosis | Choi, C. H., et al. (2018)110 | PAUF, TIR4 | EOC | 205 |
| PCDH8 | Decreased | Poor prognosis | Cao, Y., et al. (2018)111 | PCDH8 | OC | 68 |
| RIF1 | Increased | Poor prognosis | Liu, Y. B., et al. (2018)112 | RIF1 | EOC | 72 |
| FGFR2 | Increased | Poor prognosis | Li, M., et al. (2018)113 | FGFR2 | OC | 426 |
| FOXO1/PAX3 | Increased | Poor prognosis | Han, G. H., et al. (2019)114 | FOXO1 / PAX3 | EOC | 212 |
| pStat3 | Increased | Poor prognosis | Li, H., et al. (2020)115 | pStat3 | EOC | 156 |
| ATAD2 | Increased | Poor prognosis | Liu, Q., et al. (2020)116 | ATAD2 | OC | 60 |
| Cell migration | ||||||
| GRO-β | Increased | Poor prognosis | Ye, Q., et al. (2015)117 | GRO-β | OC | 136 |
| B7-H6 | Increased | Poor prognosis | Zhou, Y., et al. (2015)118 | B7-H6 | OC | 110 |
| OCT4, Notch1 and DLL4 | Increased | Poor prognosis | Yu, L., et al. (2016)119 | OCT4, Notch1 and DLL4 | EOC | 207 |
| EphA8 | Increased | Poor prognosis | Liu, X., et al. (2016)120 | EphA8 | OC | 233 |
| AGTR1 | Increased | Poor prognosis | Zhang, Q., et al. (2019)121 | AGTR1 | EOC | 902 |
| Cell invasion | ||||||
| CK2α | Increased | Poor prognosis | Ma, Z., et al. (2017)122 | CK2α | EOC | 117 |
| CEP55 | Increased | Poor prognosis | Zhang, W., et al (2017)123 | CEP55 | EOC | 213 |
| ANXA1 | Increased | Good prognosis | Manai, M., et al. (2020)124 | ANXA1 | EOC | 156 |
| Cell proliferation and migration | ||||||
| MAP3K8 | Increased | Poor prognosis | Gruosso, T., et al. (2015)125 | MAP3K8 | HGSOC | 139 |
| IL-33/ST2 axis | Increased | Poor prognosis | Tong, X., et al. (2016)126 | IL-33/ST2 axis | EOC | 152 |
| CDCP1, ADAM12 | Decreased | Good prognosis | Vlad, C., et al. (2016)127 | CDCP1, ADAM12 | SOC | 102 |
| FGFRL1 | Increased | Poor prognosis | Tai, H., et al. (2018)128 | FGFRL1 | OC | 90 |
| HSDL2 | Increased | Poor prognosis | Sun, Q., et al. (2018)129 | HSDL2 | OC | 74 |
| DUSP2 | Decreased | Poor prognosis | Liu, W., et al. (2019)130 | DUSP2 | HGSOC | 127 |
| Kallistatin (KAL) | Decreased | Poor prognosis | Wu, H., et al. (2019)131 | Kallistatin (KAL) | HGSOC | 312 |
| YTHDF1-EIF3C axis | Increased | Poor prognosis | Liu, T., et al. (2020)132 | YTHDF1-EIF3C axis | OC | 134 |
| Cell proliferation and invasion | ||||||
| IL-6R | Increased | Good prognosis | Isobe, A., et al. (2015)133 | IL-6R | OC | 94 |
| Usp7, MARCH7 | Increased | Poor prognosis | Zhang, L., et al. (2016)134 | Usp7, MARCH7 | EOC | 121 |
| PPA1 | Increased | Poor prognosis | Li, H., et al. (2017)135 | PPA1 | SOC | 139 |
| PATZ1 | Increased | Good prognosis | Zhao, C., et al. (2018)136 | PATZ1 | SOC | 208 |
| Cell migration and invasion | ||||||
| ARMC8 | Increased | Poor prognosis | Jiang, G., et al.(2015)137 | ARMC8 | OC | 247 |
| galectin-1 | Increased | Poor prognosis | Chen, L., et al. (2015)138 | galectin-1 | EOC | 110 |
| MAGE-A9 | Increased | Poor prognosis | Xu, Y., et al. (2015)139 | MAGE-A9 | EOC | 128 |
| TROP2 | Increased | Poor prognosis | Xu, N., et al. (2016)140 | TROP2 | EOC | 128 |
| GALNT6 | Increased | Poor prognosis | Lin, T. C., et al. (2017)141 | GALNT6 | EOC | 78 |
| Galectin-1 | Increased | Poor prognosis | Schulz, H., et al. (2017)142 | Galectin-1 | OC | 156 |
| Galectin-3 | Increased | Poor prognosis | Schulz, H., et al. (2017)142 | Galectin-3 | OC | 156 |
| Galectin-7 | Increased | Good prognosis | Schulz, H., et al. (2017)142 | Galectin-7 | OC | 156 |
| REDD1 | Increased | Poor prognosis | Chang, B., et al. (2018)143 | REDD1 | OC | 229 |
| RacGAP1 | Decreased | Good prognosis | Wang, C., et al. (2018)144 | RacGAP1 | EOC | 117 |
| PAI-1, PAI-RBP1 | Increased | Poor prognosis | Koensgen, D., et al. (2018)145 | PAI-1, PAI-RBP1 | OC | 156 |
| PRDX-1 | Increased | Poor prognosis | Sienko, J., et al. (2019)146 | PRDX-1 | OC | 55 |
| KAI1 | Decreased | Poor prognosis | Yu, L., et al. (2019)98 | KAI1 | SOC | 124 |
| CAV1, ATG4C | Increased | Poor prognosis | Zeng, Y., et al. (2020)147 | CAV1, ATG4C | EOC | 95 |
| Cell proliferation, migration and invasion | ||||||
| CH13L1, FKBP4 | Increased | Poor prognosis | Lawrenson, K., et al. (2015)148 | CH13L1, FKBP4 | EOC | 200 |
| REG4 | Increased | Poor prognosis | Chen, S., et al. (2015)149 | REG4 | EOC | 337 |
| Spry2 | Decreased | Poor prognosis | Masoumi-Moghaddam, S., et al. (2015)150 | Spry2 | OC | 99 |
| SWI/SNF subunits | Decreased | Poor prognosis | Abou-Taleb, H., et al. (2016)151 | SWI/SNF subunits | EOC | 152 |
| KIF2A | Decreased | Poor prognosis | Wang, D., et al. (2016)152 | KIF2A | EOC | 111 |
| Salusin-β | Increased | Poor prognosis | Zhang,Q.,et al.(2017)153 | Salusin-β | OC | 57 |
| P38α, ATF2 | Increased | Poor prognosis | Song,W.J.,et al.(2017)154 | P38α, ATF2 | OSC | 120 |
| nERβ5 | Increased | Poor prognosis | Chan, K. K. L., et al. (2017)155 | nERβ5 | OC | 106 |
| SENP3/SMT3IP1 | Increased | Poor prognosis | Cheng, J., et al. (2017)156 | SENP3/SMT3IP1 | EOC | 124 |
| BCL6, Lewis y | Increased | Poor prognosis | Zhu, L., et al. (2017)157 | BCL6, Lewis y | OC | 103 |
| CXCL11, HMGA2 | Increased | Poor prognosis | Jin, C., et al. (2018)158 | CXCL11, HMGA2 | HGSOC | 110 |
| HS3ST2 | Decreased | Poor prognosis | Huang, R.L., et al. (2018)159 | HS3ST2 | EOC | 115 |
| KIF2A | Increased | Poor prognosis | Sheng, N., et al. (2018)160 | KIF2A | OC | 108 |
| TRIM59 | Increased | Good prognosis | Wang, Y., et al. (2018)161 | TRIM59 | OC | 192 |
| S100A10 | Increased | Poor prognosis | Wang, L., et al. (2019)162 | S100A10 | OC | 138 |
| PYGB | Increased | Poor prognosis | Zhou, Y., et al. (2019)163 | PYGB | OC | 94 |
| Glycosylation disorder of protein | ||||||
| GalNAs T6, T14 | Increased | Poor prognosis | Sheta, R., et al. (2017)164 | GalNAs T6, T14 | HGSOC | 131 |
| Mitotic process | ||||||
| TOPK | Increased | Poor prognosis | Ikeda, Y., et al. (2016)165 | TOPK | EOC | 163 |
| HER2, AURKA | Increased | Poor prognosis | Li, M.J., et al. (2017)166 | HER2, AURKA | OCCC | 60 |
| KIF14 | Increased | Poor prognosis | Qiu, H. L., et al. (2017)167 | KIF14 | EOC | 170 |
| Apoptosis process | ||||||
| PDCD5 | Decreased | Poor prognosis | Gao, L., et al. (2015)168 | PDCD5 | OC | 127 |
| MDM2 | Increased | Poor prognosis | Makii, C., et al. (2016)169 | MDM2 | OCCC | 75 |
| DNA-PKcs, Akt3, p53 | Increased | Poor prognosis | Shin, K., et al. (2016)170 | DNA-PKcs, Akt3, p53 | SOC | 132 |
| Gal-1, Gal-8, Gal-9p | Increased | Poor prognosis | Labrie, M., et al. (2017)171 | Gal-1, Gal-8, Gal-9p | HGSOC | 209 |
| Cell survival (telomerase activity) | ||||||
| Phosphorylated Akt, hTERT | Increased | Poor prognosis | Lee, Y. K., et al. (2015)172 | phosphorylated Akt, hTERT | EOC | 92 |
| Chemotherapeutic sensitivity | ||||||
| JARID1B | Increased | Poor prognosis | Wang, L., et al. (2015)173 | JARID1B | EOC | 120 |
| ALDH1 | Increased | Good prognosis | Ayub, T. H., et al. (2015)174 | ALDH1 | EOC | 55 |
| PRP4K | Increased | Good prognosis | Corkery, D. P., et al. (2015)175 | PRP4K | OC | 199 |
| HtrA2 | Decreased | Poor prognosis | Miyamoto, M., et al. (2015)176 | HtrA2 | HGSOC | 142 |
| PTEN | Increased | Good prognosis | Wang, L., et al. (2015)177 | PTEN | EOC | 161 |
| NF-κBp65 | Increased | Poor prognosis | Wang, L., et al. (2015)177 | NF-κBp65 | EOC | 161 |
| eIF3a | Increased | Good prognosis | Zhang, Y., et al. (2015)178 | eIF3a | OC | 126 |
| GTF2H5 | Decreased | Good prognosis | Gayarre, J., et al. (2016)179 | GTF2H5 | HGSOC | 117 |
| POSTN | Increased | Poor prognosis | Sung, P. L., et al. (2016)180 | POSTN | EOC | 308 |
| SOX10 | Increased | Poor prognosis | Know, A.Y., et al. (2016)181 | SOX10 | EOC | 203 |
| GOLPH3L | Increased | Poor prognosis | He, S., et al. (2017)182 | GOLPH3L | OC | 177 |
| LC3A | Increased | Poor prognosis | Miyamoto, M., et al. (2017)183 | LC3A | OCCC | 117 |
| Stonin 2 (STON2) | Increased | Poor prognosis | Sun, X., et al. (2017)184 | Stonin 2 (STON2) | EOC | 89 |
| GATA3 | Increased | Poor prognosis | Chen, H. J., et al. (2018)185 | GATA3 | OC | 196 |
| EpCAM | Increased | Poor prognosis | Zhang, X., et al. (2018)186 | EpCAM | EOC | 109 |
| UBC13 | Decreased | Poor prognosis | Zhang, X., et al. (2018)187 | UBC13 | OC | 71 |
| 14-3-3ζ | Increased | Poor prognosis | Kim, H. J., et al. (2018)188 | 14-3-3ζ | OC | 88 |
| KCNN3 | Increased | Poor prognosis | Liu, X., et al. (2018)189 | KCNN3 | OC | 57 |
| HELQ | Increased | Poor prognosis | Long, J., et al. (2018)190 | HELQ | EOC | 87 |
| P15 PAF (KIAA0101) | Increased | Poor prognosis | Jin, C., et al. (2018)191 | P15 PAF (KIAA0101) | HGSOC | 118 |
| UTP23 | Decreased | Poor prognosis | Fu, Z., et al. (2019)192 | UTP23 | OC | 133 |
| ABCB9 | Decreased | Poor prognosis | Hou, L., et al. (2019)193 | ABCB9 | OC | 308 |
| PBK | Increased | Poor prognosis | Ma, H., et al. (2019)194 | PBK | HGSOC | 234 |
| Sorcin | Decreased | Good prognosis | Zhang, S., et al. (2019)195 | Sorcin | OC | 60 |
| PRC1 | Increased | Poor prognosis | Bu, H., et al. (2020)196 | PRC1 | HGSOC | 210 |
| NCALD | Decreased | Poor prognosis | Feng, L. Y. and L. Li (2020)197 | NCALD | EOC | 239 |
| Cell cycle regulation | ||||||
| CAP1 | Increased | Poor prognosis | Hua, M., et al. (2015)198 | CAP1 | EOC | 119 |
| CCNE1 | Increased | Poor prognosis | Ayhan, A., et al. (2017)199 | CCNE1 | OCCC | 120 |
| NUCKS | Increased | Poor prognosis | Shi, C., et al. (2017)200 | NUCKS | OC | 121 |
| TK1 | Increased | Poor prognosis | Wang, J., et al. (2017)201 | TK1 | SOC | 109 |
| Differentiation of cancer-associated fibroblasts (CAFs) | ||||||
| MARCKS | Increased | Poor prognosis | Doghri, R., et al. (2017)202 | MARCKS | EOC | 118 |
| Immunosuppression | ||||||
| VEGF | Increased | Poor prognosis | Horikawa, N., et al. (2017)203 | VEGF | HGSOC | 56 |
| Metabolic reprogramming | ||||||
| TBC1D8 | Increased | Poor prognosis | Chen, M., et al. (2019)204 | TBC1D8 | OC | 141 |
| Fatty acid metabolism | ||||||
| PAX2 | Increased | Poor prognosis | Feng, Y., et al. (2020)205 | PAX2 | EOC | 152 |
| Defective DNA repair | ||||||
| WRAP53β | Decreased | Poor prognosis | Hedström, E., et al. (2015)206 | WRAP53β | EOC | 151 |
| pH2AX | Increased | Poor prognosis | Mei, L., et al. (2015)207 | pH2AX | EOC | 87 |
| Others | ||||||
| SLP-2 | Increased | Poor prognosis | Sun, F., et al. (2015)208 | SLP-2 | EOC | 140 |
| CD44v8-10 | Expression | Good prognosis | Sosulski, A., et al. (2016)209 | CD44v8-10 | SOC | 210 |
| P53 | Increased | Poor prognosis | Zuo, J., et al. (2016)210 | P53 | SOC | 183 |
| Highly sulfated CS | Increased | Poor prognosis | Van der steen, S.C., et al. (2016)211 | Highly sulfated CS | EOC | 255 |
| Adiponectin receptor-1 (AdipoR1) | Increased | Good prognosis | Li, X., et al. (2017)212 | Adiponectin receptor-1 (AdipoR1) | EOC | 73 |
| TP53 | Increased | Poor prognosis | Rzepecka, I. K., et al. (2017)213 | TP53 | HGSOC | 159 |
| SMAD3 | Increased | Poor prognosis | Sakr, S., et al. (2017)214 | SMAD3 | GCT | 88 |
| ALDH5A1 | Increased | Good prognosis | Tian, X., et al. (2017)215 | ALDH5A1 | OC | 192 |
| GR | Increased | Poor prognosis | Veneris, J. T., et al. (2017)216 | GR | EOC | 341 |
| LAMP3 | Increased | Poor prognosis | Wang, D., et al. (2017)217 | LAMP3 | EOC | 135 |
| HBXIP | Increased | Poor prognosis | Wang, Y., et al. (2017)218 | HBXIP | OC | 120 |
| HSF1 pSer326 | Expression | Poor prognosis | Yasuda, K., et al. (2017)219 | HSF1 pSer326 | EOC | 122 |
| COX-1, COX-2 | Increased | Poor prognosis | Beeghly-Fadiel, A., et al. (2018)220 | COX-1, COX-2 | EOC | 190 |
| GPR30 | Expression | Poor prognosis | Zhu, C. X., et al. (2018)221 | GPR30 | EOC | 110 |
| HJURP | Increased | Poor prognosis | Li, L., et al. (2018)222 | HJURP | HGSOC | 98 |
| Galectins-8 | Increased | Good prognosis | Schulz, H., et al. (2018)223 | Galectins-8 | OC | 156 |
| HER3 | Expression | Poor prognosis | Chung, Y. W., et al. (2019)224 | HER3 | EOC | 105 |
| ANXA8 | Increased | Poor prognosis | Gou, R., et al. (2019)225 | ANXA8 | OC | 122 |
| USP10/p14ARF | Decreased | Poor prognosis | Han, G. H., et al. (2019)226 | USP10/p14ARF | EOC | 212 |
| PKP3 | Increased | Poor prognosis | Qian, H., et al. (2019)227 | PKP3 | OC | 157 |
| PDGFR-β | Expression | Good prognosis | Szubert, S., et al. (2019)228 | PDGFR-β | EOC | 52 |
| CN | Increased | Poor prognosis | Xin, B., et al. (2019)229 | CN | OC | 50 |
| TSLP | Increased | Poor prognosis | Xu, L., et al. (2019)230 | TSLP | EOC | 144 |
| BUB1B, KIF11 and KIF20A | Increased | Poor prognosis | Zhang, L., et al. (2019)231 | BUB1B, KIF11 and KIF20A | OC | 50 |
| VDR | Increased | Poor prognosis | Czogalla, B., et al. (2020)232 | VDR | EOC | 156 |
Abbreviations: TIL: tumor infiltrates lymphocytes; Gal: Galectin; OC: ovarian cancer; HGSOC: High grade serous ovarian cancer; EOC: epithelial ovarian cancer.
Tissue-based DNA biomarkers in ovarian cancer
| Expression or ratio | Potential clinical use | Example study | |||||
|---|---|---|---|---|---|---|---|
| Study | Studied biomarkers | Method | Subsite | Patients (n) | |||
| Methylation | |||||||
| MYLK3 Methylation | Increased | Good prognosis | Phelps, D.L., et al. (2017)233 | MYLK3 Methylation | Pyrosequencing | SOC | 803 |
| HNF1B | Expression | Poor prognosis | Bubancova, I., et al. (2017)234 | HNF1B | NGS, HRM, MS-PCR | OC | 64 |
| GATA4 | Expression | Good prognosis | Bubancova, I., et al. (2017)234 | GATA4 | NGS, HRM, MS-PCR | OC | 64 |
| HS3ST2 | Increased | Poor prognosis | Huang, R.L., et al. (2018)159 | HS3ST2 | TMA | EOC | 115 |
| ZNF671 | Increased | Early relapse | Mase, S., et al. (2019)235 | ZNF671 | Pyrosequencing | HGSOC | 78 |
| Structural changes of nuclear chromatin | |||||||
| Chromatin entropy nuclei | Increased | Poor prognosis | Nielsen, B. et al. (2018)236 | Chromatin entropy nuclei | Nuclear Texture analysis | OC | 246 |
| Mutation status | |||||||
| BRCA1/2 wild type | Expression | Poor prognosis | Eoh, K. J., et al. (2017)237 | BRCA1/2 wild type | Direct sequencing | EOC | 116 |
| BRCA1/2 | Expression | Good prognosis | Kim, S. I., et al. (2019)238 | BRCA1/2 | Sanger sequencing | HGSOC | 128 |
| Cell proliferation and apoptosis | |||||||
| ecDNA | Increased | Poor prognosis | Kalavska, K., et al. (2018)239 | ecDNA | RT-PCR | OC | 67 |
| Gene polymorphism | |||||||
| The AT genotype of rs189897 | Expression | Poor prognosis | Liu, J., et al. (2019)240 | The AT genotype of rs189897 | Mass ARRAY | EOC | 200 |
| rs12921862 C/C | Expression | Good prognosis | Zhang, Y., et al. (2019)241 | rs12921862 C/C | PCR-RFLP | EOC | 165 |
Abbreviations: TMA: tissue microarrays; NGS: Next Generation Sequencing; MS-PCR: Methylation-Specific PCR; RT-PCR: real time polymerase chain reaction; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
Tissue-based RNA biomarkers in ovarian cancer
| Expression or ratio | Potential clinical use | Example study | |||||
|---|---|---|---|---|---|---|---|
| Study | Studied biomarkers | Method | Subsite | Patients (n) | |||
| Cell proliferation | |||||||
| microRNA(miR)-498 | Decreased | Poor prognosis | Cong, J., et al. (2015)242 | microRNA(miR)-498 | qRT-PCR | OC | 175 |
| miR-193b | Decreased | Poor prognosis | Li, H., et al. (2015)243 | miR-193b | qRT-PCR | OC | 116 |
| miR-572 | Decreased | Good prognosis | Zhang, X., et al. (2015)244 | miR-572 | qRT-PCR | OC | 108 |
| C7 | Decreased | Poor prognosis | Ying, L., et al. (2016)245 | C7 | qRT-PCR | OC | 156 |
| HER2, STAT3 | Increased | Poor prognosis | Shang, A. Q., et al. (2017)246 | HER2, STAT3 | qRT-PCR | OC | 136 |
| SOCS3 | Decreased | Poor prognosis | Shang, A. Q., et al. (2017)246 | SOCS3 | qRT-PCR | OC | 136 |
| lncRNA RAD51-AS1 | Increased | Poor prognosis | Zhang, X., et al. (2017)247 | lncRNA RAD51-AS1 | qRT-PCR | EOC | 163 |
| lncRNA LINC 00152 | Increased | Poor prognosis | Chen, P., et al. (2018)248 | lncRNA LINC 00152 | qRT-PCR | OC | 82 |
| miR-1294 | Increased | Good prognosis | Guo, T. Y., et al. (2018)249 | miR-1294 | qRT-PCR | EOC | 76 |
| lncRNA TUG1 | Increased | Poor prognosis | Li, T. H., et al. (2018)250 | lncRNA TUG1 | qRT-PCR | EOC | 96 |
| microRNA-424-5p (miR-424-5p) | Increased | Good prognosis | Liu, J., et al. (2018)251 | microRNA-424-5p (miR-424-5p) | qRT-PCR | EOC | 83 |
| Cell migration | |||||||
| lncRNA LINC00092 | Increased | Poor prognosis | Zhao, L., et al. (2017)252 | lncRNA LINC00092 | qRT-PCR | SOC | 58 |
| lncRNA PTPRG-AS1 | Increased | Poor prognosis | Ren, X. Y., et al. (2020)253 | lncRNA PTPRG-AS1 | qRT-PCR | EOC | 184 |
| Cell invasion | |||||||
| lncRNA NEAT1 | Increased | Poor prognosis | Chen, Z. J., et al. (2016)254 | lncRNA NEAT1 | qRT-PCR | OC | 149 |
| ASAP1-IT1 | Increased | Good prognosis | Fu, Y., et al. (2016)255 | ASAP1-IT1 | qRT-PCR | EOC | 266 |
| Cell proliferation and migration | |||||||
| miR-145 | Decreased | Poor prognosis | Kim,T.H.,et al.(2015)256 | miR-145 | qRT-PCR | HGSOC | 74 |
| microRNA-196a | Increased | Poor prognosis | Fan, Y., et al. (2015)257 | microRNA-196a | qRT-PCR | EOC | 156 |
| miR-552 | Increased | Poor prognosis | Zhao, W., et al. (2019)258 | miR-552 | qRT-PCR | OC | 110 |
| Cell proliferation and invasion | |||||||
| lncRNA AB073614 | Increased | Poor prognosis | Cheng, Z., et al. (2015)259 | lncRNA AB073614 | qRT-PCR | OC | 75 |
| TBL1XR1 | Increased | Poor prognosis | Ma, M. and N. Yu (2017)260 | TBL1XR1 | qRT-PCR | SOC | 116 |
| lncRNA MNX1-AS1 | Increased | Poor prognosis | Li, A. H. and H. H. Zhang (2017)261 | lncRNA MNX1-AS1 | qRT-PCR | EOC | 177 |
| lncRNA NEAT1 | Increased | Poor prognosis | Yong, W., et al. (2018)262 | lncRNA NEAT1 | qRT-PCR | HGSOC | 75 |
| miR-532-5p | Decreased | Poor prognosis | Wei, H., et al. (2018)263 | miR-532-5p | qRT-PCR | EOC | 145 |
| Cell migration and invasion | |||||||
| ANRIL | Increased | Poor prognosis | Qiu,J.J.,et al.(2015)264 | ANRIL | qRT-PCR | SOC | 68 |
| lncRNA CCAT1 | Increased | Poor prognosis | Cao,Y.,et al.(2017)265 | lncRNA CCAT1 | qRT-PCR | EOC | 72 |
| miR-208a-5p | Increased | Good prognosis | Mei, J., et al. (2019)266 | miR-208a-5p | qRT-PCR | OC | 61 |
| STAT2 | Increased | Poor prognosis | Chen, X., et al. (2020)267 | STAT2 | RT-PCR | OC | 62 |
| lncRNA miR503HG | Decreased | Poor prognosis | Zhu, D., et al. (2020)268 | lncRNA miR503HG | qRT-PCR | OC | 61 |
| Cell proliferation, migration and invasion | |||||||
| lncRNA CCAT2 | Increased | Poor prognosis | Huang,S.,et al.(2016)269 | lncRNA CCAT2 | qRT-PCR | OC | 109 |
| GOLPH3 | Increased | Poor prognosis | Sun, J., et al. (2017)270 | GOLPH3 | qRT-PCR | EOC | 73 |
| lncRNA HOXA11as | Increased | Poor prognosis | Yim, G. W., et al. (2017)271 | lncRNA HOXA11as | qRT-PCR | SOC | 129 |
| miR-520h | Increased | Poor prognosis | Zhang, J., et al. (2018)272 | miR-520h | qRT-PCR | EOC | 116 |
| lncRNA SNHG16 | Increased | Poor prognosis | Yang, X. S., et al. (2018)273 | lncRNA SNHG16 | qRT-PCR | OC | 103 |
| lncRNA EBIC | Increased | Poor prognosis | Xu, Q. F., et al. (2018)274 | lncRNA EBIC | qRT-PCR | OC | 126 |
| lncRNA MALAT1 | Increased | Poor prognosis | Guo, C., et al. (2018)275 | lncRNA MALAT1 | qRT-PCR | OC | 60 |
| lncRNA RP11-552M11.4 | Increased | Poor prognosis | Huang, K., et al. (2018)276 | lncRNA RP11-552M11.4 | qRT-PCR | EOC | 67 |
| lncRNA OTUB1-isoform2 | Increased | Poor prognosis | Wang, S., et al. (2018)277 | lncRNA OTUB1-isoform2 | qRT-PCR | OC | 114 |
| HYOU1 | Increased | Poor prognosis | Li, X., et al. (2019)278 | HYOU1 | qRT-PCR | EOC | 127 |
| miR-203a-3p | Increased | Good prognosis | Liu, H. Y., et al. (2019)279 | miR-203a-3p | qRT-PCR | OC | 152 |
| LINC00339 | Increased | Poor prognosis | Pan, L., et al. (2019)280 | LINC00339 | qRT-PCR | OC | 75 |
| lncRNA SNHG20 | Increased | Poor prognosis | Wang, D., et al. (2019)281 | lncRNA SNHG20 | RT-PCR | EOC | 60 |
| miR-149 | Increased | Good prognosis | Zhao, L. W., et al. (2020)282 | miR-149 | qRT-PCR | OC | 72 |
| Chemotherapeutic sensitivity | |||||||
| microRNA-506 (miR-506) | Increased | Good prognosis | Liu, G., et al. (2015)283 | microRNA-506 (miR-506) | qRT-PCR | EOC | 598 |
| CHI3L1 | Increased | Poor prognosis | Chiang, Y. C., et al. (2015)284 | CHI3L1 | qRT-PCR | EOC | 180 |
| IMP3 | Increased | Poor prognosis | Hsu, K. F., et al. (2015)285 | IMP3 | qRT-PCR | EOC | 140 |
| Lin28B | Increased | Poor prognosis | Hsu, K. F., et al. (2015)285 | Lin28B | qRT-PCR | EOC | 140 |
| Tribbles 2 (TRIB2) | Decreased | Poor prognosis | Kritsch, D., et al. (2017)286 | Tribbles 2 (TRIB2) | qRT-PCR | EOC | 149 |
| let-7e | Decreased | Poor prognosis | Xiao, M., et al. (2017)287 | let-7e | qRT-PCR | EOC | 84 |
| MAL | Increased | Poor prognosis | Zanotti, L., et al. (2017)288 | MAL | qRT-PCR | HGSOC | 74 |
| miR-98-5p | Increased | Good prognosis | Wang, Y., et al. (2018)289 | miR-98-5p | qRT-PCR | EOC | 97 |
| miR-1180 | Increased | Poor prognosis | Gu, Z. W., et al. (2019)290 | miR-1180 | qRT-PCR | OC | 59 |
| lncRNA GAS5 | Increased | Good prognosis | Long, X., et al. (2019)291 | lncRNA GAS5 | qRT-PCR | EOC | 53 |
| Immune response | |||||||
| APOBEC3G | Increased | Good prognosis | Leonard, B., et al. (2016)292 | APOBEC3G | qRT-PCR | HGSOC | 354 |
| lncRNA MIR155HG | Increased | Good prognosis | Colvin, E. K., et al. (2020)293 | lncRNA MIR155HG | qRT-PCR | HGSOC | 67 |
| Chromosome structure and function | |||||||
| SMYD3 genetic polymorphisms | Expression | Poor prognosis | Liu, T. T., et al. (2016)294 | SMYD3 genetic polymorphisms | qRT-PCR | OC | 154 |
| Apoptosis process | |||||||
| CPS1-IT1 | Increased | Good prognosis | Wang, Y. S., et al. (2017)295 | CPS1-IT1 | qRT-PCR | EOC | 91 |
| Others | |||||||
| CRNDE | Increased | Poor prognosis | Szafron, L. M., et al. (2015)296 | CRNDE | qRT-PCR | OC | 135 |
| GADD45A (1506T> C) | Increased | Poor prognosis | Yuan, C., et al. (2015)297 | GADD45A (1506T> C) | qRT-PCR | OC | 258 |
| miR-510, miR-129-3P | Decreased | Poor prognosis | Zhang,X.,et al.(2015)298 | miR-510, miR-129-3P | RT-qPCR,ISH | EOC | 78 |
| FAM215A | Increased | Good prognosis | Fu, Y., et al. (2016)255 | FAM215A | qRT-PCR | EOC | 266 |
| LIN-28B/let-7a/IGF-II axis | LIN-28Blowlet-7alow or LIN-28Blowlet-7ahighIGF-IIlow | Good prognosis | Lu, L., et al. (2016)299 | LIN-28B/let-7a/IGF-II axis | qRT-PCR | EOC | 211 |
| miR-200b, miR-1274A (tRNA Lys5) and miR-141 | Decreased | Good prognosis | Halvorsen, A. R., et al. (2017)300 | miR-200b, miR-1274A (tRNA Lys5) and miR-141 | qRT-PCR | OC | 207 |
| miR-595 | Decreased | Poor prognosis | Zhou, Q. H., et al. (2017)301 | miR-595 | qRT-PCR | EOC | 166 |
| KLK11, KLK15 | Increased | Good prognosis | Geng,X.,et al.(2017)302 | KLK11, KLK15 | RT-PCR | HGSOC | 139 |
| lncRNA LINC01088 | Decreased | Poor prognosis | Ai, H., et al. (2018)303 | lncRNA LINC01088 | qRT-PCR | EOC | 184 |
| lncRNA HMMR-AS1 | Increased | Poor prognosis | Chu, Z. P., et al. (2018)304 | lncRNA HMMR-AS1 | qRT-PCR | EOC | 152 |
| circ LARP4 | Decreased | Poor prognosis | Zou, T., et al. (2018)305 | circ LARP4 | qRT-PCR | OC | 78 |
| circ HIPK3 | Increased | Poor prognosis | Liu, N., et al. (2018)306 | circ HIPK3 | qRT-PCR | EOC | 69 |
| lncRNA DGCR5 | Decreased | Poor prognosis | Chen, H., et al. (2019)307 | lncRNA DGCR5 | qRT-PCR | OC | 66 |
| FANCD2 | Increased | Poor prognosis | Moes-Sosnowska, J., et al. (2019)308 | FANCD2 | qRT-PCR | OC | 99 |
| AK7 | Decreased | Poor prognosis | Zhang, X. Y., et al. (2021)309 | AK7 | RNAseq | OC | 308 |
Abbreviations: lnc: Long non-coding RNA; circ: circular; qRT-PCR: quantitative real time polymerase chain reaction; RT-PCR: real time polymerase chain reaction; IHC: Immunohistochemistry; ISH, in situ hybridization.
Classic prognostic factors
Clinicopathologic factors and serum CA125 level are independent factors affecting the prognosis of ovarian cancer patients, which have been widely used to guide accurate and reasonable clinical treatment, so as to improve the survival rate of patients.
Clinicopathological factors
The clinicopathological factors that affect the prognosis of ovarian cancer mainly include: FIGO stage, degree of differentiation, degree of tumor reduction surgery, course of chemotherapy. Previous literature has reported the importance of ovarian cancer staging for prognosis and treatment options, ovarian cancer can be classified as stage I-IV according to FIGO staging criteria, and most patients have stage III disease. Studies have shown that patients with stage I ovarian cancer have a 5-year survival rate of more than 90%; when ovarian cancer is confined to the pelvis (stage II), the estimated 5-year survival rate is about 70%; when ovarian cancer has spread to the entire abdominal cavity (stage III) or to distant parts (stage IV), the 5-year survival rate is less than 30% [4]. The survival prognosis of patients in the early stage was significantly better than that in the late stage. Differentiation degree of ovarian cancer includes high differentiation, moderate differentiation and low differentiation (poor differentiation), there has been evidence that poor differentiation of ovarian cancer is associated with worse survival. A large sample study established a predictive model for overall survival in 1189 patients with primary ovarian epithelial carcinoma, cox regression analysis showed that the worse the differentiation, the greater the risk of death [5].
Surgery is the most effective treatment for ovarian cancer, once suspected for ovarian cancer, should be performed as early as possible. Staging surgery is performed for early stage cancer, including resection of the tumor and definite staging. Tumor cell reduction was performed for advanced cancer, and the primary tumor and all metastases were removed as far as possible to minimize the number of tumor cells. Studies have confirmed that the degree of tumor cell reduction and the number of residual lesions after the first operation are important prognostic factors for advanced ovarian cancer [6]. The research of Jing shui et al. shows that the size of residual tumor foci was negatively correlated with the survival rate of patients and those with residual tumor foci ≤ 2 cm had better prognosis [7]. It is helpful to improve the prognosis and long-term survival rate of patients by minimizing or removing residual tumor foci.
Chemotherapy is an important adjuvant treatment for ovarian cancer, and most ovarian cancer is sensitive to chemotherapy. Platinum-based drugs (cisplatin and carboplatin) and taxanes (paclitaxel and docetaxel) are chemotherapy drugs commonly used in the treatment of ovarian cancer [8]. Postoperative adjuvant chemotherapy should follow the principles of standard, early and adequate course of treatment. Currently, it is generally considered that the standard course of chemotherapy for ovarian cancer is 6 courses. Three trials of primary advanced ovarian cancer compared the efficacy of chemotherapy with cisplatin in 5-6 cycles and 8-12 cycles, and the results showed that there was no benefit after 6 cycles of chemotherapy [9]. Another study on prognostic factor analysis of 129 cases of epithelial ovarian cancer showed that the median OS of patients with postoperative chemotherapy course ≥ 6 courses was significantly higher than that of patients with less than 6 courses of chemotherapy, and the difference was statistically significant (P<0.0001). There was no statistically significant difference in median OS in patients with 6 courses of chemotherapy, 7 courses of chemotherapy, 8 courses of chemotherapy or more than 8 courses of chemotherapy (P=0.816) [10]. In summary, postoperative chemotherapy course is an important prognostic factor for ovarian cancer, and standard chemotherapy course is associated with higher overall survival.
CA125
CA125, encoded by the MUC16 gene, is a classic marker for the diagnosis of ovarian cancer and was first described in the study of Bast RC et al [11]. Serum CA125 lacks sensitivity and specificity and cannot be used as a single marker for early detection of ovarian cancer [12,13], but the CA125 value after surgery and chemotherapy plays an important role in monitoring recurrence and evaluating prognosis. Redman et al. detected the CA125 value before the third chemotherapy in 78 patients with stage II~IV ovarian cancer after the completion of two courses of chemotherapy, and the analysis showed that those with CA125 ≤ 35U/mL had a 1-year survival rate of 96%, while those with CA125>35U/mL had a 1-year survival rate of 15% [14]. The half-life of CA125 is another widely reported indicator. In some studies, CA125 was regularly detected after surgery and chemotherapy in 225 patients with advanced ovarian cancer, and the complete remission rate of patients with serum CA125 half-life <25 d was found to be 3.6 times higher than that of patients with >25 d through analysis combined with the results of secondary exploration [15]. Therefore, continuous monitoring of CA125 is of great value for efficacy evaluation and prognosis analysis of ovarian cancer patients.
Novel prognostic factors
In order to develop a powerful predictive tool with both sensitivity and specificity to monitor ovarian cancer response to treatment, the research on prognostic biomarkers for ovarian cancer is continuously advancing.
Blood-based prognostic biomarkers
Blood test is minimally invasive, simple and easy to obtain specimens, and blood test results are widely used in clinic to assist the guidance of treatment. A variety of novel prognostic biomarkers derived from blood can provide a new tool for the clinical management of ovarian cancer. A total of 43 blood based biomarker studies met our selection criteria (Table 1), of which 13 were evaluated using ELISA methods for protein biomarkers [16,18-22,30-32,34,36,37,44]. PCR technology was used for detection of DNA or RNA source biomarkers [17,33,40-42,46,47,49,53,54,57]. The 41 novel prognostic biomarkers provided by 43 studies can be classified by biological function, including cell proliferation and invasion [16-22], inflammatory response [23-29], angiogenesis [30-33], antioxidant [34], immune response [35-39], chemotherapeutic sensitivity [40-44], mitosis process [45], EMT (epithelial-to-mesenchymal transformation) and metastasis [46,47], deregulation of the cellular transport [48] and apoptosis process [31]. The following are representative novel prognostic factors reported in the literature.
A large number of studies have shown that chronic inflammation is closely related to the occurrence and development of cancer, and a variety of inflammatory cells and inflammatory factors participate in and promote the proliferation, invasion and metastasis of tumor cells, and affect the prognosis of patients [310]. Neutrophils and lymphocytes are both important cells involved in the inflammatory response process. The changes in the number of them can directly reflect the degree of inflammatory response in the body. NLR (neutrophil to lymphocyte ratio) is an important biological indicator of systemic inflammatory response, which can be obtained by calculating the ratio after the complete blood count [311]. Previous studies have shown that elevated NLR is an independent prognostic risk factor for several malignant tumors, including ovarian cancer [312-314]. The study of Stanislaus Argeny et al. found that the non-specific inflammatory response in cancerous tissues would lead to changes in the level of peripheral blood cells, mainly manifested as an increase in NLR. Studies have shown that neutrophils can alter the tumor microenvironment by producing cytokines and chemokines, they also promote the transformation of normal cells into tumor cells by secreting substances like reactive oxygen species and proteases. Moreover, the migration and diffusion ability of tumor cells can be enhanced by secreting platelet activating factor, matrix metalloproteinase and other factors related to tumor cell metastasis. In addition, lymphocytes are important components of the immune system and play an important role in immune surveillance. The decreased number of lymphocytes indicates the weakened immunity of the body and the reduced monitoring and killing effect on tumor cells, which cannot effectively prevent the proliferation and migration of tumor cells. Therefore, an elevated preoperative NLR usually indicates a poor prognosis in ovarian cancer patients [315]. The study of Zhang H et al. suggested that NLR could be used to differentiate CA125-negative ovarian cancer and was superior to CA125 in predicting patients' overall survival (OS) and progression free survival (PFS) [316]. In addition, a multivariate analysis of clinical data in 165 initial treatment ovarian cancer patients also suggested that NLR is an independent prognostic factor for PFS and OS in ovarian cancer patients [28].
Alterations in energy metabolism are a decisive biochemical feature of tumor cells, in other words, abnormal activation of glycolytic pathway still exists in tumor cells even under the condition of sufficient oxygen supply, consume large amounts of glucose and eventually produce lactic acid in order to satisfy energy supply of malignant tumor cell proliferation, this phenomenon is called aerobic glycolysis of tumors, also known as the Warburg effect [317]. In the process of glycolysis of malignant tumors, there is an important catalytic enzyme, namely lactate dehydrogenase (LDH), which mainly catalyzes the exchange of pyruvate and lactic acid, and is highly expressed in hypoxic cells, especially in tumor cells. Compared with normal tissues, the levels of glycolysis in malignant tissues were higher, and the serum LDH level of patients increased with the progression of the disease, especially in the advanced stage of the tumor [318]. A study shows that the LDH levels at different stages and grades differed significantly in ovarian cancer, survival curves revealed that higher LDH expression was correlated with shorter survival (P<0.05). In addition, SATB1 may reprogram energy metabolism in ovarian cancer by regulating LDH and MCT1 levels to promote metastasis [319]. As another marker of tissue damage and inflammation, elevated serum LDH level can promote the proliferation, metastasis and development of cancer cells, which is commonly seen in a variety of malignant tumors [320,321]. A study showed that preoperative higher LDH levels were significantly associated with poor survival in patients with high grade serous ovarian cancer through survival analysis, serum high LDH levels are a promising prognostic biomarker [26].
Mesothelial protein (MSLN) is a cell surface glycoprotein, which was found by Chang et al. [322] and is usually only expressed in mesothelial tissue of body cavity. In recent years, MSLN as a differentiation antigen has been proved to be overexpressed in malignant pleural mesothelioma, pancreatic cancer, ovarian cancer and other malignant tumors, and may through increased synthesis of cyclinD1 and suppress the degradation and forming MSLN/MUC16 complex pathways involved in tumor cell proliferation, adhesion and transfer process, it is related to transcoelomic spread of ovarian cancer cells [323]. In addition, MSLN inhibits paclitaxel-induced apoptosis through serine and threonine kinase pathways, leading to chemotherapy resistance and seriously affecting the prognosis of patients [324]. The study of Karolina Okla et al. confirmed that plasma MSLN concentration in EOC patients was significantly higher than that in benign ovarian tumor patients and healthy women. Kaplan-Meier analysis results showed that, compared with low MSLN level, only high MSLN concentration of EOC patients before treatment was significantly correlated with a shorter 5-year OS (P=0.03), which predicted poor prognosis [21]. Another study showed that MSLN can enhance the invasion of ovarian cancer by inducing MMP-7 through MAPK/ERK and JNK pathways, blocking the MSLN-related pathway may be a potential strategy to improve the prognosis of ovarian cancer patients [325].
Aurora A kinase (AAK) is encoded by the Aurka gene and is a member of the serine/threonine kinase family. And as an important mitotic regulator, it can participate in many processes of cell mitosis and maintain chromosome division and spindle stability together with centrosomes [326]. Overexpression of Aurora A has been observed in a variety of malignant tumor types and plays an important regulatory role in the key control points of the tumorigenic transformation response through p53/TP53 phosphorylation [327]. Aurora A overexpression can also lead to abnormal amplification of centrosomes, leading to multilevel allocation and instability of chromosomes during division, and then to activation of oncogenes or inactivation of tumor suppressor genes [328]. Through gene chip screening and RT-PCR, the study of Hellleman et al. confirmed that Aurora A was overexpressed in ovarian cancer tissues that did not respond to platinum therapy, compared with ovarian cancer patients who responded to platinum therapy, and patients with overexpression of Aurora A had a poor prognosis [329]. A single nucleotide polymorphism in G169A at codon 57 of Aurora A locus leads to the substitution of valine by isoleucine, leading to the production of variant II. Kimura et al. [45] showed that AAK activity was reduced by the II variant, and the inhibited AAK could lead to cell death by affecting the mitosis process. Therefore, the change of single nucleotide polymorphisms in AAK may be a protective factor for cancer risk.
Galectin is an important member of the lectin superfamily, it is widely expressed in a variety of cell types and plays an important role in apoptosis, angiogenesis, cell migration, and tumor immune escape. Dysfunction or altered expression of galectin is associated with a variety of cancer types [330]. Galectin-8 and galectin-9 both have two carbohydrate recognition domains and are tandem repeat galactosins that regulate a variety of biological functions, including cell aggregation, cell adhesion, and tumor cell apoptosis [331]. Recent studies have shown that galectin-9 promotes CD8 + T cell failure and induces proliferation of myeloid inhibitory cells by binding to T cell immunoglobulin mucin 3 (Tim-3), thereby participating in immune escape of tumor cells [332]. In addition, the expression of galectin-8 in solid tumors has also been proved to be closely related to tumor cell adhesion or metastasis [333]. Labrie M et al. showed that plasma Gal-8 and Gal-9 levels were significantly increased in HGSOC patients compared to healthy controls, and higher plasma galectin-8 and galectin-9 levels were associated with a shorter 5-year disease-free survival (DFS) and 5-year OS (P=0.005), multivariate analysis further demonstrated that both plasma galectin-8 and galectin-9 could be promising biomarkers for poor prognosis in high grade serous ovarian cancer patients [171].
Angiogenesis plays an important role in tumor growth and metastasis. Neovascularization provides oxygen and nutrients to tumor cells, which can enhance cell proliferation and invasion ability [334]. Tumor tissue can secrete a variety of proangiogenic substances to induce and regulate angiogenesis, among which vascular endothelial growth factor (VEGF) is the primary stimulator of tumor angiogenesis. VEGF family members include VEGF-A, VEGF-B, VEGF-C, VEGF-D, etc. Among them, the biologic activity of VEGF-A is the most important, which can promote neovascularization and increase vascular permeability through VEGF/VEGFR (Vascular Endothelial Growth Factor Receptor) signaling pathway [335]. Previous studies have shown that VEGF-A is closely related to the occurrence and development of cancer and some inflammatory diseases [336]. Studies have investigated the efficacy of serum VEGF-A levels as prognostic markers in Epithelial ovarian cancer (EOC) patients, the experiment confirmed that the OS of patients with high VEGF-A level was significantly lower than that of patients with low VEGF-A level, and the difference was statistically significant (P=0.015). Moreover, the VEGF-A level of patients was correlated with FIGO stage. Multivariate analysis showed that serum VEGF-A could be an independent prognostic factor for OS of patients [32]. The study of Dobrzycka B et al. showed that serum VEGF level was significantly increased in patients with serous ovarian cancer (SOC) compared with healthy control group, and higher serum VEGF level was significantly correlated with poor prognosis, and multivariate analysis confirmed that serum VEGF level was an independent risk factor for prognosis [31].
MicroRNAs (miRNAs) are a class of single-stranded small RNAs encoded by endogenous genes, which regulate the expression of target genes by acting on target mRNA to promote its degradation or inhibit its translation [337]. MiRNAs are involved in the regulation of a variety of human life activities, and studies have found that miRNAs are closely related to the occurrence and development of a variety of malignant tumors [338,339]. At present, more than 50% miRNA genes have been located in tumor-related chromosomal rearrangement regions, which have important research and application values in the diagnosis, treatment and prognosis prediction of malignant tumors. EMT is closely related to tumor invasion and metastasis, many miRNAs have been proved to directly regulate the expression of epithelial markers and indirectly regulate EMT-related growth factor signaling pathways and transcription factors to affect the EMT process [340,341]. At present, miR-200 family is the most studied miRNA related to EMT process. Gregory et al. found that TGF- Beta/ZEB/miR-200 signaling pathway can regulate the transformation of cell epithelial-mesenchymal phenotype [342]. MiR-200c and miR-141 belong to the microRNA-200 family, Gao,Y.C. et al. evaluated the value of these two miRNAs as novel prognostic biomarkers for ovarian cancer. Studies have shown that the expression levels of serum miR-200c and miR-141 in ovarian cancer patients are significantly increased compared with the normal control group, and the expression levels of the two miRNAs are correlated with different stages and pathological subtypes of ovarian cancer. Survival analysis showed that compared with the group with high serum miR-200c expression, the overall survival rate of the group with low serum miR-200c expression was significantly reduced. This is similar to the analysis results of different miR-141 expression groups, so both miR-200c and miR-141 are likely to be promising prognostic biomarkers for ovarian cancer [49]. Another study compared the expression levels of miR-200a, miR-200b and miR-200c in blood samples from 70 EOC patients and healthy controls, the results showed that these three miRNAs were significantly higher expressed in serum samples from EOC patients compared to normal controls, statistical analysis confirmed that the high expression of miR-200a, miR200b and miR-200c was significantly correlated with tumor histological subtypes, stages and lymph node metastasis, and all of them could be used as reliable indicators for predicting the prognosis of patients with EOC [46].
Tissue-based prognostic biomarkers
The overwhelming majority of selected biomarker studies investigated different tissue-based biomarkers using a variety of technical research methods. The selected tissue prognostic biomarkers can be divided into immunohistochemical biomarkers (68.77%) [59-232], DNA biomarkers (3.95%) [159,233-241] and RNA biomarkers (27.28%) [242-309]. The prognostic value of 172 protein biomarkers was evaluated by immunohistochemistry in 174 studies (Table 2). These markers are classified according to their biological functions, mainly including such functional pathways as EMT and metastasis [59-71], inflammation and immunity [72-84], antioxidant [85,86], angiogenesis [87-99], cell proliferation, migration and invasion [100-116], chemotherapeutic sensitivity [117-197] and cell cycle regulation [198-201]. The remaining 79 studies of prognostic biomarkers were based on genomic DNA or RNA (Tables 3-4), involving different functional pathways in the progression of ovarian cancer, such as gene locus methylation [159,233-235], mutation status [237,238], gene polymorphism [240,241] and the expression of non-coding RNA during cancer cell proliferation, migration and invasion [242-282].
As a new type of anti-tumor effector lymphocytes with potential therapeutic value, the correlation between TIL and patient prognosis and survival has been widely concerned. Through systematic literature retrieval, we determined that TIL is a promising prognostic biomarker, and its level can be detected by immunohistochemistry. TIL can be classified by function and location in the tumor tissue, which is generally associated with better prognosis and survival, in which the presence of CD8+ T cells is positively correlated with survival [343,344]. The presence of TIL in a variety of tumor types, including metastatic melanoma, breast cancer, colorectal cancer, and ovarian cancer, has been found to be significantly correlated with patient clinical outcomes and is an important positive prognostic factor [345-349]. There is evidence that ovarian cancer patients are usually accompanied by systemic immunosuppression. In contrast, patients with a stronger immune response have improved survival and respond better to chemotherapy [350]. Mauricio P et al. [81] evaluated TIL as a prognostic survival indicator for a group of HGSOC patients, and examined the expression of matrix and intraepithelial TIL (CD4+ and CD8+) in tissue samples. Multivariate analysis showed that intraepithelial CD4+ TIL infiltration was associated with better PFS and OS, intraepithelial CD8+ TIL infiltration was only associated with better PFS. This confirms previous studies that ovarian cancer patients with high infiltration of CD4+ and CD8+ TIL have better prognosis. As a new method for the treatment of ovarian cancer, the potential value of targeted immunotherapy is an important research direction, which can be used to guide clinical practice, reduce recurrence and improve the long-term survival rate of patients.
Mitochondrial superoxide dismutase (MnSOD or SOD2) is the most important antioxidant enzyme in mitochondria, which protects cells from oxidative damage induced by reactive oxygen species (ROS) and lipid peroxidation by converting endogenous superoxide to hydrogen peroxide [351]. Studies have demonstrated that SOD2 overexpression can enhance the invasion and metastasis of tumor cells by increasing the expression of matrix metalloproteinases (MMP) family members or activating Redox sensitive signaling pathways [352]. New evidence suggests that inhibition of SOD2 activity in tumor cells leads to increased apoptosis, inhibition of proliferation and increased sensitivity to chemotherapeutics [353]. There is growing evidence that SOD2 overexpression is associated with poor prognosis in a variety of cancer types, including renal clear cell carcinoma and ovarian cancer [354-356]. A study based on SOD2 immunohistochemical staining confirmed the correlation between SOD2 expression and patient prognosis in the endometriosis-associated ovarian cancer (EAOC) case group. Kaplan-Meier analysis showed that high SOD2 expression was associated with shorter PFS (P=0.0669) and poorer OS (P=0.0405), and increased SOD2 expression was a predictive biomarker for poor prognosis in EAOC [86].
Genome-wide analysis has confirmed that epigenetic changes are common events in many cancers, cellular genomic epigenetic disorders are important causes of many diseases, including cancer and autoimmune diseases. Epigenetic changes in human malignancies mainly include DNA methylation, nucleosomal remodeling histone modification and non-coding RNA dysregulation [357]. Numerous studies have confirmed that abnormal methylation of multiple genes involved in DNA repair, Akt /mTOR, Redox response, apoptosis, cell adhesion and cancer stem cell signaling pathways are associated with poor prognosis in ovarian cancer patients [358]. Mase et al. [235] confirmed that the DNA methylation status of ZNF671 was closely related to the recurrence and prognosis of patients with serous ovarian cancer. Multiple analysis methods combined showed that the methylation status of ZNF671 was an independent factor to predict the early recurrence of patients and patients with DNA methylation of ZNF671 had poor prognosis (P<0.05). A subsequent study validated the prognostic significance of HS3ST2 methylation in patients with advanced EOC in three separate dataset of TSGH, AOCS, and TCGA, studies have confirmed that HS3ST2 inhibits the malignant phenotype of ovarian cancer by interfering with various carcinogenic ligand signals, such as IL-6, FGF2 and EGF, and patients with low HS3ST2 expression accompanied by high expression of carcinogenic cytokines or growth factors have the worst prognosis [159]. In conclusion, abnormal DNA methylation in tumor cells can be used as an effective prognostic marker for ovarian cancer. Non-coding RNA is an important part of epigenetic changes, among which long non-coding RNA (lncRNA) is an emerging regulatory RNA that is involved in the regulation of a variety of physiological and pathological processes and is abnormally expressed in a variety of types of cancers. It has been reported that the differential expression of lncRNA in ovarian cancer, lung cancer, gastric cancer and liver cancer is related to the prognosis of patients [359]. Cao Y et al. [265] confirmed that the expression of lncRNA CCAT1 was up-regulated in EOC tissues, and the high expression of lncRNA CCAT1 could promote the process of EMT of EOC cells, and enhance the migration and invasion ability of cells. Furthermore, high lncRNA CCAT1 expression was associated with FIGO stage, histological grade, lymph node metastasis and poor survival. Multivariate cox regression analysis showed that CCAT1 expression was an independent prognostic factor. In addition, it has been demonstrated that silencing of lncRNA CCAT2 in cancer cells significantly inhibits cell proliferation, migration and invasion through the Wnt/β-catenin signaling pathway, and the results of subsequent survival analysis showed that high CCAT2 expression was associated with shorter OS or DFS, cox proportional risk regression model analysis showed that CCAT2 expression level was an independent prognostic indicator for overall survival, and these data results confirmed that lncRNA CCAT2 was a reliable prognostic marker for ovarian cancer [269].
Conclusion
Ovarian cancer is the most fatal gynecological malignancy with high incidence and low survival rate. By exploring the prognostic biomarkers associated with ovarian cancer recurrence and progression, independent risk factors affecting patient prognosis were identified, which laid a solid foundation for the development of novel treatment strategies and the improvement of patient treatment outcomes. This review searched the literature and database for the relevant reports on prognostic biomarkers of ovarian cancer, reviewed the classic clinical prognostic biomarkers, and focused on the recently discovered various prognostic markers. Advances in genomics, proteomics and metabolomics have provided favorable conditions for the discovery of novel prognostic biomarkers that have identified a variety of promising prognostic biomarkers, including miRNA, lncRNA and TIL, these biomarkers can affect the prognosis of patients through a variety of biological functional pathways. TCGA data sets and public databases can provide data information for large patient cohort genome studies, the application of bioinformatics modeling and high-throughput molecular analysis techniques has greatly enriched the knowledge related to biological processes such as cancer progression. The prognostic value of a variety of novel biomarkers was evaluated by integrating genomic, proteomic and metabolomic data and clinical information with a multivariate analysis model. The effectiveness of these novel prognostic biomarkers still needs to be further validated in large clinical trials. By studying the functional pathways of regulation of these molecular markers, the potential molecular mechanisms are revealed, so as to identify new therapeutic targets. This is a high-precision medical method, which may promote personalized treatment of ovarian cancer patients and improve their prognosis.
Supplementary Material
Supplementary materials.
Acknowledgements
Contributions
Shuna Liu and Ming Wu did the literature search and analysed and interpreted data. Shuna Liu wrote the manuscript. Ming Wu prepared the Tables and Figures. Fang Wang designed and supervised the study. We both reviewed and approved the final manuscript before submission.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020Jan;70(1):7-30
2. Tian F, Jia L, Chu Z, Han H, Zhang Y, Cai J. MicroRNA-519a inhibits the proliferation and promotes the apoptosis of ovarian cancer cells through targeting signal transducer and activator of transcription 3. Exp Ther Med. 2018;15(2):1819-1824
3. BAKER Vicki V. Treatment Options for Ovarian Cancer. Clinical Obstetrics & Gynecology. 2001;44(3):522-530
4. Clark T G, Stewart M E, Altman D G. et al. A prognostic model for ovarian cancer. British Journal of Cancer. 2001;85(7):944-952
5. Zheng Q, Wang P, Hui R. et al. Cox regression analysis of prognostic factors in ovarian cancer patients. Cancer. 2009;16(02):99-102
6. Jing S, Chen Z, Xie F. et al. Analysis of prognostic factors of 57 cases of ovarian cancer. Chinese Journal of Anatomy and Clinical Sciences. 2013;18(2):136-138
7. Parmar MK, Ledermann JA, Colombo N. et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361(9375):2099-2106
8. Bertelsen K, Grenman S, Rustin GJ. How long should first-line chemotherapy continue? Ann Oncol. 1999;10(Suppl 1):17-20
9. Liu C, LI Li, Zhao B. et al. Ⅲ C ~ Ⅳ stage epithelial ovarian cancer prognosis factors analysis. International Journal of Gynecology & Obstetrics. 2018;45(06):50-53 +59
10. Bast RC Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68(5):1331-1337
11. Bell J, Brady MF, Young RC. et al. Randomized phase III trial of three versus six cycles of adjuvant carboplatin and paclitaxel in early stage epithelial ovarian carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2006;102(3):432-439
12. Zurawski VR Jr, Orjaseter H, Andersen A, Jellum E. Elevated serum CA 125 levels prior to diagnosis of ovarian neoplasia: relevance for early detection of ovarian cancer. Int J Cancer. 1988;42(5):677-680
13. Redman CW, Blackledge GR, Kelly K, Powell J, Buxton EJ, Luesley DM. Early serum CA125 response and outcome in epithelial ovarian cancer. Eur J Cancer. 1990;26(5):593-596
14. Gadducci A, Zola P, Landoni F. et al. Serum half-life of CA 125 during early chemotherapy as an independent prognostic variable for patients with advanced epithelial ovarian cancer: results of a multicentric Italian study. Gynecol Oncol. 1995;58(1):42-47
15. Kato S, Abarzua-Catalan L, Trigo C. et al. Leptin stimulates migration and invasion and maintains cancer stem-like properties in ovarian cancer cells: an explanation for poor outcomes in obese women. Oncotarget. 2015;6(25):21100-21119
16. Lyu N, Wang Y, Wang J, Zhang Z, Kong W. Study on early diagnosis of epithelial ovarian cancer by analysis of plasma septin-9 and clusterin level. J Cancer Res Ther. 2018;14(Supplement):S444-S449
17. Meng X, Joosse SA, Müller V. et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113(9):1358-1366
18. Cheon DJ, Li AJ, Beach JA. et al. ADAM12 is a prognostic factor associated with an aggressive molecular subtype of high-grade serous ovarian carcinoma. Carcinogenesis. 2015;36(7):739-747
19. Lyu N, Wang Y, Wang J, Zhang Z, Kong W. Study on early diagnosis of epithelial ovarian cancer by analysis of plasma septin-9 and clusterin level. J Cancer Res Ther. 2018;14(Supplement):S444-S449
20. Cymbaluk-Płoska A, Chudecka-Głaz A, Pius-Sadowska E, Machaliński B, Menkiszak J, Sompolska-Rzechuła A. Suitability assessment of baseline concentration of MMP3, TIMP3, HE4 and CA125 in the serum of patients with ovarian cancer. J Ovarian Res. 2018;11(1):1
21. Okła K, Surówka J, Frąszczak K. et al. Assessment of the clinicopathological relevance of mesothelin level in plasma, peritoneal fluid, and tumor tissue of epithelial ovarian cancer patients. Tumour Biol. 2018;40(10):1010428318804937
22. Jin C, Yang M, Han X. et al. Evaluation of the value of preoperative CYFRA21-1 in the diagnosis and prognosis of epithelial ovarian cancer in conjunction with CA125. J Ovarian Res. 2019;12(1):114
23. Feng Z, Wen H, Bi R. et al. Preoperative Neutrophil-to-Lymphocyte Ratio as a Predictive and Prognostic Factor for High-Grade Serous Ovarian Cancer. PLoS One. 2016;11(5):e0156101
24. Li Z, Hong N, Robertson M, Wang C, Jiang G. Preoperative red cell distribution width and neutrophil-to-lymphocyte ratio predict survival in patients with epithelial ovarian cancer. Sci Rep. 2017;7:43001
25. Liu Y, Chen S, Zheng C. et al. The prognostic value of the preoperative c-reactive protein/albumin ratio in ovarian cancer. BMC Cancer. 2017;17(1):285
26. Pinto MP, Balmaceda C, Bravo ML. et al. Patient inflammatory status and CD4+/CD8+ intraepithelial tumor lymphocyte infiltration are predictors of outcomes in high-grade serous ovarian cancer. Gynecol Oncol. 2018;151(1):10-17
27. Yu W, Ye Z, Fang X, Jiang X, Jiang Y. Preoperative albumin-to-fibrinogen ratio predicts chemotherapy resistance and prognosis in patients with advanced epithelial ovarian cancer. J Ovarian Res. 2019;12(1):88
28. Ceran MU, Tasdemir U, Colak E, Güngör T. Can complete blood count inflammatory parameters in epithelial ovarian cancer contribute to prognosis? - a survival analysis. J Ovarian Res. 2019;12(1):16
29. Nomelini RS, Carrijo Chiovato AF, Abdulmassih FBF, da Silva RC, Tavares-Murta BM, Murta EFC. Neutrophil-to-lymphocyte ratio and platelet count as prognostic factors in ovarian malignancies. J Cancer Res Ther. 2019;15(6):1226-1230
30. Chen J, Liu Z, Fang S. et al. Fibulin-4 is associated with tumor progression and a poor prognosis in ovarian carcinomas. BMC Cancer. 2015;15:91
31. Dobrzycka B, Mackowiak-Matejczyk B, Terlikowska KM, Kulesza-Bronczyk B, Kinalski M, Terlikowski SJ. Prognostic significance of pretreatment VEGF, survivin, and Smac/DIABLO serum levels in patients with serous ovarian carcinoma. Tumour Biol. 2015;36(6):4157-4165
32. Komatsu H, Oishi T, Itamochi H. et al. Serum Vascular Endothelial Growth Factor-A as a Prognostic Biomarker for Epithelial Ovarian Cancer. Int J Gynecol Cancer. 2017;27(7):1325-1332
33. Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY, Hua KQ. Exosomal Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes Angiogenesis and Predicts Poor Prognosis in Epithelial Ovarian Cancer. Int J Biol Sci. 2018;14(14):1960-1973
34. Pylväs-Eerola M, Karihtala P, Puistola U. Preoperative serum 8-hydroxydeoxyguanosine is associated with chemoresistance and is a powerful prognostic factor in endometrioid-type epithelial ovarian cancer. BMC Cancer. 2015;15:493
35. Hao CJ, Li J, Liu P. et al. Effects of the balance between type 1 and type 2 T helper cells on ovarian cancer. Genet Mol Res. 2016;15(2):10.4238 /gmr.15027936
36. Chatterjee J, Dai W, Aziz NHA. et al. Clinical Use of Programmed Cell Death-1 and Its Ligand Expression as Discriminatory and Predictive Markers in Ovarian Cancer. Clin Cancer Res. 2017;23(13):3453-3460
37. De La Motte Rouge T, Corné J, Cauchois A. et al. Serum CD95L Level Correlates with Tumor Immune Infiltration and Is a Positive Prognostic Marker for Advanced High-Grade Serous Ovarian Cancer. Mol Cancer Res. 2019;17(12):2537-2548
38. Lee YJ, Chung YS, Lee JY. et al. Pretreatment lymphocytopenia is an adverse prognostic biomarker in advanced-stage ovarian cancer. Cancer Med. 2019;8(2):564-571
39. Waki K, Kawano K, Tsuda N, Komatsu N, Yamada A. CD4/CD8 ratio is a prognostic factor in IgG nonresponders among peptide vaccine-treated ovarian cancer patients. Cancer Sci. 2020;111(4):1124-1131
40. Konopka B, Szafron LM, Kwiatkowska E. et al. The significance of c.690G>T polymorphism (rs34529039) and expression of the CEBPA gene in ovarian cancer outcome. Oncotarget. 2016;7(41):67412-67424
41. Feng Z, Wen H, Bi R, Duan Y, Yang W, Wu X. Thrombocytosis and hyperfibrinogenemia are predictive factors of clinical outcomes in high-grade serous ovarian cancer patients. BMC Cancer. 2016;16:43
42. Chebouti I, Kuhlmann JD, Buderath P. et al. ERCC1-expressing circulating tumor cells as a potential diagnostic tool for monitoring response to platinum-based chemotherapy and for predicting post-therapeutic outcome of ovarian cancer. Oncotarget. 2017;8(15):24303-24313
43. Fukagawa S, Miyata K, Yotsumoto F. et al. MicroRNA-135a-3p as a promising biomarker and nucleic acid therapeutic agent for ovarian cancer. Cancer Sci. 2017;108(5):886-896
44. Labrie M, De Araujo LOF, Communal L, Mes-Masson AM, St-Pierre Y. Tissue and plasma levels of galectins in patients with high grade serous ovarian carcinoma as new predictive biomarkers. Sci Rep. 2017;7(1):13244
45. Niu H, Shin H, Gao F. et al. Aurora A Functional Single Nucleotide Polymorphism (SNP) Correlates With Clinical Outcome in Patients With Advanced Solid Tumors Treated With Alisertib, an Investigational Aurora A Kinase Inhibitor. EBioMedicine. 2017;25:50-57
46. Zuberi M, Mir R, Das J. et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17(10):779-787
47. Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923-16935
48. Huang L, Zhou Y, Cao XP. et al. KPNA2 is a potential diagnostic serum biomarker for epithelial ovarian cancer and correlates with poor prognosis. Tumour Biol. 2017;39(6):1010428317706289
49. Gao YC, Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015;36(6):4843-4850
50. Chen Y, Zhang L, Liu WX, Liu XY. Prognostic significance of preoperative anemia, leukocytosis and thrombocytosis in chinese women with epithelial ovarian cancer. Asian Pac J Cancer Prev. 2015;16(3):933-939
51. Kurosaki A, Hasegawa K, Kato T. et al. Serum folate receptor alpha as a biomarker for ovarian cancer: Implications for diagnosis, prognosis and predicting its local tumor expression. Int J Cancer. 2016;138(8):1994-2002
52. Živný JH, Leahomschi S, Klener P Jr, Živný J, Haluzík M, Cibula D. Comparison of Plasma Osteopontin Levels between Patients with Borderline Ovarian Tumours and Serous Ovarian Carcinoma. Folia Biol (Praha). 2016;62(6):258-262
53. Zuberi M, Khan I, Mir R, Gandhi G, Ray PC, Saxena A. Utility of Serum miR-125b as a Diagnostic and Prognostic Indicator and Its Alliance with a Panel of Tumor Suppressor Genes in Epithelial Ovarian Cancer. PLoS One. 2016;11(4):e0153902
54. Zhu T, Gao W, Chen X. et al. A Pilot Study of Circulating MicroRNA-125b as a Diagnostic and Prognostic Biomarker for Epithelial Ovarian Cancer. Int J Gynecol Cancer. 2017;27(1):3-10
55. Montavon Sartorius C, Schoetzau A, Kettelhack H. et al. ABO blood groups as a prognostic factor for recurrence in ovarian and vulvar cancer. PLoS One. 2018;13(3):e0195213
56. He W, Zhang P, Ye M. et al. Polymorphisms of the Ras-Association Domain Family 1 Isoform A (RASSF1A) Gene are Associated with Ovarian Cancer, and with the Prognostic Factors of Grade and Stage, in Women in Southern China. Med Sci Monit. 2018;24:2360-2367
57. Link T, Kuhlmann JD, Kobelt D. et al. Clinical relevance of circulating MACC1 and S100A4 transcripts for ovarian cancer. Mol Oncol. 2019;13(5):1268-1279
58. Lu X, Li Y, Xia B. et al. Selection of small plasma peptides for the auxiliary diagnosis and prognosis of epithelial ovarian cancer by using UPLC/MS-based nontargeted and targeted analyses. Int J Cancer. 2019;144(8):2033-2042
59. Hou M, Cheng Z, Shen H. et al. High expression of CTHRC1 promotes EMT of epithelial ovarian cancer (EOC) and is associated with poor prognosis. Oncotarget. 2015;6(34):35813-35829
60. Prislei S, Martinelli E, Zannoni GF. et al. Role and prognostic significance of the epithelial-mesenchymal transition factor ZEB2 in ovarian cancer. Oncotarget. 2015;6(22):18966-18979
61. Tjhay F, Motohara T, Tayama S. et al. CD44 variant 6 is correlated with peritoneal dissemination and poor prognosis in patients with advanced epithelial ovarian cancer. Cancer Sci. 2015;106(10):1421-1428
62. Sun Y, Hu L, Zheng H. et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol. 2015;235(1):25-36
63. Kwon M, Kim JH, Rybak Y. et al. Reduced expression of FILIP1L, a novel WNT pathway inhibitor, is associated with poor survival, progression and chemoresistance in ovarian cancer. Oncotarget. 2016;7(47):77052-77070
64. Nakamura H, Nagasaka K, Kawana K. et al. Expression of Par3 polarity protein correlates with poor prognosis in ovarian cancer. BMC Cancer. 2016;16(1):897
65. Vos MC, Hollemans E, Ezendam N. et al. MMP-14 and CD44 in Epithelial-to-Mesenchymal Transition (EMT) in ovarian cancer. J Ovarian Res. 2016;9(1):53
66. Wang Y, Zhou X, Xu M. et al. OTUB1-catalyzed deubiquitination of FOXM1 facilitates tumor progression and predicts a poor prognosis in ovarian cancer. Oncotarget. 2016;7(24):36681-36697
67. Chen L, Yao Y, Sun L. et al. Snail Driving Alternative Splicing of CD44 by ESRP1 Enhances Invasion and Migration in Epithelial Ovarian Cancer. Cell Physiol Biochem. 2017;43(6):2489-2504
68. Chen Y, Wang DD, Wu YP. et al. MDM2 promotes epithelial-mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br J Cancer. 2017;117(8):1192-1201
69. Nakamura K, Terai Y, Tanabe A. et al. CD24 expression is a marker for predicting clinical outcome and regulates the epithelial-mesenchymal transition in ovarian cancer via both the Akt and ERK pathways. Oncol Rep. 2017;37(6):3189-3200
70. Xu Y, Zhang Q, Miao C. et al. CCNG1 (Cyclin G1) regulation by mutant-P53 via induction of Notch3 expression promotes high-grade serous ovarian cancer (HGSOC) tumorigenesis and progression. Cancer Med. 2019;8(1):351-362
71. Ramalho S, Andrade LAA, Filho CC. et al. Role of discoidin domain receptor 2 (DDR2) and microRNA-182 in survival of women with high-grade serous ovarian cancer. Tumour Biol. 2019;41(1):1010428318823988
72. Knutson KL, Maurer MJ, Preston CC. et al. Regulatory T cells, inherited variation, and clinical outcome in epithelial ovarian cancer. Cancer Immunol Immunother. 2015;64(12):1495-1504
73. Darb-Esfahani S, Kunze CA, Kulbe H. et al. Prognostic impact of programmed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor-infiltrating lymphocytes in ovarian high grade serous carcinoma. Oncotarget. 2016;7(2):1486-1499
74. Lundgren S, Berntsson J, Nodin B, Micke P, Jirström K. Prognostic impact of tumour-associated B cells and plasma cells in epithelial ovarian cancer. J Ovarian Res. 2016;9:21
75. James FR, Jiminez-Linan M, Alsop J. et al. Association between tumour infiltrating lymphocytes, histotype and clinical outcome in epithelial ovarian cancer. BMC Cancer. 2017;17(1):657
76. Xu Y, Chen L, Xu B. et al. Higher Numbers of T-Bet+ Tumor-Infiltrating Lymphocytes Associate with Better Survival in Human Epithelial Ovarian Cancer. Cell Physiol Biochem. 2017;41(2):475-483
77. Zhu J, Wen H, Bi R, Wu Y, Wu X. Prognostic value of programmed death-ligand 1 (PD-L1) expression in ovarian clear cell carcinoma. J Gynecol Oncol. 2017;28(6):e77
78. Carter JH, Deddens JA, Mueller G. et al. Transcription factors WT1 and p53 combined: a prognostic biomarker in ovarian cancer. Br J Cancer. 2018;119(4):462-470
79. Nakagawa S, Serada S, Kakubari R. et al. Intratumoral Delivery of an Adenoviral Vector Carrying the SOCS-1 Gene Enhances T-Cell-Mediated Antitumor Immunity By Suppressing PD-L1. Mol Cancer Ther. 2018;17(9):1941-1950
80. Kim KH, Choi KU, Kim A. et al. PD-L1 expression on stromal tumor-infiltrating lymphocytes is a favorable prognostic factor in ovarian serous carcinoma. J Ovarian Res. 2019;12(1):56
81. Martinelli-Kläy CP, Lombardi T, Mendis B. et al. Tissue eosinophilia in oral intraepithelial neoplasia as a probable indicator of invasion. Oral Dis. 2018;24(1-2):103-108
82. Szubert S, Jozwicki W, Wicherek L, Koper K. Cytoplasmic and membranous receptor-binding cancer antigens expressed on SiSo cells (RCAS1) immunoreactivity in epithelial ovarian cancer cells represent differing biological function of RCAS1. Folia Histochem Cytobiol. 2019;57(3):116-126
83. Zong L, Zhou Y, Zhang M, Chen J, Xiang Y. VISTA expression is associated with a favorable prognosis in patients with high-grade serous ovarian cancer. Cancer Immunol Immunother. 2020;69(1):33-42
84. Jäntti T, Luhtala S, Mäenpää J, Staff S. Characterization of immunoreactivity with whole-slide imaging and digital analysis in high-grade serous ovarian cancer. Tumour Biol. 2020;42(11):1010428320971404
85. Liew PL, Hsu CS, Liu WM, Lee YC, Lee YC, Chen CL. Prognostic and predictive values of Nrf2, Keap1, p16 and E-cadherin expression in ovarian epithelial carcinoma. Int J Clin Exp Pathol. 2015;8(5):5642-5649
86. Amano T, Chano T, Isono T, Kimura F, Kushima R, Murakami T. Abundance of mitochondrial superoxide dismutase is a negative predictive biomarker for endometriosis-associated ovarian cancers. World J Surg Oncol. 2019;17(1):24
87. Kinose Y, Sawada K, Makino H. et al. IKKβ Regulates VEGF Expression and Is a Potential Therapeutic Target for Ovarian Cancer as an Antiangiogenic Treatment. Mol Cancer Ther. 2015;14(4):909-919
88. Corvigno S, Wisman GB, Mezheyeuski A. et al. Markers of fibroblast-rich tumor stroma and perivascular cells in serous ovarian cancer: Inter- and intra-patient heterogeneity and impact on survival. Oncotarget. 2016;7(14):18573-18584
89. Skirnisdottir I, Seidal T, Åkerud H. The relationship of the angiogenesis regulators VEGF-A, VEGF-R1 and VEGF-R2 to p53 status and prognostic factors in epithelial ovarian carcinoma in FIGO-stages I-II. Int J Oncol. 2016;48(3):998-1006
90. Onisim A, Iancu M, Vlad C. et al. Expression of Nestin and CD133 in serous ovarian carcinoma. J BUON. 2016;21(5):1168-1175
91. Huang B, Yin M, Li X. et al. Migration-inducing gene 7 promotes tumorigenesis and angiogenesis and independently predicts poor prognosis of epithelial ovarian cancer. Oncotarget. 2016;7(19):27552-27566
92. Shen W, Li HL, Liu L, Cheng JX. Expression levels of PTEN, HIF-1α, and VEGF as prognostic factors in ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2596-2603
93. Yu X, Wang Y, Qiu H. et al. AEG-1 Contributes to Metastasis in Hypoxia-Related Ovarian Cancer by Modulating the HIF-1alpha/NF-kappaB/VEGF Pathway. Biomed Res Int. 2018;2018:3145689
94. Chen Y, Zhang L, Liu WX, Wang K. VEGF and SEMA4D have synergistic effects on the promotion of angiogenesis in epithelial ovarian cancer. Cell Mol Biol Lett. 2018;23:2
95. Yang Z, Shen H, He W. et al. Expression of TBC1D16 Is Associated with Favorable Prognosis of Epithelial Ovarian Cancer. Tohoku J Exp Med. 2018;245(3):141-148
96. Meng Q, Duan P, Li L, Miao Y. Expression of Placenta Growth Factor Is Associated with Unfavorable Prognosis of Advanced-Stage Serous Ovarian Cancer. Tohoku J Exp Med. 2018;244(4):291-296
97. Sopo M, Anttila M, Hämäläinen K. et al. Expression profiles of VEGF-A, VEGF-D and VEGFR1 are higher in distant metastases than in matched primary high grade epithelial ovarian cancer. BMC Cancer. 2019;19(1):584
98. Yu L, Mao X, Jiao Y, Song W, Wang D. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2019;44(12):1344-1352.
99. Sopo M, Sallinen H, Hämäläinen K. et al. High expression of Tie-2 predicts poor prognosis in primary high grade serous ovarian cancer. PLoS One. 2020;15(11):e0241484
100. Cai Y, Wang J, Zhang L. et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Med Oncol. 2015;32(1):391. Turcotte M, Spring K, Pommey S, et al. CD73 is associated with poor prognosis in high-grade serous ovarian cancer. Cancer Res. 2015;75(21):4494-4503
101. Mehner C, Oberg AL, Kalli KR. et al. Serine protease inhibitor Kazal type 1 (SPINK1) drives proliferation and anoikis resistance in a subset of ovarian cancers. Oncotarget. 2015;6(34):35737-35754
102. Zhao H, Guo E, Hu T. et al. KCNN4 and S100A14 act as predictors of recurrence in optimally debulked patients with serous ovarian cancer. Oncotarget. 2016;7(28):43924-43938
103. Xu L, Wu H, Jiang C. et al. Dacomitinib, a new pan-EGFR inhibitor, is effective in killing ovarian cancer cells. Discov Med. 2016;22(122):297-309
104. Hu L, Liu R. Expression of Gab1 Is Associated with Poor Prognosis of Patients with Epithelial Ovarian Cancer. Tohoku J Exp Med. 2016;239(3):177-184
105. Chang L, Guo R, Yuan Z. IL-36α suppresses proliferation of ovarian cancer cells. Tumour Biol. 2017;39(6):1010428317706918
106. Zhang X, Liu D, Li M. et al. Prognostic and therapeutic value of disruptor of telomeric silencing-1-like (DOT1L) expression in patients with ovarian cancer. J Hematol Oncol. 2017;10(1):29
107. Ricciardelli C, Lokman NA, Pyragius CE. et al. Keratin 5 overexpression is associated with serous ovarian cancer recurrence and chemotherapy resistance. Oncotarget. 2017;8(11):17819-17832
108. Hiramatsu K, Serada S, Enomoto T. et al. LSR Antibody Therapy Inhibits Ovarian Epithelial Tumor Growth by Inhibiting Lipid Uptake. Cancer Res. 2018;78(2):516-527
109. Choi CH, Kang TH, Song JS. et al. Elevated expression of pancreatic adenocarcinoma upregulated factor (PAUF) is associated with poor prognosis and chemoresistance in epithelial ovarian cancer. Sci Rep. 2018;8(1):12161
110. Cao Y, Yu Y, Chen X. et al. Low Expression of Protocadherin-8 Promotes the Progression of Ovarian Cancer. Int J Gynecol Cancer. 2018;28(2):346-354
111. Liu YB, Mei Y, Tian ZW, Long J, Luo CH, Zhou HH. Downregulation of RIF1 Enhances Sensitivity to Platinum-Based Chemotherapy in Epithelial Ovarian Cancer (EOC) by Regulating Nucleotide Excision Repair (NER) Pathway. Cell Physiol Biochem. 2018;46(5):1971-1984
112. Li M, Qian Z, Ma X. et al. MiR-628-5p decreases the tumorigenicity of epithelial ovarian cancer cells by targeting at FGFR2. Biochem Biophys Res Commun. 2018;495(2):2085-2091
113. Han GH, Chay DB, Nam S, Cho H, Chung JY, Kim JH. Prognostic implications of forkhead box protein O1 (FOXO1) and paired box 3 (PAX3) in epithelial ovarian cancer. BMC Cancer. 2019;19(1):1202
114. Li H, Qian Y, Wang X, Pi R, Zhao X, Wei X. Targeted activation of Stat3 in combination with paclitaxel results in increased apoptosis in epithelial ovarian cancer cells and a reduced tumour burden. Cell Prolif. 2020;53(1):e12719
115. Liu Q, Liu H, Li L. et al. ATAD2 predicts poor outcomes in patients with ovarian cancer and is a marker of proliferation. Int J Oncol. 2020;56(1):219-231
116. Ye Q, Zhai X, Wang W. et al. Overexpression of Growth-Related Oncogene-β Is Associated with Tumorigenesis, Metastasis, and Poor Prognosis in Ovarian Cancer. Dis Markers. 2015;2015:387382
117. Zhou Y, Xu Y, Chen L, Xu B, Wu C, Jiang J. B7-H6 expression correlates with cancer progression and patient's survival in human ovarian cancer. Int J Clin Exp Pathol. 2015;8(8):9428-9433
118. Yu L, Jiao YJ, Zhou L, Song WQ, Wu SW, Wang DN. Nan Fang Yi Ke Da Xue Xue Bao. 2016;37(4):444-450.
119. Liu X, Xu Y, Jin Q. et al. EphA8 is a prognostic marker for epithelial ovarian cancer. Oncotarget. 2016;7(15):20801-20809
120. Zhang Q, Yu S, Lam MMT. et al. Angiotensin II promotes ovarian cancer spheroid formation and metastasis by upregulation of lipid desaturation and suppression of endoplasmic reticulum stress. J Exp Clin Cancer Res. 2019;38(1):116
121. Ma Z, Wang X, He J, Xia J, Li Y. Increased expression of protein kinase CK2α correlates with poor patient prognosis in epithelial ovarian cancer. PLoS One. 2017;12(3):e0174037
122. Zhang W, Niu C, He W. et al. Upregulation of centrosomal protein 55 is associated with unfavorable prognosis and tumor invasion in epithelial ovarian carcinoma. Tumour Biol. 2016;37(5):6239-6254
123. Manai M, Doghri R, Finetti P. et al. Overexpression of Annexin A1 Is an Independent Predictor of Longer Overall Survival in Epithelial Ovarian Cancer. In Vivo. 2020;34(1):177-184
124. Gruosso T, Garnier C, Abelanet S. et al. MAP3K8/TPL-2/COT is a potential predictive marker for MEK inhibitor treatment in high-grade serous ovarian carcinomas. Nat Commun. 2015;6:8583
125. Tong X, Barbour M, Hou K. et al. Interleukin-33 predicts poor prognosis and promotes ovarian cancer cell growth and metastasis through regulating ERK and JNK signaling pathways. Mol Oncol. 2016;10(1):113-125
126. Vlad C, Kubelac P, Onisim A. et al. Expression of CDCP1 and ADAM12 in the ovarian cancer microenvironment. J BUON. 2016;21(4):973-978
127. Tai H, Wu Z, Sun S, Zhang Z, Xu C. FGFRL1 Promotes Ovarian Cancer Progression by Crosstalk with Hedgehog Signaling. J Immunol Res. 2018;2018:7438608
128. Sun Q, Zhang Y, Su J, Li T, Jiang Y. Role of Hydroxysteroid Dehydrogenase-Like 2 (HSDL2) in Human Ovarian Cancer. Med Sci Monit. 2018;24:3997-4008
129. Liu W, Tian X, Ding X, Zhang L. Expression of Dual-Specificity Phosphatase 2 (DUSP2) in Patients with Serous Ovarian Carcinoma and in SKOV3 and OVCAR3 Cells In Vitro. Med Sci Monit. 2019;25:10180-10189
130. Wu H, Li R, Zhang Z. et al. Kallistatin inhibits tumour progression and platinum resistance in high-grade serous ovarian cancer. J Ovarian Res. 2019;12(1):125
131. Liu T, Wei Q, Jin J. et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816-3831
132. Isobe A, Sawada K, Kinose Y. et al. Interleukin 6 receptor is an independent prognostic factor and a potential therapeutic target of ovarian cancer. PLoS One. 2015;10(2):e0118080
133. Zhang L, Wang H, Tian L, Li H. Expression of USP7 and MARCH7 Is Correlated with Poor Prognosis in Epithelial Ovarian Cancer. Tohoku J Exp Med. 2016;239(3):165-175
134. Li H, Xiao N, Li Z, Wang Q. Expression of Inorganic Pyrophosphatase (PPA1) Correlates with Poor Prognosis of Epithelial Ovarian Cancer. Tohoku J Exp Med. 2017;241(2):165-173
135. Zhao C, Yan M, Li C, Feng Z. POZ/BTB and AT-Hook-Containing Zinc Finger Protein 1 (PATZ1) Suppresses Progression of Ovarian Cancer and Serves as an Independent Prognosis Factor. Med Sci Monit. 2018;24:4262-4270
136. Jiang G, Yang D, Wang L. et al. A novel biomarker ARMc8 promotes the malignant progression of ovarian cancer. Hum Pathol. 2015;46(10):1471-1479
137. Chen L, Yao Y, Sun L. et al. Clinical implication of the serum galectin-1 expression in epithelial ovarian cancer patients. J Ovarian Res. 2015;8:78
138. Xu Y, Wang C, Zhang Y, Jia L, Huang J. Overexpression of MAGE-A9 Is Predictive of Poor Prognosis in Epithelial Ovarian Cancer. Sci Rep. 2015;5:12104
139. Xu N, Zhang Z, Zhu J. et al. Overexpression of trophoblast cell surface antigen 2 as an independent marker for a poor prognosis and as a potential therapeutic target in epithelial ovarian carcinoma. Int J Exp Pathol. 2016;97(2):150-158
140. Lin TC, Chen ST, Huang MC. et al. GALNT6 expression enhances aggressive phenotypes of ovarian cancer cells by regulating EGFR activity. Oncotarget. 2017;8(26):42588-42601
141. Schulz H, Schmoeckel E, Kuhn C. et al. Galectins-1, -3, and -7 Are Prognostic Markers for Survival of Ovarian Cancer Patients. Int J Mol Sci. 2017;18(6):1230
142. Chang B, Meng J, Zhu H. et al. Overexpression of the recently identified oncogene REDD1 correlates with tumor progression and is an independent unfavorable prognostic factor for ovarian carcinoma. Diagn Pathol. 2018;13(1):87
143. Wang C, Wang W, Liu Y, Yong M, Yang Y, Zhou H. Rac GTPase activating protein 1 promotes oncogenic progression of epithelial ovarian cancer. Cancer Sci. 2018;109(1):84-93
144. Koensgen D, Stope MB, Tuerbachova I. et al. Expression, Intracellular Localization, and Prognostic Value of Plasminogen Activator Inhibitor 1 and PAI-1 RNA-Binding Protein 1 in Primary and Recurrent Ovarian Cancer: A Study of the Tumor Bank Ovarian Cancer Network. Gynecol Obstet Invest. 2018;83(5):508-514
145. Sieńko J, Teliga-Czajkowska J, Przytula E, Czajkowski K, Smolarczyk R, Nowis
146. D. Peroxiredoxin-1 as a prognostic factor in patients with ovarian cancer. Ann Agric Environ Med. 2019;26(3):415-419
147. Zeng Y, Chen M, Ganesh S, Hu S, Chen H. Clinicopathological and prognostic significance of caveolin-1 and ATG4C expression in the epithelial ovarian cancer. PLoS One. 2020;15(5):e0232235
148. Lawrenson K, Mhawech-Fauceglia P, Worthington J. et al. Identification of novel candidate biomarkers of epithelial ovarian cancer by profiling the secretomes of three-dimensional genetic models of ovarian carcinogenesis. Int J Cancer. 2015;137(8):1806-1817
149. Chen S, Gou WF, Zhao S. et al. The role of the REG4 gene and its encoding product in ovarian epithelial carcinoma. BMC Cancer. 2015;15:471
150. Masoumi-Moghaddam S, Amini A, Wei AQ, Robertson G, Morris DL. Sprouty 2 protein, but not Sprouty 4, is an independent prognostic biomarker for human epithelial ovarian cancer. Int J Cancer. 2015;137(3):560-570
151. Abou-Taleb H, Yamaguchi K, Matsumura N. et al. Comprehensive assessment of the expression of the SWI/SNF complex defines two distinct prognostic subtypes of ovarian clear cell carcinoma. Oncotarget. 2016;7(34):54758-54770
152. Wang D, Zhu H, Ye Q, Wang C, Xu Y. Prognostic Value of KIF2A and HER2-Neu Overexpression in Patients With Epithelial Ovarian Cancer. Medicine (Baltimore). 2016;95(8):e2803
153. Zhang Q, Chen WM, Zhang XX. et al. Overexpression of salusin-β is associated with poor prognosis in ovarian cancer. Oncol Rep. 2017;37(3):1826-1832
154. Song WJ, Dong Y, Luo C, Chen YY. p38MAPK family isoform p38α and activating transcription factor 2 are associated with the malignant phenotypes and poor prognosis of patients with ovarian adenocarcinoma. Pathol Res Pract. 2017;213(10):1282-1288
155. Chan KKL, Siu MKY, Jiang YX. et al. Differential expression of estrogen receptor subtypes and variants in ovarian cancer: effects on cell invasion, proliferation and prognosis. BMC Cancer. 2017;17(1):606
156. Cheng J, Su M, Jin Y. et al. Upregulation of SENP3/SMT3IP1 promotes epithelial ovarian cancer progression and forecasts poor prognosis. Tumour Biol. 2017;39(3):1010428317694543
157. Zhu L, Feng H, Jin S. et al. High expressions of BCL6 and Lewis y antigen are correlated with high tumor burden and poor prognosis in epithelial ovarian cancer. Tumour Biol. 2017;39(7):1010428317711655
158. Jin C, Liu Z, Li Y. et al. PCNA-associated factor P15PAF, targeted by FOXM1, predicts poor prognosis in high-grade serous ovarian cancer patients. Int J Cancer. 2018;143(11):2973-2984
159. Huang RL, Chen HJ, Chen LY. et al. Epigenetic loss of heparan sulfate 3-O-sulfation sensitizes ovarian carcinoma to oncogenic signals and predicts prognosis. Int J Cancer. 2018;143(8):1943-1953
160. Sheng N, Xu YZ, Xi QH. et al. Overexpression of KIF2A is Suppressed by miR-206 and Associated with Poor Prognosis in Ovarian Cancer. Cell Physiol Biochem. 2018;50(3):810-822
161. Wang Y, Zhou Z, Wang X. et al. TRIM59 Is a Novel Marker of Poor Prognosis and Promotes Malignant Progression of Ovarian Cancer by Inducing Annexin A2 Expression. Int J Biol Sci. 2018;14(14):2073-2082
162. Wang L, Yan W, Li X. et al. S100A10 silencing suppresses proliferation, migration and invasion of ovarian cancer cells and enhances sensitivity to carboplatin. J Ovarian Res. 2019;12(1):113
163. Zhou Y, Jin Z, Wang C. Glycogen phosphorylase B promotes ovarian cancer progression via Wnt/β-catenin signaling and is regulated by miR-133a-3p. Biomed Pharmacother. 2019;120:109449
164. Sheta R, Bachvarova M, Plante M. et al. Altered expression of different GalNAc-transferases is associated with disease progression and poor prognosis in women with high-grade serous ovarian cancer. Int J Oncol. 2017;51(6):1887-1897
165. Ikeda Y, Park JH, Miyamoto T. et al. T-LAK Cell-Originated Protein Kinase (TOPK) as a Prognostic Factor and a Potential Therapeutic Target in Ovarian Cancer. Clin Cancer Res. 2016;22(24):6110-6117
166. Li MJ, Li HR, Cheng X. et al. Clinical significance of targeting drug-based molecular biomarkers expression in ovarian clear cell carcinoma. Zhonghua Fu Chan Ke Za Zhi. 2017;52(12):835-843
167. Qiu HL, Deng SZ, Li C. et al. High expression of KIF14 is associated with poor prognosis in patients with epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21(2):239-245
168. Gao L, Ye X, Ma RQ. et al. Low programmed cell death 5 expression is a prognostic factor in ovarian cancer. Chin Med J (Engl). 2015;128(8):1084-1090
169. Makii C, Oda K, Ikeda Y. et al. MDM2 is a potential therapeutic target and prognostic factor for ovarian clear cell carcinomas with wild type TP53. Oncotarget. 2016;7(46):75328-75338
170. Shin K, Kim KH, Yoon MS. et al. Expression of Interactive Genes Associated with Apoptosis and Their Prognostic Value for Ovarian Serous Adenocarcinoma. Adv Clin Exp Med. 2016;25(3):513-521
171. Labrie M, De Araujo LOF, Communal L, Mes-Masson AM, St-Pierre Y. Tissue and plasma levels of galectins in patients with high grade serous ovarian carcinoma as new predictive biomarkers. Sci Rep. 2017;7(1):13244
172. Lee YK, Chung HH, Kim JW, Song YS, Park NH. Expression of phosphorylated Akt and hTERT is associated with prognosis of epithelial ovarian carcinoma. Int J Clin Exp Pathol. 2015;8(11):14971-14976
173. Wang L, Wang C, Jin S, Qu D, Ying H. Expression of NF-κB and PTEN in primary epithelial ovarian carcinoma and the correlation with chemoresistance. Int J Clin Exp Pathol. 2015;8(9):10953-10963
174. Ayub TH, Keyver-Paik MD, Debald M. et al. Accumulation of ALDH1-positive cells after neoadjuvant chemotherapy predicts treatment resistance and prognosticates poor outcome in ovarian cancer. Oncotarget. 2015;6(18):16437-16448
175. Corkery DP, Le Page C, Meunier L, Provencher D, Mes-Masson AM, Dellaire G. PRP4K is a HER2-regulated modifier of taxane sensitivity. Cell Cycle. 2015;14(7):1059-1069
176. Miyamoto M, Takano M, Iwaya K. et al. High-temperature-required protein A2 as a predictive marker for response to chemotherapy and prognosis in patients with high-grade serous ovarian cancers. Br J Cancer. 2017;116(6):e2
177. Wang L, Wang C, Jin S, Qu D, Ying H. Expression of NF-κB and PTEN in primary epithelial ovarian carcinoma and the correlation with chemoresistance. Int J Clin Exp Pathol. 2015;8(9):10953-10963
178. Zhang Y, Yu JJ, Tian Y. et al. eIF3a improve cisplatin sensitivity in ovarian cancer by regulating XPC and p27Kip1 translation. Oncotarget. 2015;6(28):25441-25451
179. Gayarre J, Kamieniak MM, Cazorla-Jiménez A. et al. The NER-related gene GTF2H5 predicts survival in high-grade serous ovarian cancer patients. J Gynecol Oncol. 2016;27(1):e7
180. Sung PL, Jan YH, Lin SC. et al. Periostin in tumor microenvironment is associated with poor prognosis and platinum resistance in epithelial ovarian carcinoma. Oncotarget. 2016;7(4):4036-4047
181. Kwon AY, Heo I, Lee HJ. et al. Sox10 expression in ovarian epithelial tumors is associated with poor overall survival. Virchows Arch. 2016;468(5):597-605
182. He S, Niu G, Shang J. et al. The oncogenic Golgi phosphoprotein 3 like overexpression is associated with cisplatin resistance in ovarian carcinoma and activating the NF-κB signaling pathway. J Exp Clin Cancer Res. 2017;36(1):137
183. Miyamoto M, Takano M, Aoyama T. et al. Inhibition of autophagy protein LC3A as a therapeutic target in ovarian clear cell carcinomas. J Gynecol Oncol. 2017;28(3):e33
184. Sun X, Zhang W, Li H. et al. Stonin 2 Overexpression is Correlated with Unfavorable Prognosis and Tumor Invasion in Epithelial Ovarian Cancer. Int J Mol Sci. 2017;18(8):1653
185. Chen HJ, Huang RL, Liew PL. et al. GATA3 as a master regulator and therapeutic target in ovarian high-grade serous carcinoma stem cells. Int J Cancer. 2018;143(12):3106-3119
186. Zhang X, Li H, Yu X. et al. Analysis of Circulating Tumor Cells in Ovarian Cancer and Their Clinical Value as a Biomarker. Cell Physiol Biochem. 2018;48(5):1983-1994
187. Zhang X, Feng Y, Wang XY. et al. The inhibition of UBC13 expression and blockage of the DNMT1-CHFR-Aurora A pathway contribute to paclitaxel resistance in ovarian cancer. Cell Death Dis. 2018;9(2):93
188. Kim HJ, Sung SH, Kim CY. et al. 14-3-3ζ Overexpression is Associated with Poor Prognosis in Ovarian Cancer. Yonsei Med J. 2018;59(1):51-56
189. Liu X, Wei L, Zhao B, Cai X, Dong C, Yin F. Low expression of KCNN3 may affect drug resistance in ovarian cancer. Mol Med Rep. 2018;18(2):1377-1386
190. Long J, Zhu JY, Liu YB. et al. Helicase POLQ-like (HELQ) as a novel indicator of platinum-based chemoresistance for epithelial ovarian cancer. Gynecol Oncol. 2018;149(2):341-349
191. Jin C, Liu Z, Li Y. et al. PCNA-associated factor P15PAF, targeted by FOXM1, predicts poor prognosis in high-grade serous ovarian cancer patients. Int J Cancer. 2018;143(11):2973-2984
192. Fu Z, Wang C, Chen Y, Zhang X, Wang X, Xie X. Down-regulation of UTP23 promotes paclitaxel resistance and predicts poorer prognosis in ovarian cancer. Pathol Res Pract. 2019;215(11):152625
193. Hou L, Zhang X, Jiao Y. et al. ATP binding cassette subfamily B member 9 (ABCB9) is a prognostic indicator of overall survival in ovarian cancer. Medicine (Baltimore). 2019;98(19):e15698
194. Ma H, Li Y, Wang X. et al. PBK, targeted by EVI1, promotes metastasis and confers cisplatin resistance through inducing autophagy in high-grade serous ovarian carcinoma. Cell Death Dis. 2019;10(3):166
195. Zhang S, Deng M, Wang Q, Jiang Y, Xu Q, Cao L. Level of Sorcin expression influences chemoresistance and overall survival in patients with ovarian cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2019;44(10):1113-1119
196. Bu H, Li Y, Jin C. et al. Overexpression of PRC1 indicates a poor prognosis in ovarian cancer. Int J Oncol. 2020;56(3):685-696
197. Feng LY, Li L. Low expression of NCALD is associated with chemotherapy resistance and poor prognosis in epithelial ovarian cancer. J Ovarian Res. 2020;13(1):35
198. Hua M, Yan S, Deng Y. et al. CAP1 is overexpressed in human epithelial ovarian cancer and promotes cell proliferation. Int J Mol Med. 2015;35(4):941-949
199. Ayhan A, Kuhn E, Wu RC. et al. CCNE1 copy-number gain and overexpression identify ovarian clear cell carcinoma with a poor prognosis. Mod Pathol. 2017;30(2):297-303
200. Shi C, Qin L, Gao H. et al. NUCKS nuclear elevated expression indicates progression and prognosis of ovarian cancer. Tumour Biol. 2017;39(9):1010428317714631
201. Wang J, Liu Q, Zhou X. et al. Thymidine kinase 1 expression in ovarian serous adenocarcinoma is superior to Ki-67: A new prognostic biomarker. Tumour Biol. 2017;39(6):1010428317706479
202. Doghri R, Manai M, Finetti P. et al. Stromal Expression of MARCKS Protein in Ovarian Carcinomas Has Unfavorable Prognostic Value. Int J Mol Sci. 2017;19(1):41
203. Horikawa N, Abiko K, Matsumura N. et al. Expression of Vascular Endothelial Growth Factor in Ovarian Cancer Inhibits Tumor Immunity through the Accumulation of Myeloid-Derived Suppressor Cells. Clin Cancer Res. 2017;23(2):587-599
204. Chen M, Sheng XJ, Qin YY. et al. TBC1D8 Amplification Drives Tumorigenesis through Metabolism Reprogramming in Ovarian Cancer. Theranostics. 2019;9(3):676-690
205. Feng Y, Tang Y, Mao Y. et al. PAX2 promotes epithelial ovarian cancer progression involving fatty acid metabolic reprogramming. Int J Oncol. 2020;56(3):697-708
206. Hedström E, Pederiva C, Farnebo J. et al. Downregulation of the cancer susceptibility protein WRAP53β in epithelial ovarian cancer leads to defective DNA repair and poor clinical outcome. Cell Death Dis. 2015;6(10):e1892
207. Mei L, Hu Q, Peng J. et al. Phospho-histone H2AX is a diagnostic and prognostic marker for epithelial ovarian cancer. Int J Clin Exp Pathol. 2015;8(5):5597-5602
208. Sun F, Ding W, He JH, Wang XJ, Ma ZB, Li YF. Stomatin-like protein 2 is overexpressed in epithelial ovarian cancer and predicts poor patient survival. BMC Cancer. 2015;15:746
209. Sosulski A, Horn H, Zhang L. et al. CD44 Splice Variant v8-10 as a Marker of Serous Ovarian Cancer Prognosis. PLoS One. 2016;11(6):e0156595
210. Zuo J, Song Y, Li Z, Wu LY. Relationship between P53 Protein Expression and Prognosis of Advanced Ovarian Serous Adenocarcinoma. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2016;38(2):169-174
211. van der Steen SC, van Tilborg AA, Vallen MJ, Bulten J, van Kuppevelt TH, Massuger LF. Prognostic significance of highly sulfated chondroitin sulfates in ovarian cancer defined by the single chain antibody GD3A11. Gynecol Oncol. 2016;140(3):527-536
212. Li X, Yu Z, Fang L, Liu F, Jiang K. Expression of Adiponectin Receptor-1 and Prognosis of Epithelial Ovarian Cancer Patients. Med Sci Monit. 2017;23:1514-1521
213. Rzepecka IK, Szafron LM, Stys A. et al. Prognosis of patients with BRCA1-associated ovarian carcinomas depends on TP53 accumulation status in tumor cells. Gynecol Oncol. 2017;144(2):369-376
214. Sakr S, Abdulfatah E, Thomas S. et al. Granulosa Cell Tumors: Novel Predictors of Recurrence in Early-stage Patients. Int J Gynecol Pathol. 2017;36(3):240-252
215. Tian X, Han Y, Yu L. et al. Decreased expression of ALDH5A1 predicts prognosis in patients with ovarian cancer. Cancer Biol Ther. 2017;18(4):245-251
216. Veneris JT, Darcy KM, Mhawech-Fauceglia P. et al. High glucocorticoid receptor expression predicts short progression-free survival in ovarian cancer. Gynecol Oncol. 2017;146(1):153-160
217. Wang D, Cao X, Zhang Y. et al. LAMP3 expression correlated with poor clinical outcome in human ovarian cancer. Tumour Biol. 2017;39(3):1010428317695014
218. Wang Y, Sun J, Li N. et al. HBXIP overexpression is correlated with the clinical features and survival outcome of ovarian cancer. J Ovarian Res. 2017;10(1):26
219. Yasuda K, Hirohashi Y, Mariya T. et al. Phosphorylation of HSF1 at serine 326 residue is related to the maintenance of gynecologic cancer stem cells through expression of HSP27. Oncotarget. 2017;8(19):31540-31553
220. Beeghly-Fadiel A, Wilson AJ, Keene S. et al. Differential cyclooxygenase expression levels and survival associations in type I and type II ovarian tumors. J Ovarian Res. 2018;11(1):17
221. Zhu CX, Xiong W, Wang ML. et al. Nuclear G protein-coupled oestrogen receptor (GPR30) predicts poor survival in patients with ovarian cancer. J Int Med Res. 2018;46(2):723-731
222. Li L, Li X, Meng Q, Khan AQ, Chen X. Increased Expression of Holliday Junction-Recognizing Protein (HJURP) as an Independent Prognostic Biomarker in Advanced-Stage Serous Ovarian Carcinoma. Med Sci Monit. 2018;24:3050-3055
223. Schulz H, Kuhn C, Hofmann S. et al. Overall Survival of Ovarian Cancer Patients Is Determined by Expression of Galectins-8 and -9. Int J Mol Sci. 2018;19(1):323
224. Chung YW, Kim S, Hong JH. et al. Overexpression of HER2/HER3 and clinical feature of ovarian cancer. J Gynecol Oncol. 2019;30(5):e75
225. Gou R, Zhu L, Zheng M. et al. Annexin A8 can serve as potential prognostic biomarker and therapeutic target for ovarian cancer: based on the comprehensive analysis of Annexins. J Transl Med. 2019;17(1):275
226. Han GH, Chay DB, Yi JM, Cho H, Chung JY, Kim JH. Loss of Both USP10 and p14ARF Protein Expression Is an Independent Prognostic Biomarker for Poor Prognosis in Patients With Epithelial Ovarian Cancer. Cancer Genomics Proteomics. 2019;16(6):553-562
227. Qian H, Yuan D, Bao J. et al. Increased expression of plakophilin 3 is associated with poor prognosis in ovarian cancer. Medicine (Baltimore). 2019;98(10):e14608
228. Szubert S, Moszynski R, Szpurek D. et al. The expression of Platelet-derived Growth factor receptors (PDGFRs) and their correlation with overall survival of patients with ovarian cancer. Ginekol Pol. 2019;90(5):242-249
229. Xin B, Ji KQ, Liu YS, Zhao XD. Higher expression of calcineurin predicts poor prognosis in unique subtype of ovarian cancer. J Ovarian Res. 2019;12(1):75
230. Xu L, Guo Y, Xu N. et al. Overexpression of thymic stromal lymphopoietin is correlated with poor prognosis in epithelial ovarian carcinoma. Biosci Rep. 2019;39(5):BSR20190116
231. Zhang L, Sun L, Zhang B, Chen L. Identification of Differentially Expressed Genes (DEGs) Relevant to Prognosis of Ovarian Cancer by Use of Integrated Bioinformatics Analysis and Validation by Immunohistochemistry Assay. Med Sci Monit. 2019;25:9902-9912
232. Czogalla B, Deuster E, Liao Y. et al. Cytoplasmic VDR expression as an independent risk factor for ovarian cancer. Histochem Cell Biol. 2020;154(4):421-429
233. Phelps DL, Borley JV, Flower KJ. et al. Methylation of MYLK3 gene promoter region: a biomarker to stratify surgical care in ovarian cancer in a multicentre study. Br J Cancer. 2017;116(10):1287-1293
234. Bubancova I, Kovarikova H, Laco J. et al. Next-Generation Sequencing Approach in Methylation Analysis of HNF1B and GATA4 Genes: Searching for Biomarkers in Ovarian Cancer. Int J Mol Sci. 2017;18(2):474
235. Mase S, Shinjo K, Totani H. et al. ZNF671 DNA methylation as a molecular predictor for the early recurrence of serous ovarian cancer. Cancer Sci. 2019;110(3):1105-1116
236. Nielsen B, Kleppe A, Hveem TS. et al. Association Between Proportion of Nuclei With High Chromatin Entropy and Prognosis in Gynecological Cancers. J Natl Cancer Inst. 2018;110(12):1400-1408
237. Eoh KJ, Park HS, Park JS. et al. Comparison of Clinical Outcomes of BRCA1/2 Pathologic Mutation, Variants of Unknown Significance, or Wild Type Epithelial Ovarian Cancer Patients. Cancer Res Treat. 2017;49(2):408-415
238. Kim SI, Lee M, Kim HS. et al. Effect of BRCA mutational status on survival outcome in advanced-stage high-grade serous ovarian cancer. J Ovarian Res. 2019;12(1):40
239. Kalavska K, Minarik T, Vlkova B. et al. Prognostic value of various subtypes of extracellular DNA in ovarian cancer patients. J Ovarian Res. 2018;11(1):85
240. Liu J, Liu T, Liang L. et al. Clinical relationships between the rs2212020 and rs189897 polymorphisms of the ITGA9 gene and epithelial ovarian cancer. J Genet. 2019;98:28
241. Zhang Y, Wang Y, Huang X. et al. Association between AXIN1 gene polymorphisms and epithelial ovarian cancer in Chinese population. Biomark Med. 2019;13(6):445-455
242. Cong J, Liu R, Wang X, Wang J, Wang H, Hou J. Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2015;19(24):4762-4765
243. Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a Novel Biomarker for Patients with Ovarian Cancer. Med Sci Monit. 2015;21:3929-3934
244. Zhang X, Liu J, Zang D. et al. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6(17):15180-15193
245. Ying L, Zhang F, Pan X. et al. Complement component 7 (C7), a potential tumor suppressor, is correlated with tumor progression and prognosis. Oncotarget. 2016;7(52):86536-86546
246. Shang AQ, Wu J, Bi F. et al. Relationship between HER2 and JAK/STAT-SOCS3 signaling pathway and clinicopathological features and prognosis of ovarian cancer. Cancer Biol Ther. 2017;18(5):314-322
247. Zhang X, Liu G, Qiu J, Zhang N, Ding J, Hua K. E2F1-regulated long non-coding RNA RAD51-AS1 promotes cell cycle progression, inhibits apoptosis and predicts poor prognosis in epithelial ovarian cancer. Sci Rep. 2017;7(1):4469
248. Chen P, Fang X, Xia B, Zhao Y, Li Q, Wu X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018;7(9):4530-4541
249. Guo TY, Xu HY, Chen WJ, Wu MX, Dai X. Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(22):7646-7652
250. Li TH, Zhang JJ, Liu SX, Chen Y. Long non-coding RNA taurine-upregulated gene 1 predicts unfavorable prognosis, promotes cells proliferation, and inhibits cells apoptosis in epithelial ovarian cancer. Medicine (Baltimore). 2018;97(19):e0575
251. Liu J, Gu Z, Tang Y, Hao J, Zhang C, Yang X. Tumour-suppressive microRNA-424-5p directly targets CCNE1 as potential prognostic markers in epithelial ovarian cancer. Cell Cycle. 2018;17(3):309-318
252. Zhao L, Ji G, Le X. et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017;77(6):1369-1382
253. Ren XY, Yang WB, Tian Y. Overexpression of long noncoding RNA PTPRG-AS1 is associated with poor prognosis in epithelial ovarian cancer. Rev Assoc Med Bras (1992). 2020;66(7):948-953
254. Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20(16):3373-3377
255. Fu Y, Biglia N, Wang Z. et al. Long non-coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer. Gynecol Oncol. 2016;143(3):642-649
256. Kim TH, Song JY, Park H. et al. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015;356(2 Pt B):937-945
257. Fan Y, Fan J, Huang L. et al. Increased expression of microRNA-196a predicts poor prognosis in human ovarian carcinoma. Int J Clin Exp Pathol. 2015;8(4):4132-4137
258. Zhao W, Han T, Li B, Ma Q, Yang P, Li H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J Ovarian Res. 2019;12(1):121
259. Cheng Z, Guo J, Chen L, Luo N, Yang W, Qu X. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget. 2015;6(28):25381-25389
260. Ma M, Yu N. Over-Expression of TBL1XR1 Indicates Poor Prognosis of Serous Epithelial Ovarian Cancer. Tohoku J Exp Med. 2017;241(3):239-247
261. Li AH, Zhang HH. Overexpression of lncRNA MNX1-AS1 is associated with poor clinical outcome in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21(24):5618-5623
262. Yong W, Yu D, Jun Z. et al. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018;9(9):861
263. Wei H, Tang QL, Zhang K, Sun JJ, Ding RF. miR-532-5p is a prognostic marker and suppresses cells proliferation and invasion by targeting TWIST1 in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(18):5842-5850
264. Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46(6):2497-2505
265. Cao Y, Shi H, Ren F, Jia Y, Zhang R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp Cell Res. 2017;359(1):185-194
266. Mei J, Huang Y, Hao L. et al. DAAM1-mediated migration and invasion of ovarian cancer cells are suppressed by miR-208a-5p. Pathol Res Pract. 2019;215(7):152452
267. Chen X, Huang J, Lü Y. High expression of STAT2 in ovarian cancer and its effect on metastasis of ovarian cancer cells. Nan Fang Yi Ke Da Xue Xue Bao. 2020;40(1):34-41
268. Zhu D, Huang X, Liang F, Zhao L. LncRNA miR503HG interacts with miR-31-5p through multiple ways to regulate cancer cell invasion and migration in ovarian cancer. J Ovarian Res. 2020;13(1):3
269. Huang S, Qing C, Huang Z, Zhu Y. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11(1):49
270. Sun J, Yang X, Zhang R. et al. GOLPH3 induces epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway in epithelial ovarian cancer. Cancer Med. 2017;6(4):834-844
271. Yim GW, Kim HJ, Kim LK. et al. Long Non-coding RNA HOXA11 Antisense Promotes Cell Proliferation and Invasion and Predicts Patient Prognosis in Serous Ovarian Cancer. Cancer Res Treat. 2017;49(3):656-668
272. Zhang J, Liu W, Shen F. et al. The activation of microRNA-520h-associated TGF-β1/c-Myb/Smad7 axis promotes epithelial ovarian cancer progression. Cell Death Dis. 2018;9(9):884
273. Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(3):616-622
274. Xu QF, Tang YX, Wang X. LncRNA EBIC promoted proliferation, metastasis and cisplatin resistance of ovarian cancer cells and predicted poor survival in ovarian cancer patients. Eur Rev Med Pharmacol Sci. 2018;22(14):4440-4447
275. Guo C, Wang X, Chen LP. et al. Long non-coding RNA MALAT1 regulates ovarian cancer cell proliferation, migration and apoptosis through Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3703-3712
276. Huang K, Geng J, Wang J. Long non-coding RNA RP11-552M11.4 promotes cells proliferation, migration and invasion by targeting BRCA2 in ovarian cancer. Cancer Sci. 2018;109(5):1428-1446
277. Wang S, Ning Y, Wei P. et al. The non-coding RNA OTUB1-isoform2 promotes ovarian tumour progression and predicts poor prognosis. J Cell Mol Med. 2018;22(10):4794-4806
278. Li X, Zhang NX, Ye HY. et al. HYOU1 promotes cell growth and metastasis via activating PI3K/AKT signaling in epithelial ovarian cancer and predicts poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23(10):4126-4135
279. Liu HY, Zhang YY, Zhu BL. et al. MiR-203a-3p regulates the biological behaviors of ovarian cancer cells through mediating the Akt/GSK-3β/Snail signaling pathway by targeting ATM. J Ovarian Res. 2019;12(1):60
280. Pan L, Meng Q, Li H, Liang K, Li B. LINC00339 promotes cell proliferation, migration, and invasion of ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed Pharmacother. 2019;120:109423
281. Wang D, Dai J, Hou S, Qian Y. LncRNA SNHG20 predicts a poor prognosis and promotes cell progression in epithelial ovarian cancer. Biosci Rep. 2019;39(4):BSR20182186
282. Zhao LW, Yu AJ, Zhang YJ, Wang XC, Han B, Wang XH. MicroRNA-149 suppresses the malignant phenotypes of ovarian cancer via downregulation of MSI2 and inhibition of PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):55-64
283. Liu G, Xue F, Zhang W. miR-506: a regulator of chemo-sensitivity through suppression of the RAD51-homologous recombination axis. Chin J Cancer. 2015;34(11):485-487
284. Chiang YC, Lin HW, Chang CF. et al. Overexpression of CHI3L1 is associated with chemoresistance and poor outcome of epithelial ovarian carcinoma. Oncotarget. 2015;6(37):39740-39755
285. Hsu KF, Shen MR, Huang YF. et al. Overexpression of the RNA-binding proteins Lin28B and IGF2BP3 (IMP3) is associated with chemoresistance and poor disease outcome in ovarian cancer. Br J Cancer. 2015;113(3):414-424
286. Kritsch D, Hoffmann F, Steinbach D. et al. Tribbles 2 mediates cisplatin sensitivity and DNA damage response in epithelial ovarian cancer. Int J Cancer. 2017;141(8):1600-1614
287. Xiao M, Cai J, Cai L. et al. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J Ovarian Res. 2017;10(1):24
288. Zanotti L, Romani C, Tassone L. et al. MAL gene overexpression as a marker of high-grade serous ovarian carcinoma stem-like cells that predicts chemoresistance and poor prognosis. BMC Cancer. 2017;17(1):366
289. Wang Y, Bao W, Liu Y. et al. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018;9(5):447
290. Gu ZW, He YF, Wang WJ, Tian Q, Di W. MiR-1180 from bone marrow-derived mesenchymal stem cells induces glycolysis and chemoresistance in ovarian cancer cells by upregulating the Wnt signaling pathway. J Zhejiang Univ Sci B. 2019;20(3):219-237
291. Long X, Song K, Hu H. et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38(1):345
292. Leonard B, Starrett GJ, Maurer MJ. et al. APOBEC3G Expression Correlates with T-Cell Infiltration and Improved Clinical Outcomes in High-grade Serous Ovarian Carcinoma. Clin Cancer Res. 2016;22(18):4746-4755
293. Colvin EK, Howell VM, Mok SC, Samimi G, Vafaee F. Expression of long noncoding RNAs in cancer-associated fibroblasts linked to patient survival in ovarian cancer. Cancer Sci. 2020;111(5):1805-1817
294. Liu TT, Xu H, Gao WP. et al. SET and MYND Domain-Containing Protein 3 (SMYD3) Polymorphism as a Risk Factor for Susceptibility and Poor Prognosis in Ovarian Cancer. Med Sci Monit. 2016;22:5131-5140
295. Wang YS, Ma LN, Sun JX, Liu N, Wang H. Long non-coding RNA CPS1-IT1 is a positive prognostic factor and inhibits epithelial ovarian cancer tumorigenesis. Eur Rev Med Pharmacol Sci. 2017;21(14):3169-3175
296. Szafron LM, Balcerak A, Grzybowska EA. et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget. 2015;6(41):43897-43910
297. Yuan C, Liu X, Liu X. et al. The GADD45A (1506T>C) Polymorphism Is Associated with Ovarian Cancer Susceptibility and Prognosis. PLoS One. 2015;10(9):e0138692
298. Zhang X, Guo G, Wang G. et al. Profile of differentially expressed miRNAs in high-grade serous carcinoma and clear cell ovarian carcinoma, and the expression of miR-510 in ovarian carcinoma. Mol Med Rep. 2015;12(6):8021-8031
299. Lu L, Katsaros D, Canuto EM, Biglia N, Risch HA, Yu H. LIN-28B/let-7a/IGF-II axis molecular subtypes are associated with epithelial ovarian cancer prognosis. Gynecol Oncol. 2016;141(1):121-127
300. Halvorsen AR, Kristensen G, Embleton A. et al. Evaluation of Prognostic and Predictive Significance of Circulating MicroRNAs in Ovarian Cancer Patients. Dis Markers. 2017;2017:3098542
301. Zhou QH, Zhao YM, Jia LL, Zhang Y. Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21(19):4278-4282
302. Geng X, Liu Y, Diersch S. et al. Clinical relevance of kallikrein-related peptidase 9, 10, 11, and 15 mRNA expression in advanced high-grade serous ovarian cancer. PLoS One. 2017;12(11):e0186847
303. Ai H, Xie W, Xiu AH. et al. The down-regulation of long non-coding RNA LINC01088 is associated with the poor prognosis of epithelial ovarian cancer patients. Eur Rev Med Pharmacol Sci. 2018;22(18):5836-5841
304. Chu ZP, Dai J, Jia LG. et al. Increased expression of long noncoding RNA HMMR-AS1 in epithelial ovarian cancer: an independent prognostic factor. Eur Rev Med Pharmacol Sci. 2018;22(23):8145-8150
305. Zou T, Wang PL, Gao Y, Liang WT. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22(21):7178-7182
306. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713-3718
307. Chen H, Tian X, Luan Y, Lu H. Downregulated Long Noncoding RNA DGCR5 Acts as a New Promising Biomarker for the Diagnosis and Prognosis of Ovarian Cancer. Technol Cancer Res Treat. 2019;18:1533033819896809
308. Moes-Sosnowska J, Rzepecka IK, Chodzynska J. et al. Clinical importance of FANCD2, BRIP1, BRCA1, BRCA2 and FANCF expression in ovarian carcinomas. Cancer Biol Ther. 2019;20(6):843-854
309. Zhang XY, Zhou LL, Jiao Y. et al. Adenylate kinase 7 is a prognostic indicator of overall survival in ovarian cancer. Medicine (Baltimore). 2021;100(1):e24134
310. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860-867.311
311. Zahorec R. Ratio of neutrophil to lymphocyte counts-rapid and simple parameter of systemic inflammation and stress in critically ill. Bratisl Lek Listy. 2001;102(1):5-14
312. Guthrie GJ, Charles KA, Roxburgh CS, Horgan PG, McMillan DC, Clarke SJ. The systemic inflammation-based neutrophil-lymphocyte ratio: experience in patients with cancer. Crit Rev Oncol Hematol. 2013;88(1):218-230
313. Wang Y, Liu P, Xu Y. et al. Preoperative neutrophil-to-lymphocyte ratio predicts response to first-line platinum-based chemotherapy and prognosis in serous ovarian cancer. Cancer Chemother Pharmacol. 2015;75(2):255-262
314. Huang QT, Zhou L, Zeng WJ. et al. Prognostic Significance of Neutrophil-to-Lymphocyte Ratio in Ovarian Cancer: A Systematic Review and Meta-Analysis of Observational Studies. Cell Physiol Biochem. 2017;41(6):2411-2418
315. Mallappa S, Sinha A, Gupta S, Chadwick SJ. Preoperative neutrophil to lymphocyte ratio >5 is a prognostic factor for recurrent colorectal cancer. Colorectal Dis. 2013;15(3):323-328
316. Zhang H, Huo Q, Huang L, Cheng Y, Liu Y, Bao H. Neutrophil-to-Lymphocyte Ratio in Ovarian Cancer Patients with Low CA125 Concentration. Biomed Res Int. 2019;2019:8107906
317. Bhattacharya B, Mohd Omar MF, Soong R. The Warburg effect and drug resistance. Br J Pharmacol. 2016;173(6):970-979
318. Deme D, Telekes A. A laktátdehidrogenáz (LDH) prognosztikai jelentősége az onkol ó gi á ban [Prognostic importance of lactate dehydrogenase (LDH) in oncology]. Orv Hetil. 2017;158(50):1977-1988
319. Xiang J, Zhou L, Zhuang Y. et al. Lactate dehydrogenase is correlated with clinical stage and grade and is downregulated by si-SAΤB1 in ovarian cancer. Oncol Rep. 2018;40(5):2788-2797
320. Valvona CJ, Fillmore HL, Nunn PB, Pilkington GJ. The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor. Brain Pathol. 2016;26(1):3-17
321. Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 2013;65(11):904-910
322. Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996;93(1):136-140
323. He X, Wang L, Riedel H. et al. Mesothelin promotes epithelial-to-mesenchymal transition and tumorigenicity of human lung cancer and mesothelioma cells. Mol Cancer. 2017;16(1):63
324. Coelho R, Marcos-Silva L, Ricardo S. et al. Peritoneal dissemination of ovarian cancer: role of MUC16-mesothelin interaction and implications for treatment. Expert Rev Anticancer Ther. 2018;18(2):177-186
325. Chang MC, Chen CA, Chen PJ. et al. Mesothelin enhances invasion of ovarian cancer by inducing MMP-7 through MAPK/ERK and JNK pathways. Biochem J. 2012;442(2):293-302
326. Yan M, Wang C, He B. et al. Aurora-A Kinase: A Potent Oncogene and Target for Cancer Therapy. Med Res Rev. 2016;36(6):1036-1079
327. Liu Q, Kaneko S, Yang L. et al. Mitotic kinase Aurora-A phosphorylation and inactivation of p53. Cancer Research. 2004 64
328. Lentini L, Amato A, Schillaci T, Di Leonardo A. Simultaneous Aurora-A/STK15 overexpression and centrosome amplification induce chromosomal instability in tumour cells with a MIN phenotype. BMC Cancer. 2007;7:212
329. Matulonis UA, Sharma S, Ghamande S. et al. Phase II study of MLN8237 (alisertib), an investigational Aurora A kinase inhibitor, in patients with platinum-resistant or -refractory epithelial ovarian, fallopian tube, or primary peritoneal carcinoma. Gynecol Oncol. 2012;127(1):63-69
330. Thijssen VL, Heusschen R, Caers J, Griffioen AW. Galectin expression in cancer diagnosis and prognosis: A systematic review. Biochim Biophys Acta. 2015;1855(2):235-247
331. Schulz H, Kuhn C, Hofmann S. et al. Overall Survival of Ovarian Cancer Patients Is Determined by Expression of Galectins-8 and -9. Int J Mol Sci. 2018;19(1):323
332. Nagahara K, Arikawa T, Oomizu S. et al. Galectin-9 increases Tim-3+ dendritic cells and CD8+ T cells and enhances antitumor immunity via galectin-9-Tim-3 interactions. Journal of Immunology. 2008;181(11):7660-7669
333. Bidon-Wagner N, Le Pennec JP. Human galectin-8 isoforms and cancer. Glycoconj J. 2002;19(7-9):557-563
334. Viallard C, Larriv é e B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409-426
335. Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4-10
336. Matsumoto K, Ema M. Roles of VEGF-A signalling in development, regeneration, and tumours. J Biochem. 2014;156(1):1-10
337. Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12-23
338. Meng F, Henson R, Lang M. et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130(7):2113-2129
339. Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29(15):2161-2164
340. Naishadham D. Cancer statistics, 2012. Ca A Cancer Journal for Clinicians. 2015;63(1):11
341. Chung TK, Cheung TH, Huen NY. et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124(6):1358-1365
342. Gregory PA, Bert AG, Paterson EL. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593-601
343. Sato E, Olson SH, Ahn J. et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102(51):18538-18543
344. Hamanishi J, Mandai M, Iwasaki M. et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A. 2007;104(9):3360-3365
345. Clemente CG, Mihm MC Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77(7):1303-1310
346. Pagès F, Galon J, Dieu-Nosjean MC, Tartour E, Sautès-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29(8):1093-1102
347. Santoiemma PP, Powell DJ Jr. Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol Ther. 2015;16(6):807-820
348. Tuthill RJ, Unger JM, Liu PY, Flaherty LE, Sondak VK; Southwest Oncology Group. Risk assessment in localized primary cutaneous melanoma: a Southwest Oncology Group study evaluating nine factors and a test of the Clark logistic regression prediction model. Am J Clin Pathol. 2002;118(4):504-511
349. Zhang L, Conejo-Garcia JR, Katsaros D. et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203-213
350. Turner TB, Buchsbaum DJ, Straughn JM Jr, Randall TD, Arend RC. Ovarian cancer and the immune system - The role of targeted therapies. Gynecol Oncol. 2016;142(2):349-356
351. Miao L, St Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med. 2009;47(4):344-356
352. Bartling TR, Subbaram S, Clark RR, Chandrasekaran A, Kar S, Melendez JA. Redox-sensitive gene-regulatory events controlling aberrant matrix metalloproteinase-1 expression. Free Radic Biol Med. 2014;74:99-107
353. Cui Y, She K, Tian D, Zhang P, Xin X. miR-146a Inhibits Proliferation and Enhances Chemosensitivity in Epithelial Ovarian Cancer via Reduction of SOD2. Oncol Res. 2016;23(6):275-282
354. Chang B, Yang H, Jiao Y. et al. SOD2 deregulation enhances migration, invasion and has poor prognosis in salivary adenoid cystic carcinoma. Sci Rep. 2016;6:25918
355. Xu Z, Chen Y, Gu D. et al. SOD2 rs4880 CT/CC genotype predicts poor survival for Chinese gastric cancer patients received platinum and fluorouracil based adjuvant chemotherapy. Am J Transl Res. 2015;7(2):401-410
356. Isono T, Chano T, Yonese J, Yuasa T. Therapeutic inhibition of mitochondrial function induces cell death in starvation-resistant renal cell carcinomas. Sci Rep. 2016;6:25669
357. Kanwal R, Gupta K, Gupta S. Cancer epigenetics: an introduction. Methods Mol Biol. 2015;1238:3-25
358. Huang RL, Chen HJ, Chen LY. et al. Epigenetic loss of heparan sulfate 3-O-sulfation sensitizes ovarian carcinoma to oncogenic signals and predicts prognosis. Int J Cancer. 2018;143(8):1943-1953
359. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77(15):3965-3981
Author contact
![]() Corresponding author: Fang Wang, E-mail: wangfangedu.cn, Tel./Fax: +86 2586862814.
Corresponding author: Fang Wang, E-mail: wangfangedu.cn, Tel./Fax: +86 2586862814.

 Global reach, higher impact
Global reach, higher impact