Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(14):4218-4228. doi:10.7150/jca.47506 This issue Cite
Research Paper
The Overexpression of NMHC IIA Promoted Invasion and Metastasis of Nasopharyngeal Carcinoma Cells
1. Medical Laboratory of The Third Affiliated Hospital of ShenZhen university, Shenzhen, 518001, China.
2. Department of pathology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China.
3. Department of Otolaryngology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China.
4. Guangzhou Key Laboratory of Otorhinolaryngology, Guangzhou, China.
#These authors contributed equally to this paper.
Received 2020-4-27; Accepted 2021-5-1; Published 2021-5-17
Abstract
Background: Nasopharyngeal carcinoma (NPC) is a kind of head and neck squamous cell carcinoma (HNSCC) with a strong tendency for metastasis and recurrence. Non-muscle myosin heavy chain IIA (NMHC IIA) plays important roles in recurrence and metastasis of cancers. However, the function and mechanism of NMHC IIA expression in NPC remain unclear.
Methods: A receiver operating characteristic (ROC) curve was constructed for 141 specimens of HNSCC tissues and 44 control samples from The Cancer Genome Atlas (TCGA) database. Co-expressed genes with MYH9 were identified using LinkedOmics. Transcription factors (TFs) and miRNA regulation network were constructed using Networkanalyst. The migration and invasion ability of nasopharyngeal carcinoma cells were evaluated by in vitro migration and matrigel invasion assays, respectively.
Results: The public microarray results showed that MYH9 expression levels were upregulated in HNSCC tissues compared with the matched adjacent normal tissues in this study (p<0.0001). The AUC of MYH9 reached up to 0.8303 at a cutoff value of 175.2, with a sensitivity and specificity of 70.21% and 86.36%, respectively. MYH9 expression was increased in lymph node metastasis HNSCC tumors compared with that in tumors without lymph node metastasis (p<0.05) and showed a strong positive association with expression of FLNA. High MYH9 and FLNA expression were related with poorer overall survival in HNSCC. MYH9 with positively associated genes regulated focal adhesion, cell-substrate junction assembly and cell morphogenesis were involved in differentiation using GO and KEGG analysis. MYH9 was correlated with a network of TFs including SP1, SRF, JUN and FOS in HNSCC. The suppression of endogenous NMHC IIA decreased cellular migration and invasion in HNE1 cells and reduced the expression of phosphorylation of EGFR, AKT and ERK. The over-expression of NMHC IIA increased cellular migration and invasion in COS-7 cells and increased the expression of phosphorylation of EGFR, AKT and ERK.
Conclusion: Expression of NMHC IIA mRNA was higher in HNSCC than in the adjacent normal tissues. NMHC IIA expression was increased in lymph node metastasis HNSCC tumors compared with tumors without lymph node metastasis. High MYH9 was association with poorer overall survival in HNSCC. NMHC IIA expression increased the invasion and metastasis abilities of the nasopharyngeal cancer cell line in vitro by augmenting the expression of phosphorylation of EGFR, AKT and ERK. These findings will be beneficial for providing an effectively therapeutic strategy for NPC.
Keywords: NMHC IIA, head and neck squamous cell carcinoma, nasopharyngeal cancer, invasion, metastasis
Introduction
Head and neck squamous cell carcinoma (HNSCC) contains a group of cancers arising in the oropharynx, oral cavity, nasal cavity and larynx, hypopharynx and paranasal sinuses [1]. Nasopharyngeal carcinoma (NPC) is a kind of HNSCC with high incidence Epstein-Barr virus (EBV)-related epithelial malignancy, which of remarkable regional and ethnic specificity that can lead to serious health problems in south China and southeastern Asia compared with the Western world [2]. According to the literature, nearly 80,000 people are diagnosed with NPC every year around the world, ranking 23rd in new-onset malignant tumors [3]. Approximately 50,000 people die annually of NPC [4]. Most patients have reached the advanced stage at their first visit, with local lymph node and/or distant metastasis [5]; therefore, early diagnosis is a major clinical problem. Currently, radiotherapy and cisplatin concurrent radiotherapy are the standard treatments for primary nasopharyngeal carcinoma [6]. However, 10%-36% of patients with NPC still experience local recurrence after standard treatment [7]. Patients with recurrence or metastasis after treatment often have a poor prognosis, which is the main reason for the failure of treatment and the poor survival rate of NPC patients. Therefore, screening biomarkers of NPC for early detection, the prediction of prognosis and the monitoring of recurrence are of great significance for the clinical diagnosis and treatment of NPC.
Non-muscle myosin heavy chain IIA (NMHC IIA), which is encoded by MYH9 located at chromosome 22q11.2, is a dominating subunit of the actomyosin cytoskeleton and is generally considered to contribute to contraction of the cell posterior during migration [8, 9]. Many articles have reported that NMHC IIA plays an important role in tumor cell invasion and metastasis. A recent article reported that MYH9 promoted growth and metastasis in colorectal cancer [10]. One study showed that the long noncoding RNA PTCSC2 binding to the MYH9 protein was more likely to lead to thyroid cancer [11]. Another study showed that miR-647 inhibited the invasion and metastasis of gastric cancer cells by targeting the SRF/MYH9 axis [12]. The S100A4-MYH9 axis has been reported to promote the migration and invasion of gastric cancer cells by TGF-β-mediated epithelial-mesenchymal transition [13]. However, NMHC IIA has been identified as a tumor suppressor of squamous cell carcinomas [14]. So, the function and mechanism of NMHC IIA in NPC development is worth studying.
In the present study, we explored the significance of NMHC IIA in NPC tumorigenesis and invasion. A receiver operating characteristic (ROC) curve was constructed by plotting the data pairs for sensitivity and (1 - specificity) using The Cancer Genome Atlas (TCGA) database. Subsequently, co-expressed genes with MYH9 were predicted by LinkedOmics [15]. Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed by Metascape to explore the potential regulatory functions of MYH9 [16]. Transcription factors (TFs) and miRNA regulation network were constructed using Networkanalyst [17]. The migration and invasion abilities of nasopharyngeal carcinoma cells were evaluated by in vitro migration and matrigel invasion assays, respectively. Thus, our results could provide new evidence for understanding the molecular mechanisms of MYH9 in the invasion and metastasis of NPC cells.
Materials and methods
HNSCC patients and mRNA expression profiles
The RNA sequencing data of HNSCC and normal control samples were obtained from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) database. Patient exclusion criteria were as follows: i) first histologic diagnosis was not HNSCC; ii) the presence of another malignant tumor in addition to HNSCC; iii) incomplete data for analysis; and iv) received chemotherapy or radiotherapy. Overall, 141 HNSCC patient samples and 44 normal control samples were included in this study. Table 1 shows information on the tumor stage of these tissues.
The pathological stage information of HNSCC cancer tissues and adjacent normal tissues from TCGA database in this study.
| HNSCC tissues | |
|---|---|
| Stage | Number |
| NA | 10 |
| Stage I | 15 |
| Stage II | 40 |
| Stage III | 30 |
| Stage IVa | 43 |
| Stage IVb | 2 |
| Stage IVc | 1 |
LinkedOmics analysis
The LinkedOmics database (http://www.linkedomics.org/login.php) contains multi-omics data and clinical data for 32 cancer types. It was used to study differentially expressed genes in correlation with MYH9 in the RNAseq data type of TCGA HNSC cohort. MYH9 co-expression was analyzed statistically using Pearson's correlation coefficient, presenting in volcano plots and heat maps.
Functional Enrichment Analysis
The Metascape (http://metascape.org) online database was used to perform a functional enrichment analysis, which included GO and the KEGG analysis. The terms with minimum count of 3, p-value < 0.01 and enrichment factor of >1.5 were set as the cutoff criterion. Protein-protein Interaction Enrichment analysis was performed using Molecular Complex Detection (MCODE) algorithm.
Prediction of transcription factors and miRNAs
NetworkAnalyst (http://www.networkanalyst.ca) is an online visual analytics platform for comprehensive gene expression profiling. Transcription factors (TFs) and miRNAs were predicted by TF-miRNA coregulatory interactions function.
The Kaplan-Meier Plotter Analysis
The prognostic value of the mRNA expression of MYH9 with HNSCC was evaluated using the Kaplan-Meier plotter (www.kmplot.com) [18]. The overall survival of patients was determined by dividing the patient samples into two groups based on median expression (high vs. low expression). Log-rank p value < 0.05 was considered statistically significant.
Cell culture
NPEC2 Bmi-1 cells grown in keratinocyte/serum-free (KSF) medium (Invitrogen) are immortalized nasopharyngeal epithelial cells induced by oncogene Bmi-1. All human EBV-negative NPC cell lines (SUNE1, SUNE2, 6-10B, 5-8F, CNE2, HONE1 and HNE1) and the COS-7 cell line, an African green monkey kidney fibroblast-like cell line maintained in our laboratory, were cultured in RPMI 1640 medium (GIBCO) supplemented with 5% fetal bovine serum (FBS, GIBCO). All cells were grown in a humidified 5% CO2 incubator at 37 °C and passaged using standard cell culture techniques.
Cell transfection
Cell transfection was performed with PEI (Polysciences) according to the manufacturer's instructions. For the overexpression experiments, COS-7 cells were plated at a density of 4-6×104 cells per well into 24-well plates. Sixteen hours after seeding, the cells were grown to approximately 40% confluence, and each well received 1.5 ml Fugene HD and 0.75 mg of the indicated plasmids. For the siRNA experiments, the siRNAs that targeted NMHC-IIA were obtained from Guangzhou RiboBio Co., Ltd., and a total of 2.75 × 105 HNE1 cells were seeded into six-well plates. A final concentration of 50 nM siRNA duplex was reversely transfected with Lipofectamine RNAiMAX unless otherwise indicated. The sequences of the siRNAs used are as follows: NMHC-IIA siRNA #1 GCAACATCGTCTTCAAGAA and #2 ACACGGAGCTGATCAACGA.
Knockdown Assays
The shRNAs that targeted NMHC-IIA were obtained from Sigma. Lentivirus particles were generated by transfecting HEK-293T cells with pMD2.G and psPAX2 packaging plasmids and the NMHC-IIA shRNA-encoding plasmids using PEI (Polysciences) according to the manufacturer's instructions. Viruses were collected 48 h post-transfection and filtered. A total of 2×105 HNE1 cells were seeded into six-well plates. Sixteen hours after seeding, the cells were grown to approximately 40% confluence and infected with fresh lentivirus overnight in the presence of 10 µg/mL polybrene. Cells were screened by 1 µg/ml puro for 3 days. An HNE1 cell line stably silenced with NMHC-IIA was successfully established. The sequences of the short hairpin RNAs (shRNAs) used are as follows: NMHC-IIA shRNA #1 CCGGCCGCGAAGTCAGCTCCCTAAACTCGAGTTTAGGGAGCTGACTTCGCGGTTTTTG, #2 CCGGACGGAGATGGAGGACCTTATGCTCGAGCATAAGGTCCTCCATCTCCGTTTTTTG, #3 CCGGGACAGCAATCTGTACCGCATTCTCGAGAATGCGGTACAGATTGCTGTCTTTTTG and #5 CC GGGCCAAGCTCAAGAACAAGCATCTCGAGATGCTTGTTCTTGAGCTTGGCTTTTTG.
Quantitative real-time PCR and Western blot analysis
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen, Grand Island, NY) and reversely transcribed applying a reverse transcriptase kit (Invitrogen). The mRNA level was evaluated by real-time PCR using Power SYBR Green qPCR SuperMix-UDG (Invitrogen) and carried out using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). The procedure of the real-time PCR reaction as below: (1) 50 °C for 120 s; (2) pre-denaturation at 95 °C for 10 min; (3) denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 60 s, 40 cycles. All gene expression levels were normalized to that of the housekeeping gene GAPDH as an internal standard. The forward primer for MYH9 was 5'-CCATCACAGACACCGCCTACAG-3', and the reverse primer was 5'-CTTCTTGGTGTTCTCCGTCTTGC-3'. The forward primer for GAPDH was 5'-CGAGGTCATAGTTCCTGTTGGTG-3', and the reverse primer was 5'-CCCAATACGACCAAATCCGTT-3'.
Western blot analysis was carried out as previously described [19]. The Western blot was probed with polyclonal rabbit NMHC-IIA (ab13849, Abcam, 1:1,000 dilution), phospho-AKT1-Ser473 (#4060, Cell Signaling Technology, CST, 1:1,000 dilution), rabbit polyclonal anti-AKT (KC-5A01, Kangcheng Biotech., 1:1,000 dilution), p-EGFR (Tyr1068, #3777, CST, 1:1,000 dilution), EGFR (#4267, CST, 1:1,000 dilution), phospho-ERK1/2 (Thr202/Tyr204, #4376, CST, 1:1,000 dilution), p44/42 MAPK (Erk1/2) (#4695, CST, 1:1,000 dilution) and rabbit anti-GAPDH (BD, Transduction Laboratories, Lexington, UK) antibodies.
In vitro matrigel invasion and migration assays
In vitro invasion assays were performed to measure the ability of cells to invade a matrigel matrix overlying a membrane containing an 8 µm pore size polycarbonate filter. Cells (5 × 104) were seeded into medium containing 1% FBS in the top chamber coated with matrigel (BD, Transduction Laboratories, Lexington, UK), whereas medium containing 10% FBS was added to the bottom chamber. After cultivation for 48 h, the chambers were fixed with 1% paraformaldehyde and stained with hematoxylin. Invasive cells were plotted as the average number in ten random fields of view at 200× magnification for each filter. The data are expressed as the average number of cells migrating through the filters. The procedures for the migration assay were similar to those described for the matrigel invasion assay except that no matrigel was used, and the incubation time was 16 h.
Statistical analysis
Statistical analyses were performed using the statistical software package SPSS 16.0. Differences among variables were analyzed by 2-tailed Student's t tests. Data are presented as the mean ± SD unless otherwise indicated. p values ≤0.05 were considered statistically significant.
Results
Expression of NMHC IIA mRNA in HNSCC and matched adjacent normal tissues from the TCGA database
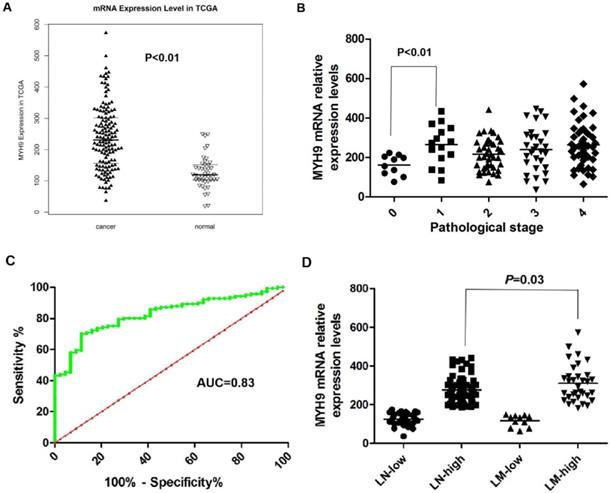
We previously reported that NMHC-IIA is an important factor for EBV infection in nasopharyngeal epithelial cells [20]. The public mRNA microarray results showed that the NMHC IIA mRNA level was upregulated in HNSCC tissues compared with that in the matched adjacent normal tissues (p<0.01, Figure 1A). Moreover, high expression levels of NMHC IIA mRNA were observed in high clinical pathological stage (I-IV) compared with zero clinical pathological stage in HNSCC (p<0.01, Figure 1B). A ROC curve was constructed on 141 HNSCC tissue samples and 44 control specimens. The results showed that the area under the ROC curve (AUC) reached up to 0.8303 (95% confidence interval (CI)=0.770-0.891; p<0.001, Figure 1C). The optimal cutoff value was 175.2, with a sensitivity and specificity of 70.21% and 86.36%, respectively. A high level of NMHC IIA in HNSCC was significantly correlated with lymph node metastasis (p < 0.05) (Figure 1D).
NMHC IIA mRNA expression levels in HNSCC cancer tissues and adjacent normal tissues. A. Enhanced expression of NMHC IIA mRNA expression levels in HNSCC cancer tissues compared with adjacent normal tissues (normal=44, cancer=141, p<0.01). B. NMHC IIA mRNA expression levels in HNSCC cancer of different pathological stages (T=141, p<0.01). C. The ROC curve for differentiating HNSCC tissues from controls. D. NMHC IIA had increased expression in lymph-node metastasis HNSCC compared with lymph-node non-metastasis tumors.
GO and KEGG pathway analyses of co-expression genes correlated with MYH9
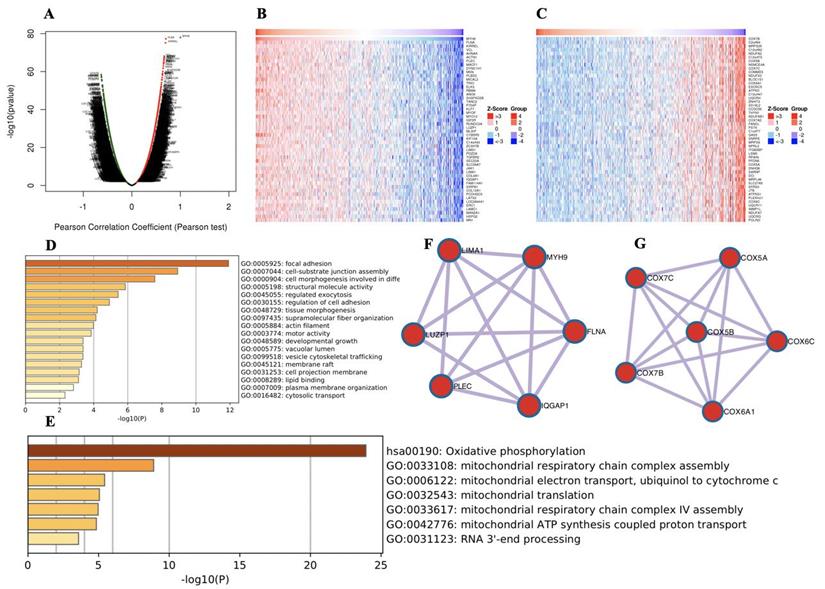
To further explore the functions of MYH9 in HNSCC, we analyzed the mRNA sequencing data from 520 HNSCC patients in the TCGA through LinkedOmics, as shown in the volcano plot (Figure 2A). The 50 significant genes set positively and negatively correlated with MYH9 as shown in the heat map (Figure 2B, 2C). MYH9 expression showed a strong positive association with expression of FLNA (positive rank #1, Pearson correlation = 0.70 p = 5.138e-78) and KIRREL (positive rank #2, Pearson correlation = 0.69, p = 6.868e-76). FLNA acts as a scaffold for the binding of more than 90 partners, which is critical for cell migration and adhesion[21]. GO and KEGG analysis by Metascape software showed that the top 50 significant genes set positively correlated with MYH9 were mainly enriched in focal adhesion, cell-substrate junction assembly and cell morphogenesis involved in differentiation (Figure 2D). The top 50 significant genes set negatively correlated with MYH9 were enrichment in oxidative phosphorylation, mitochondrial respiratory chain complex assembly and mitochondrial electron transport, ubiquinol to cytochrome c (Figure 2E). Subsequently, one core module was obtained from positive and negative gene PPI network using the MCODE clustering algorithm, respectively. Proteins involved in positive cluster were LIMA1, LUZP1, PLEC, IQGAP1, FLNA and MYH9 (Figure 2F). The enriched “Biological Process” GO terms contained regulation of actin filament-based process (4 genes), blood vessel development (3 genes). REACTOME pathway analysis showed that Cell-Cell communication (3 genes) was involved. Proteins involved in negative cluster were COX7C, COX7B, COX5B, COX6A1, COX6C and COX5A, which were mainly associated with mitochondrial electron transport, cytochrome c to oxygen and proton transmembrane transport in terms of Biological Process (Figure 2G).
Genes differentially expressed in correlation with MYH9 in HNSCC. A. A Pearson test was used to analyze correlations between MYH9 and genes expressed in HNSCC. B-C. Heat maps showing genes positively and negatively correlated with MYH9 (TOP 50). Red indicates positively correlated genes and green indicates negatively correlated genes. D-E. The significantly enriched GO annotations and KEGG pathways of MYH9 of top 50 significant gene sets (D) positively and (E) negatively correlated with MYH9. F-G. Network module obtained from the (F) positively and (G) negatively genes protein-protein interaction network.
Transcription factors, miRNA and survival analysis of FLNA and MYH9
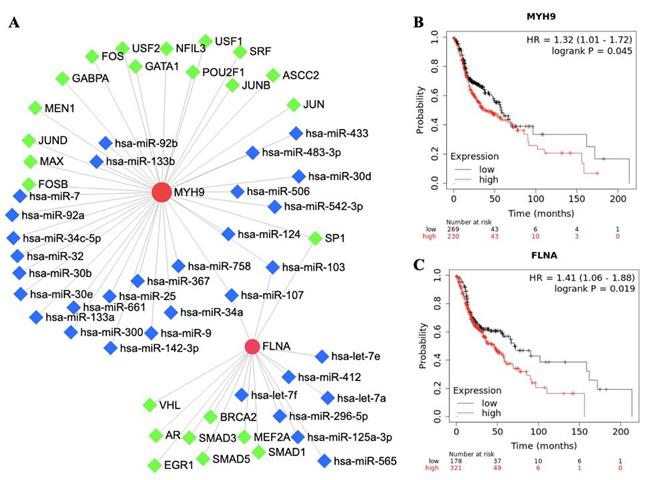
The roles of FLNA and MYH9 in tumorigenesis and development have been reported in cancer. To further understand the regulatory network of FLNA and MYH9, TFs and miRNA were predicted by NetworkAnalyst. FLNA was regulated by 9 TFs and MYH9 was regulated by 15 TFs. SP1, hsa-miR-103 and hsa-miR-107 were found to regulate FLNA and MYH9 (Figure 3A). The prognostic value of FLNA and MYH9 was obtained from Kaplan-Meier plotter. High expression of MYH9 (p = 0.045, HR = 1.32) and FLNA (p = 0.019, HR = 1.41) predicted poor overall survival (Figure 3B, 3C).
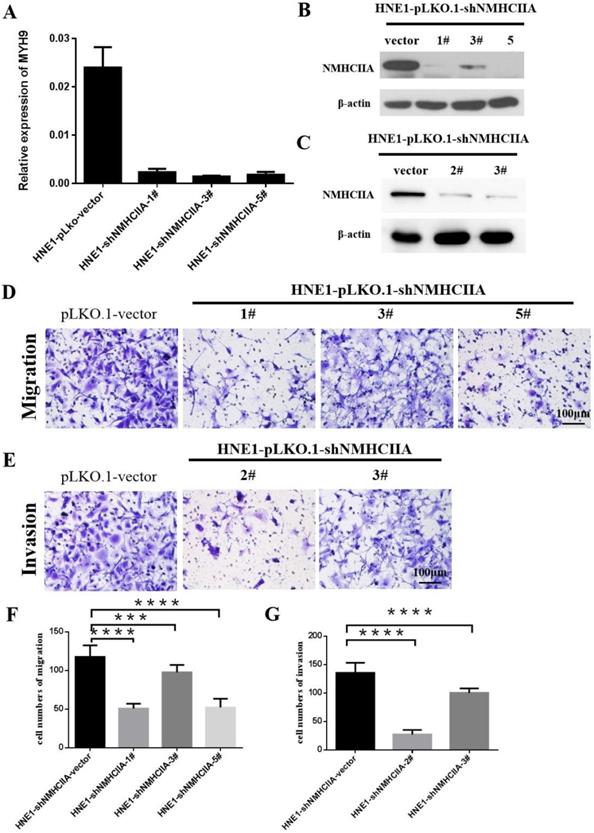
Silencing endogenous NMHC IIA reduces the migration and invasion of nasopharyngeal cancer cells
Based on the above findings, we next investigated the impact of NMHC IIA on migration and invasion in nasopharyngeal cancer cells. Four short hairpin RNAs for NMHC IIA were generated to reduce NMHC IIA expression in the HNE1 cell line, as shown in Figure 4A-C. The migration assay suggested that after the knockdown of NMHC IIA, the migration capability of HNE1 cells was obviously lower than that of the control group (Figure 4D, 4F). Boyden chamber invasion assays revealed that the invasiveness of HNE1 cells was dramatically decreased by the ablation of NMHC IIA using the shRNA-mediated silencing of endogenous NMHC IIA protein (Figure 4E, 4G). Moreover, the NMHC IIA siRNA was used to decrease NMHC IIA expression in HNE1 cells, and a similar result was obtained, as shown in Supplementary Figure 1. These findings indicate that silencing endogenous NMHC IIA reduces the invasion and metastatic abilities of nasopharyngeal cancer cells.
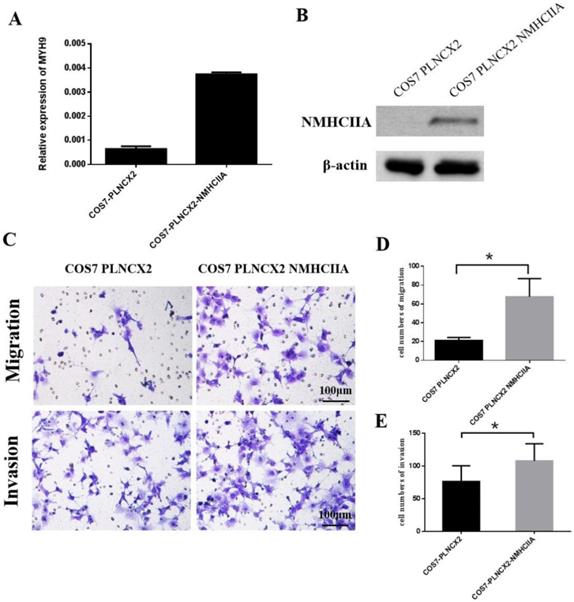
Overexpression of NMHC IIA enhances the migration and invasion of COS-7 cells in vitro
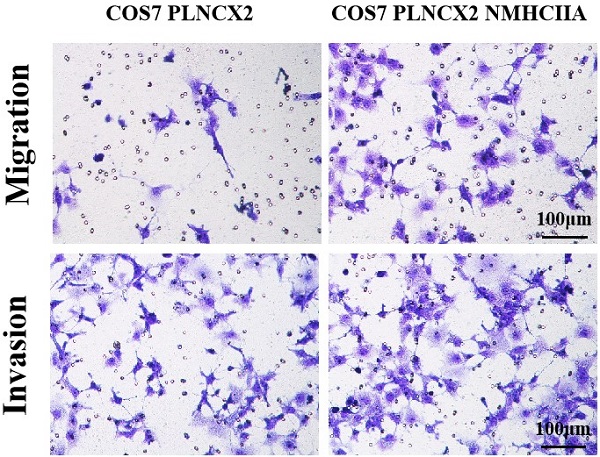
To further verify the influence of NMHC IIA on migration and invasion in cells, the NMHC IIA expression plasmid was stably transfected into the COS-7 cell line, which is derived from kidney tissue of the African green monkey (Cercopithecus aethiops) and does not express the NMHC IIA protein. Figure 5A and Figure 5B show that restored NMHC IIA expression was confirmed by real-time PCR and western blot assays in COS-7 cells. Migration and matrigel invasion chamber assays were then performed to determine the effect of NMHC IIA on the migration and invasion of COS-7 cells. The results showed that the overexpression of NMHC IIA notably increased COS-7 cell migration and invasion compared to that observed in control cells (Figure 5C-E).
The network and survival analysis of FLNA and MYH9. A. Transcription factors and miRNA of FLNA and MYH9. B-C. Kaplan-Meier analysis of overall survival in (B) MYH9 and (C) FLNA.
Suppression of endogenous NMHC IIA reduced cellular migration and invasion in HNE1. A-C. NMHC IIA expression was confirmed by Quantitative real-time PCR and western-blot in HNE1 cells expressing scrambled shRNA or NMHC IIA shRNA. D-E. The analysis of the migration and invasive properties in HNE1 cells expressing scrambled shRNA or NMHC IIA shRNA. The migratory or invasive cells were stained by crystal violet and then photographed by fluorescence inversion microscope system. Original magnification ×200. F. Migratory cells were plotted as the average number of cells per field of view from ten random fields. G. Invasive cells were plotted as the average number of cells per field of view from ten random fields.
Potential mechanism induced by NMHC IIA in NPC progression
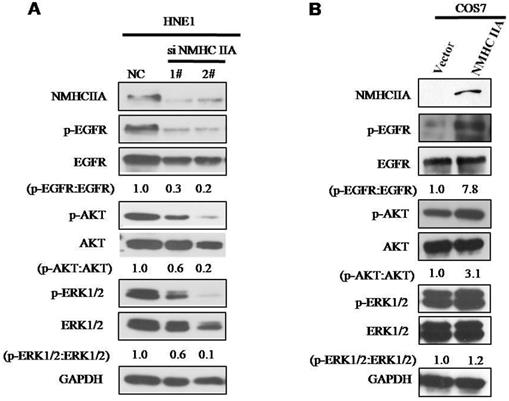
To explore the molecular mechanism underlying the effect of NMHC IIA on the migration and invasion of nasopharyngeal cells, we carried out Western blot analysis to examine the phosphorylation status of proteins associated with cancer signaling. EGFR, as well as its critical downstream signaling components AKT and ERK, is aberrantly expressed in NPC [21]. As shown in Figure 6, silencing endogenous NMHC IIA in HNE1 cells decreased the expression and phosphorylation of EGFR, AKT and ERK. Overexpression of NMHC IIA in COS7 cells enhances the expression and phosphorylation of EGFR, AKT and ERK. These results suggest that increasing NMHC IIA expression could be responsible for enhancing NPC progression by inducing EGFR, AKT and ERK phosphorylation.
The exogenous expression of NMHC IIA enhanced the migration and invasiveness in COS7 cells. A. Quantitative real time PCR analysis of MYH9 gene in COS7 cells expressing control vector PLNCX2 or PLNCX2/NMHC IIA. The relative fold increase of transcripts was normalized to the amount of RNA harvested from cells expressing control vector PLNCX2. GAPDH served as the internal control. The data were presented as the mean ± SD (n = 3). B. Western blot analysis of NMHC IIA protein in COS7 cells expressing control vector PLNCX2 or PLNCX2/NMHC IIA. β-actin was used as a loading control. C. The migratory and invasive abilities induced by NMHC IIA were analyzed in COS7 cells expressing control vector PLNCX2 or PLNCX2/NMHC IIA. The migratory or invasive cells were stained by crystal violet and then photographed by fluorescence inversion microscope system (200×). E. Migratory cells or invasive cells were plotted as the average number of cells per field of view from ten random fields.
NMHC IIA have effect on the expression of phosphorylation of EGFR, AKT and ERK. A. Silencing endogenous NMHC IIA in HNE1 cells decreased expression of phosphorylation of EGFR, AKT and ERK. B. Overexpression of NMHC IIA in COS7 cells enhances the expression and phosphorylation of EGFR, AKT and ERK.
Discussion
NPC is a squamous cell carcinoma that usually occurs around the osculum of the Eustachian tube in the lateral wall of the nasopharynx [22]. The environmental, genetic, and viral causative factors, either acting alone or in combination, would generate multiple epigenetic and genetic alterations [23]. The development of NPC involves the accumulation of multiple epigenetic and genetic changes, leading to the evolution of clonal cell populations, which have growth advantages over other cells [2]. In our study, we demonstrated a significant increase in NMHC IIA expression at the mRNA level between HNSCC cancer and adjacent normal HNSCC tissues using a public data. This result was consistent with those of previous studies of other human cancers [24]. Our results showed that the AUC of MYH9 reached up to 0.8303. The optimal cutoff value was 175.2, with a sensitivity and specificity of 70.21% and 86.36%, respectively. This result indicated that MYH9 may be as new candidate biomarkers for HNSCC patients by bioinformatics analysis.
To gain detailed insights into the potential functions of MYH9 in HNSCC and its regulatory network, we performed bioinformatics analysis of TCGA HNSCC RNAseq sequencing data to guide MYH9 research. We found that MYH9 expression showed a strong positive association with expression of FLNA. Filamin A (FLNa), encoded by the FLNA gene, crosslinks various signaling molecules and membrane receptors. Previous studies identified the FLNA as determinant in cancer progression and metastasis through affecting cancer cell growth and migration [25, 26], but the role of FLNA in NPC is still unknown. In our study, high expression of MYH9 and FLNA predicted poor overall survival obtained from Kaplan-Meier plotter. Moreover, FLNA and MYH9 are substrates of Mechanistic Target of Rapamycin Complex 2 (mTORC2), which is a key regulator in tumorigenesis, promoting cell growth, supporting their irregular or metastatic ability for diseased cells [27]. We suspected that FLNA and MYH9 are effectors of mTORC2 controlling the motility and invasion of NPC cells.
To further explore the co-expression genes correlated with MYH9, we constructed the PPI network using top 50 significant genes positively and negatively correlated with MYH9. The positive core cluster is consisted of LIMA1, LUZP1, PLEC, IQGAP1, FLNA and MYH9. LUZP1 was identified as a FLNA binding partner by Wang et al. [28]. Knockdown of LIMA1 has been demonstrated to enhance cancer cell invasion [29]. IQGAP1 as a scaffold connects phosphoinositide signaling to cytoskeletal reorganization [30]. Those results show that six hub genes might be essential in the function of NPC cell development. Interestingly, negative core cluster mainly regulate mitochondrial function. Cytochrome c oxidase (COX) is one of the principle enzymes in mitochondria-mediated apoptosis and cell respiration. The expression alteration of those genes may provide novel insights into the development of NPC from the perspective of energy metabolite.
We found that MYH9 in HNSCC was associated with a network of TFs including SP1, SRF, JUN and FOS. SP1 was verified as a direct functional target of miR-24, which enhances cell viability and the radiosensitivity of NPC cells [31]. SRF regulates the transcriptional activity of SNAIL in NPC cells [32]. Jun proteins (c-Jun, JunB, JunD) and Fos proteins (c-Fos, FosB, Fra-1, Fra-2) are belongs to Activator protein 1 (AP-1), which played an important role in the prognosis of oral squamous cell carcinoma. hsa-miR-103 and hsa-miR-107 were regulators of FLNA and MYH9. Studies have elucidated that hsa-miR-103 and hsa-miR-107 were involved in Wnt3a/β-catenin/ATF6 signaling pathway and were critical to the progression of colorectal cancer, Alzheimer's disease, breast cancer and cardiac function [33-36]. The TFs and miRNA regulation network may promote understanding of the molecular mechanisms of NPC development.
More than 90% of cancer deaths are caused by metastases, not the primary tumors from which these malignant lesions arise [24, 37]. Therefore, it is necessary to elucidate the potential factors of tumorigenicity, invasion and metastasis of NPC which was warranted to develop novel treatments and cures. In our study, the intense expression of NMHC IIA in HNSCC was correlated with lymph node metastasis using a bioinformatics data, while silencing endogenous NMHC IIA reduced the migration and invasion of nasopharyngeal cancer cell lines in vitro. This result is similar to a recent study that reported that the endogenous overexpression of NMHC IIA enhanced cell migration in vitro [10]. Our study indicated that NMHC IIA expression was potentially responsible for enhancing NPC progression by inducing EGFR, AKT and ERK phosphorylation. Additional work is required to investigate the mechanism by which NMHC IIA regulates these pathways in the invasion and metastasis of NPC. Therefore, exploring the function and mechanism of these pathways can help us better understand the pathogenicity of NPC cells and aid in the development of more sensitive and effective diagnostic targets.
Conclusion
In conclusion, our study revealed MYH9 as new candidate biomarkers for HNSCC patients by bioinformatics analysis. MYH9 expression was strong positive association with expression of FLNA using TCGA HNSCC RNAseq sequencing data. High MYH9 and FLNA expression were associated with the poorer overall survival in HNSCC.MYH9 was positively correlated genes regulate focal adhesion, cell-substrate junction assembly and cell morphogenesis involved in differentiation by performing GO and KEGG analysis. MYH9 was associated with a network of TFs including SP1, SRF, JUN and FOS in HNSCC. Moreover, silencing endogenous NMHC IIA reduced the migration and invasion of the nasopharyngeal cancer cell line in vitro by inhibiting the phosphorylation of EGFR, AKT and ERK, while overexpression of NMHC IIA notably increased cells migration and invasion by enhancing the phosphorylation of EGFR, AKT and ERK. These results indicated that the transcript levels of MYH9 may be a suitable biomarker of NPC.
Abbreviations
HNSCC: Head and neck squamous cell carcinoma; NPC: Nasopharyngeal carcinoma; NMHC IIA: Non-muscle myosin heavy chain IIA; ROC: A receiver operating characteristic; TFs: Transcription factors; PPI: protein-protein interaction network.
Supplementary Material
Supplementary figure S1.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant NO. 81772921, 81502344). A grant from the Science and Technology Planning Project of Shenzhen City of China (NO. JCYJ20180306172209668). The discipline construction ability promotion project of Shenzhen health and population family planning commission (NO. SZXJ2017018). Sanming Project of Medicine in Shenzhen (NO. SZSM201601062). The Natural Science Foundation of Guangdong Province (NO. 2016A030313257). The Science and Technology Planning Project of Guangdong Province (NO.2014A020212141).
Authorship
X Zhang, J Li, D Xiong and D Liu established and supervised the study. D Xiong and D Liu designed analysis framework. W Wu and D Chen analyzed data. X Dou and X Ji performed the literature research and statistical analysis. X Zhang, J Li, D Xiong, D Chen and D Liu made manuscript revisions. All authors reviewed the results and approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Economopoulou P, de Bree R, Kotsantis I, Psyrri A. Diagnostic Tumor Markers in Head and Neck Squamous Cell Carcinoma (HNSCC) in the Clinical Setting. Front Oncol. 2019;9:827
2. Lo KW, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:451-62
3. Shivappa N, Hébert JR, Zucchetto A, Montella M, Libra M, Garavello W. et al. Increased Risk of Nasopharyngeal Carcinoma with Increasing Levels of Diet-Associated Inflammation in an Italian Case-Control Study. Nutr Cancer. 2016;68:1123-30
4. Mahdavifar N, Ghoncheh M, Mohammadian-Hafshejani A, Khosravi B, Salehiniya H. Epidemiology and Inequality in the Incidence and Mortality of Nasopharynx Cancer in Asia. Osong Public Health Res Perspect. 2016;7:360-72
5. Janvilisri T. Omics-based identification of biomarkers for nasopharyngeal carcinoma. Dis Markers. 2015;2015:762128
6. Bhattacharyya T, Babu G, Kainickal CT. Current Role of Chemotherapy in Nonmetastatic Nasopharyngeal Cancer. J Oncol. 2018;2018:3725837
7. Yu YH, Xia WX, Shi JL, Ma WJ, Li Y, Ye YF. et al. A model to predict the risk of lethal nasopharyngeal necrosis after re-irradiation with intensity-modulated radiotherapy in nasopharyngeal carcinoma patients. Chin J Cancer. 2016;35:59
8. Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G. et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704-9
9. Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D. et al. Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69:530-9
10. Wang B, Qi X, Liu J, Zhou R, Lin C, Shangguan J. et al. MYH9 Promotes Growth and Metastasis via Activation of MAPK/AKT Signaling in Colorectal Cancer. J Cancer. 2019;10:874-84
11. Wang Y, He H, Li W, Phay J, Shen R, Yu L. et al. MYH9 binds to lncRNA gene PTCSC2 and regulates FOXE1 in the 9q22 thyroid cancer risk locus. Proc Natl Acad Sci U S A. 2017;114:474-9
12. Ye G, Huang K, Yu J, Zhao L, Zhu X, Yang Q. et al. MicroRNA-647 Targets SRF-MYH9 Axis to Suppress Invasion and Metastasis of Gastric Cancer. Theranostics. 2017;7:3338-53
13. Li F, Shi J, Xu Z, Yao X, Mou T, Yu J. et al. S100A4-MYH9 Axis Promote Migration and Invasion of Gastric Cancer Cells by Inducing TGF-β-Mediated Epithelial-Mesenchymal Transition. J Cancer. 2018;9:3839-49
14. Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D. et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309-13
15. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956-d63
16. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523
17. Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234-w41
18. Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725-31
19. Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX. et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol. 2018;3:1-8
20. Xiong D, Du Y, Wang HB, Zhao B, Zhang H, Li Y. et al. Nonmuscle myosin heavy chain IIA mediates Epstein-Barr virus infection of nasopharyngeal epithelial cells. Proc Natl Acad Sci U S A. 2015;112:11036-41
21. Shao QQ, Zhang TP, Zhao WJ, Liu ZW, You L, Zhou L. et al. Filamin A: Insights into its Exact Role in Cancers. Pathol Oncol Res. 2016;22:245-52
22. Sham JS, Wei WI, Zong YS, Choy D, Guo YQ, Luo Y. et al. Detection of subclinical nasopharyngeal carcinoma by fibreoptic endoscopy and multiple biopsy. Lancet. 1990;335:371-4
23. Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423-8
24. Kas SM, de Ruiter JR, Schipper K, Annunziato S, Schut E, Klarenbeek S. et al. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet. 2017;49:1219-30
25. Cheng M, Jiang Y, Yang H, Zhao D, Li L, Liu X. FLNA promotes chemoresistance of colorectal cancer through inducing epithelial-mesenchymal transition and smad2 signaling pathway. Am J Cancer Res. 2020;10:403-23
26. Wang K, Zhu TN, Zhao RJ. Filamin A regulates EGFR/ERK/Akt signaling and affects colorectal cancer cell growth and migration. Mol Med Rep. 2019;20:3671-8
27. Chantaravisoot N. Characterizing the Mechanistic Target of Rapamycin Complex 2 Pathway in Glioblastoma Multiforme. Dissertations & Theses - Gradworks. 2015
28. Wang J, Nakamura F. Identification of Filamin A Mechanobinding Partner II: Fimbacin Is a Novel Actin Cross-Linking and Filamin A Binding Protein. Biochemistry. 2019;58:4737-43
29. Ohashi T, Idogawa M, Sasaki Y, Tokino T. p53 mediates the suppression of cancer cell invasion by inducing LIMA1/EPLIN. Cancer Lett. 2017;390:58-66
30. Yerramilli VS, Ross AH, Reisinger J, Plante K, Gericke A. IQGAP1 Scaffolding Connects EGFR and Phosphoinositide Signaling to Cytoskeletal Reorganization. Biophysical Journal. 2020;118:246a
31. Kang M, Xiao J, Wang J, Zhou P, Wei T, Zhao T. et al. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 2016;5:1163-73
32. Ren X, Yang X, Cheng B, Chen X, Zhang T, He Q. et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun. 2017;8:14053
33. Wang J, Chen C, Zhang Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer's disease. J Clin Lab Anal. 2020;34:e23006
34. Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J. et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631-41
35. Rech M, Kuhn AR, Lumens J, Carai P, van Leeuwen R, Verhesen W. et al. AntagomiR-103 and -107 Treatment Affects Cardiac Function and Metabolism. Mol Ther Nucleic Acids. 2019;14:424-37
36. Triulzi T, Iorio MV, Tagliabue E, Casalini P. microRNA: New Players in Metastatic Process. 2013.
37. Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895-904
Author contact
![]() Corresponding authors: E-mail: zxm0760com; or, lijianentcom.
Corresponding authors: E-mail: zxm0760com; or, lijianentcom.

 Global reach, higher impact
Global reach, higher impact