Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(15):4585-4594. doi:10.7150/jca.59933 This issue Cite
Research Paper
Hallmark microRNA signature in liquid biopsy identifies hepatocellular carcinoma and differentiates it from liver metastasis
1. OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China.
2. Department of Clinical Oncology, LKS Faculty of Medicine, The Hong Kong Special Administrative Region, People's Republic of China.
* Contributed equally to this work.
Received 2021-3-1; Accepted 2021-5-19; Published 2021-6-1
Abstract
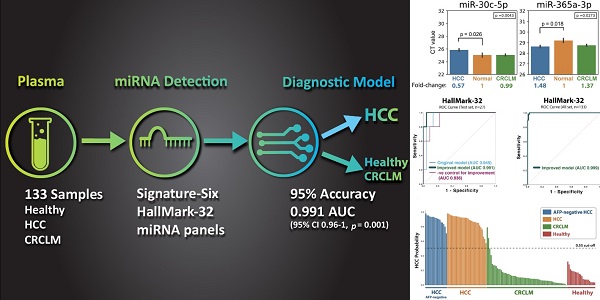
Purpose: This study aims to develop a liquid biopsy assay to identify HCC and differentially diagnose hepatocellular carcinoma (HCC) from colorectal carcinoma (CRC) liver metastasis. Methods: Thirty-two microRNAs (“HallMark-32” panel) were designed to target the ten cancer hallmarks in HCC. Quantitative PCR and supervised machine learning models were applied to develop an HCC-specific diagnostic model. One hundred thirty-three plasma samples from intermediate-stage HCC patients, colorectal cancer (CRC) patients with liver metastasis, and healthy individuals were examined. Results: Six differentially expressed microRNAs (“Signature-Six” panel) were identified after comparing HCC and healthy individuals. The microRNA miR-221-3p, miR-223-3p, miR-26a-5p, and miR-30c-5p were significantly down-regulated in the plasma of HCC samples, while miR-365a-3p and miR-423-3p were significantly up-regulated. Machine learning models combined with HallMark-32 and Signature-Six panels demonstrated promising performance with an AUC of 0.85-0.96 (p ≤ 0.018) and 0.84-0.93 (p ≤ 0.021), respectively. Further modeling improvement by adjusting sample quality variation in the HallMark-32 panel boosted the accuracy to 95% ± 0.01 and AUC to 0.991 (95% CI 0.96-1, p = 0.001), respectively. Even in alpha fetoprotein (AFP)-negative (< 20ng/mL) HCC samples, HallMark-32 still achieved 100% sensitivity in identifying HCC. The Cancer Genome Atlas (TCGA, n=372) analysis demonstrated a significant association between HallMark-32 and HCC patient survival. Conclusion: To the best of our knowledge, this is the first report to utilize circulating miRNAs and machine learning to differentiate HCC from CRC liver metastasis. In this setting, HallMark-32 and Signature-Six are promising non-invasive tests for HCC differential diagnosis and distinguishing HCC from healthy individuals.
Keywords: Hepatocellular carcinoma, miRNA signature, Liquid biopsy, Machine learning, HCC diagnosis, HCC screening
1. Introduction
Hepatocellular carcinoma (HCC) is a highly fatal cancer with a death toll over 810,000 deaths every year worldwide [1]. Most HCC patients are detected only in advanced stages with a dismal prognosis of less than one-year overall survival [2-4]. Diagnostic and treatment delays are often linked to worse survival outcomes in HCC [5, 6]. Nearly 20% of patients wait for more than three months from clinical presentation to diagnosis, which is close to the tumor volume doubling time of HCC [6, 7]. Therefore, early diagnosis is the pivotal key to improving outcomes for HCC. Over the last decade, the advent of biotechnology has been expediting liquid biopsy application in various clinical settings.
Developing a liquid biopsy assay for HCC diagnosis is clinically useful for several reasons. First, liquid biopsy could potentially complement conventional tissue biopsy for HCC diagnosis. Tissue biopsy has common pitfalls including sampling bias and inaccessibility for safe biopsy. For instance, invasive biopsy procedures are sometimes not feasible for HCC patients with tumors located adjacent to major blood vessels or patients with coagulopathy due to imposing high intra-abdominal bleeding risk [8, 9]. In contrast, liquid biopsy technology is non-invasive, repeatable and thus can provide a safe and timely diagnosis of early HCC.
Second, liquid biopsy could potentially complement the detection limit in the traditional approaches for non-invasive HCC surveillance and diagnosis. For screening early-stage HCC, liver ultrasound combined with AFP testing could only achieve 63% sensitivity [10]. The sensitivity could be further reduced by 19 - 56% in obesity and chronic liver disease conditions [11]. Computed tomography (CT) and Magnetic Resonance Imaging (MRI) sensitivities for 1-2 cm HCC are 65% and 80%-92%, respectively, but the sensitivities plummet to 10% and 34%-71% for early HCC tumors <1 cm [12]. An additional liquid biopsy technology may improve the performances of current HCC surveillance strategy.
Third, liquid biopsy could aid the differential diagnosis of liver nodules, especially those with equivocal imaging features and those not accessible to biopsy. Since the liver is a common site for metastasis, differentiating HCC from liver metastasis patients is essential for treatment guidance. For instance, gastrointestinal cancers are known to have a high tendency to metastasize to the liver through the portal vein. Furthermore, patients with underlying chronic liver diseases may have a higher risk of HCC and CRC [13], making the differential diagnosis more essential. More confusingly, a subset of CRC is indistinguishable from HCC with respect to the serological AFP and carcinoembryonic antigen (CEA) levels. Approximately 2.6% (5/193) of CRC patients are positive in the AFP test [14] and 45% (9/20) of AFP-positive CRC patients are negative in the CEA test [15]. Therefore, it is an unmet need to develop a liquid biopsy test to differentiate HCC from colorectal cancer liver metastasis (CRCLM).
This study aims to develop a non-invasive liquid biopsy microRNA (miRNA) assay for HCC diagnosis. We began by selecting 32 miRNAs (“HallMark-32” panel) known to regulate the ten hallmarks in HCC. Subsequently, we identified six signature miRNAs (“Signature-Six” panel) based on their expression profiles. HCC-specific diagnostic models were then developed by supervised machine learning. The model diagnostic performances were evaluated using 133 plasma samples from HCC, CRCLM, and healthy individuals. The goal of this study is to develop a reliable liquid biopsy for HCC identification and differential diagnosis.
2. Methodology
2.1. Patient & blood collection
Patients diagnosed with HCC and CRCLM from July 2015 to Dec 2019 at Queen Mary Hospital were recruited. All HCC samples were from patients diagnosed with Barcelona Clinic Liver Cancer (BCLC) intermediate stage B. All CRCLM samples were from patients diagnosed with CRC having synchronous liver metastasis. All blood samples were obtained after written informed consent. A total of 133 samples were collected including 22 healthy individuals, 63 CRCLM, and 48 HCC patients. Eight milliliters of peripheral blood samples were collected in the Streck Cell-Free DNA tubes (Streck Inc., USA). The study was approved by the Institutional Review Board of the University of Hong Kong.
2.2. Sample preparation and miRNA isolation
Blood sample were spun down at 2,000g for 20 minutes at 18°C, followed by 16,000rpm centrifugation for 10 minutes at 10°C. The plasma was then transferred to a new 1.5mL centrifuge tube and stored at -20°C. After centrifugation, the supernatant was further precipitated by isopropanol, followed by RNA purification using a column-based silica membrane technology (MACHEREY-NAGEL, Germany). The eluted RNA was subjected to PolyA tailing reaction with PolyA polymerase and ATP and incubated for 60 minutes at 37°C, followed by 5-minute deactivation at 70°C. Subsequently, cDNA synthesis was performed by adding oligo-dT adapter primer, miRNA-specific forward primer (Table S1), reverse transcriptase and then the reaction was incubated for 20 minutes at 42°C, followed by 5 minutes at 85°C.
2.3. miRNA analysis
Roche LightCycler 480 was used for qPCR reactions. The CT values for microRNA targets were generated by the second derivative maximum of the fluorescence curve. For the miRNAs that have no CT value after calculation, a CT value of 45 was filled to indicate low expression level for the subsequent diagnostic modeling, and the CT values were converted to fold differences by the equation 2^(-ΔCt). Samples missing more than 50% of miRNA CT values were excluded from this study. Analysis of variance (ANOVA) and Student's T-test was applied to examine the difference of miRNA expression between HCC, healthy, and CRCLM groups using the statistic package in python. The p-values <0.05 were considered statistically significant. The seaborn and matplotlib package in python were used for results visualization.
2.4. Data preprocessing and machine learning models
Data were split into training (n=106) and test (n=27) sets with an 80% splitting ratio. Samples were balanced by using the imblearn package and Scikit-learn library in the python environment. Four supervised classifier algorithms: Artificial Neural Network, Random Forest, Gradient Boosting Classifier, and Logistic Regression were tested for their performances to distinguish HCC samples from healthy and CRCLM samples. The diagnostic model prediction outcome is binary; HCC samples were assigned as one, while CRCLM and healthy individuals were assigned as zero. For model improvement, the sample quality data included RNA concentration and their 260/280 and 260/230 absorbance ratios, cDNA concentration and their 260/280 and 260/230 absorbance ratios, and the specificity of each miRNA melting curve. All features were standardized by the StandardScaler package in python. For each algorithm, 30 models were first developed, and their mean accuracy was taken for evaluation. To further examine the performance of the algorithms, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), area under curve (AUC), receiver-operating-characteristic (ROC) curves were computed in python. All experiments, analysis, and machine learning modelling for the healthy, HCC, and CRCLM were repeated at least three times.
2.5. Establishing HallMark-32 panel
To select the miRNA candidates for HCC detection, we reviewed literature that reported miRNA in HCC and then selected miRNAs accordingly to ten hallmark properties proposed by Hanahan and Weinberg [16]. The hallmarks included sustaining proliferation signaling, evading growth suppressor, avoiding immune destruction, enabling replicative immortality, tumor-promoting inflammation, activating invasion and metastasis, inducing angiogenesis, genome instability, resisting cell death, and deregulating cellular energetics. All hallmarks of HCC were covered by at least one of the miRNAs in the HallMark-32 panel (Table 1) (Table S2).
2.6. TCGA miRNA expression and survival data analysis
The liver hepatocellular carcinoma (LIHC) dataset (version 2016-01-28, n=372) from TCGA database was downloaded. The vital status (dead vs. alive), survival days (days to death or days to last follow-up), and miRNA expressions (reads per million miRNA mapped) were analyzed. Kaplan-Meier (KM) survival curve analysis was performed in SPSS software. Cox proportional hazards analysis was computed in python.
3. Results
3.1. Discovery of HCC-specific Signature-Six miRNA panel
To identify signature miRNAs that express differently between HCC and healthy individuals for HCC detection, we examined the expression profiles of 32 miRNAs by qPCR in 70 plasma samples (48 HCC and 22 healthy individuals) (Table S3 and Figure S1). Six signature miRNAs (miR-221-3p, miR-223-3p, miR-26a-5p, miR-30c-5p, miR-365a-3p and miR-423-3p) were identified. The level of miR-365a-3p and miR-423-3p was significantly elevated in the plasma of HCC samples compared to healthy samples, while miR-221-3p, miR-223-3p, miR-26a-5p, and miR-30c-5p were significantly down-regulated (Table 2 and Figure 1). These miRNAs were then consolidated to be the “Signature-Six” panel.
HallMark-32 panel design based on the published miRNA HCC studies.
| HCC Hallmarks | MicroRNA Candidates [References are shown in Supplementary Table 3] |
|---|---|
| Sustaining Proliferative Signaling | 150-5p, 125b-5p, 101-3p, 1246, 21-5p, 145-5p, 214-3p, 320d, 18a-5p, 26a-5p, 193a-5p, 19a-3p, 222-3p, 486-5p, 223-3p, 374a-5p, 424-5p, 122-5p, 29a-3p, 451a |
| Evading Growth Suppressors | 574-3p, 125b-5p, 23a-3p, 145-5p, 214-3p, 423-3p, 423-5p, 424-5p |
| Avoiding Immune Destruction | 23a-3p, 423-5p, 424-5p |
| Enabling Replicative Immortality | 1246, 21-5p, 192-5p, 148a-3p |
| Tumor Promoting Inflammation | 148a-3p, 30c-5p |
| Activating Invasion And Metastasis | 150-5p, 125b-5p, 191-5p, 101-3p, 1246, 21-5p, 145-5p, 125a-5p, 214-3p, 26a-5p, 19a-3p, 148a-3p, 486-5p, 374a-5p, 221-3p, 424-5p, 122-5p, 29a-3p, 451a |
| Inducing Angiogenesis | 148a-3p, 423-3p, 424-5p |
| Genome Instability And Mutation | 374a-5p, 221-3p |
| Resisting Cell Death | 125b-5p, 101-3p, 1246, 145-5p, 125a-5p, 192-5p, 18a-5p, 26a-5p, 193a-5p, 222-3p, 223-3p, 423-5p |
| Deregulating Cellular Energetics | 101-3p, 22-5p |
Expression level of miRNAs in Signature-Six, fold-change relative to healthy, and the p-value results in t-test and ANOVA analyses.
| miRNA | Mean (CT values) | SEM (CT values) | Fold-Change relative to Healthy | HCC vs. CRCLM (p-value) | HCC vs. Healthy (p-value) | HCC vs. CRCLM vs. Healthy (p-value) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCC | Healthy | CRCLM | HCC | Healthy | CRCLM | HCC | Healthy | CRCLM | ||||
| 221-3p | 28.15 | 27.27 | 26.29 | 0.21 | 0.36 | 0.25 | 0.55 | 1 | 1.97 | 0.000** | 0.042* | 0.000** |
| 223-3p | 25.72 | 24.14 | 22.81 | 0.25 | 0.73 | 0.28 | 0.33 | 1 | 2.51 | 0.000** | 0.045* | 0.000** |
| 26a-5p | 28.14 | 26.77 | 26.1 | 0.26 | 0.62 | 0.25 | 0.39 | 1 | 1.6 | 0.000** | 0.046* | 0.000** |
| 30c-5p | 25.85 | 25.04 | 25.05 | 0.17 | 0.31 | 0.17 | 0.57 | 1 | 0.99 | 0.001** | 0.026* | 0.004** |
| 365a-3p | 28.63 | 29.2 | 28.75 | 0.12 | 0.2 | 0.09 | 1.48 | 1 | 1.37 | 0.474 | 0.018* | 0.027* |
| 423-3p | 29.51 | 30.35 | 29.54 | 0.23 | 0.23 | 0.19 | 1.79 | 1 | 1.75 | 0.917 | 0.012* | 0.062 |
Notes: * p-value ≤ 0.05; ** p-value ≤ 0.01; High CT value means low miRNA expression.
Bar chart showing the expression of Signature-Six in HCC, Healthy and CRCLM specimens, and their fold change relative to healthy. The p-values at the upper right corner represent p-value in ANOVA test comparing the CT values in three groups (HCC, Healthy and CRCLM). The p-values at the top of the bars represent p-value in t-test comparing HCC and healthy individuals.
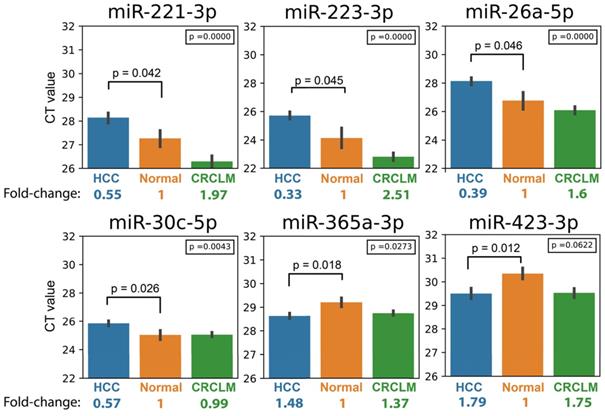
Next, we investigated whether Signature-Six expressed differently among three groups of specimens (HCC, CRCLM, and Healthy) using ANOVA analysis. Five miRNAs in Signature-Six consistently showed statistical significance (Table 2). The expression of miR-223-3p in the Signature-Six panel achieved the highest significance (p-value=8.56 x 10-9). Furthermore, when comparing the miRNA expressions between HCC and CRCLM, four miRNAs in Signature-Six were significantly different (Table 2). As expected, the internal control miR-451a exhibited no difference in all statistical tests (Table S3). Taken together, the differential expression profile of miRNAs provided a strong foundation for building the HCC diagnostic model.
3.2. HCC Diagnostic Model development with Signature-Six and HallMark-32
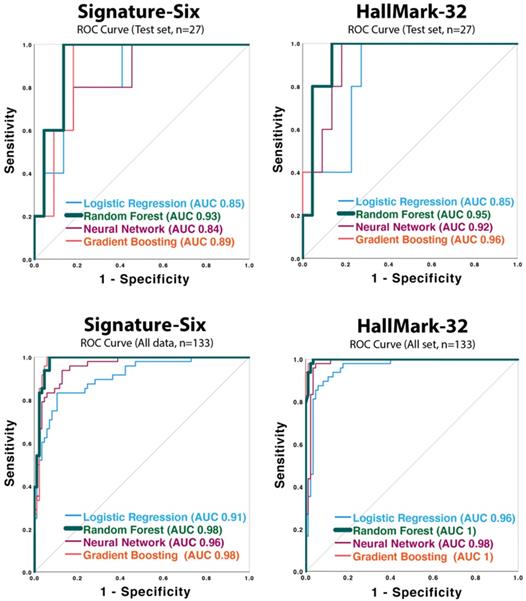
To develop an HCC diagnostic model, four supervised machine learning classification models (Neural Network, Random Forest, Gradient Boosting Classifier, and Logistic Regression) were applied. One hundred thirty-three plasma samples (48 HCC, 63 CRCLM, 22 healthy individuals) were utilized. Each machine learning model was developed using a training dataset (n=106), and then the performance was validated in a test dataset (n=27). Performance indices such as accuracy, AUC score, p-value for AUC, sensitivity, specificity, PPV, and NPV were evaluated for each model and are summarized in Table 3. ROC curves are shown in Figure 2. All the AUC scores in both HallMark-32 and Signature-Six are statistically significant (p ≤ 0.021) (Table 3 and Figure 2). In HallMark-32, the average AUCs from the four algorithms in the test set (n=27) and the whole dataset (n=133) were 0.92 and 0.99 (Table 3A), which were slightly higher than that in Signature-Six (Table 3B, average AUC is 0.88 and 0.96). In both HallMark-32 and Signature-Six, the Random Forest model outperformed other machine learning models based on mean accuracy. In HallMark-32, the Random Forest model provided 91% accuracy compared to 86%, 89%, 84% accuracy in Neural Network, Gradient Boosting Classifier, and Logistic Regression models, respectively (Table 3). The Signature-Six Random Forest model exhibited an AUC score of 0.93 (95% CI: 0.83-1, p-value = 0.003), 100% sensitivity, and 86% specificity; the HallMark-32 Random Forest model exhibited an AUC score of 0.95 (95% CI: 0.86-1, p-value = 0.002), 91% sensitivity, and 91% specificity. Taken together, the HallMark-32 Random Forest model provided the best performance for proceeding further model improvement.
Receiver operating characteristic (ROC) curves of the neural network, random forest, gradient boost classifier, and logistic regression models applied to Signature-Six and HallMark-32 panel in test set (n=27) and the whole dataset (n=133).
3.3. Model improvement by sample quality adjustment
Sample quality variation may influence the miRNA detection and, thus, the result of the diagnostic model. To further improve the HallMark-32 HCC diagnostic model, we integrated the sample quality variation into the models. The sample quality features involved 38 parameters covering RNA concentration, RNA 260/280 and 260/230 absorbance ratios, cDNA concentration, cDNA 260/280 and 260/230 absorbance ratios, and the specificity of melting curves (Table S4). After combining these sample quality parameters and the miRNA expression levels, the improved HallMark-32 model contained 70 features in total. After model improvement, the accuracy and AUC of the HallMark-32 Random forest model were increased from 91% to 95% and from 0.945 (95% CI 0.86-1) to 0.991 (95% CI 0.96-1), respectively (Table 4B and Figure 3A). The AUC score of the improved model is more significant than the model before improvement, with a p-value of 0.001 and 0.002, respectively. To prove that the success of model improvement was not a random chance, we generated a negative control for the improvement strategy, by replacing the sample quality features with random numbers. As expected, the negative control did not improve accuracy, but conversely, reduced the accuracy from 95% to 84% (Table 4B). The result demonstrated that the integration of sample quality parameters is instrumental and specific for the HCC identification. In summary, with a 0.55 probability cut-off, the test set validation (n=27) demonstrated an AUC value of 0.991 (95% CI 0.96-1, p = 0.001), 100% sensitivity, 91% specificity, 0.92 PPV, 1 NPV, and 95% accuracy. By applying the whole dataset (n=133; Table 4A and Figure 3A) to the improved HallMark-32 model, the HCC diagnostic model demonstrated an AUC value of 0.999 (95% CI 0.99-1, p=0.000), 100% sensitivity, 98% specificity, 0.96 PPV, and 1 NPV.
3.4. Identifying HCC in alpha-fetoprotein-negative samples
Although AFP has been widely used for facilitating HCC screening and diagnosis, the sensitivity only ranged from 41% to 65% [14]. Therefore, we interrogated whether HallMark-32 could identify HCC in AFP-negative (< 20ng/mL) samples. To test this, we examined the predicted HCC probability in the AFP-negative samples available in our HCC dataset (n = 17; Figure 3B). The AFP-negative HCC samples were predicted with a high HCC probability (0.76 - 1) by the HallMark-32 model. The improved HallMark-32 Random forest model could identify HCC in AFP-negative samples with 100% sensitivity with a 0.55 probability cut-off (Figure 3B). Collectively, the HallMark-32 model can also identify HCC in AFP-negative samples, suggesting a potential application of HallMark-32 for HCC identification irrespective of serum AFP level.
3.5. Clinical association between HallMark-32 panel and patient survival
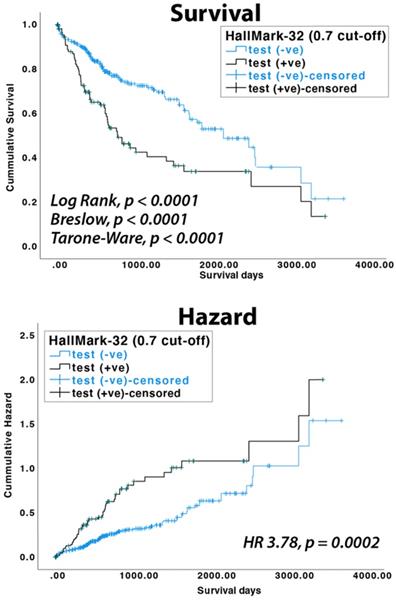
To test if the HallMark-32 panel is associated with HCC patient survival, we analyzed miRNA expression data available in the TCGA database. In the TCGA LIHC dataset, 100, 90, 90, and 10 HCC patients belonged to the pathological stages I, II, III, and IV, respectively. Using random forest and a probability cut-off of 0.55, HCC patients with positive prediction results showed a lower survival rate (p<0.004 in log-rank test). Using a probability cut-off of 0.7, the clinical association with survival rate is more significant (p<0.0001 in log-rank, Breslow, and Tarone-Ware test, Figure 4). HCC patients with positive results have a significantly higher risk of death than patients with negative results (HR = 3.78, 95% CI = 1.70-5.43, p = 0.0002 in Cox proportional hazards test, Figure 4). The median survival days for HCC patients with positive and negative results are 770 days (SE 133.7 days) and 2131 days (SE 314.8 days), respectively.
4. Discussion
In summary, this study highlighted five key findings: 1) Six signature miRNAs (miR-221-3p, miR-223-3p, miR-26a-5p, miR-30c-5p, miR-365a-3p, and miR-423-3p) were identified. Their expression profiles differed significantly in HCC and healthy individuals, providing a good diagnostic model for non-invasive HCC identification. 2) We reported a strategy to optimize the miRNA-based diagnostic model performance by adjusting sample quality variation. 3) The improved HallMark-32 random forest model demonstrated outstanding performance for identifying HCC, as well as differentiating HCC from CRCLM and healthy individuals. 4) HallMark-32 allowed HCC identification even in AFP-negative samples. 5) TCGA analysis illustrated the clinical association between the HallMark-32 panel and HCC patient survival.
Performance of four machine learning algorithms in HallMark-32 and Signature-Six assays.
| A) | HallMark-32 | ||||||||||||
| Test set (n = 27) | All data (n=133) | ||||||||||||
| Model | Mean Accuracy ± SD | AUC (95% CI) | p-value for AUC | Sensitivity | Specificity | PPV | NPV | AUC (95% CI) | p-value for AUC | Sensitivity | Specificity | PPV | NPV |
| Neural Network | 0.86 ± 0.04 | 0.92 (0.811-1) | 0.004 | 1 | 0.86 | 0.88 | 1 | 0.98 (0.96-1) | 0.000 | 0.96 | 0.95 | 0.92 | 0.98 |
| Random Forest | 0.91 ± 0 | 0.95 (0.86-1) | 0.002 | 0.91 | 0.91 | 0.91 | 0.91 | 1 (0.99-1) | 0.000 | 0.98 | 0.98 | 0.96 | 0.99 |
| Gradient Boosting | 0.89 ± 0 | 0.96 (0.88-1) | 0.002 | 0.91 | 0.86 | 0.87 | 0.9 | 1 (0.996-1) | 0.000 | 0.98 | 0.96 | 0.94 | 0.99 |
| Logistic Regression | 0.84 ± 0 | 0.85 (0.69-1) | 0.018 | 0.91 | 0.77 | 0.8 | 0.89 | 0.96 (0.93-1) | 0.000 | 0.92 | 0.88 | 0.81 | 0.95 |
| B) | Signature-Six | ||||||||||||
| Test set (n = 27) | All data (n=133) | ||||||||||||
| Model | Mean Accuracy ± SD | AUC (95% CI) | p-value for AUC | Sensitivity | Specificity | PPV | NPV | AUC (95% CI) | p-value for AUC | Sensitivity | Specificity | PPV | NPV |
| Neural Network | 0.82 ± 0.04 | 0.84 (0.66-1) | 0.021 | 0.73 | 0.86 | 0.84 | 0.76 | 0.96 (0.92-1) | 0.000 | 0.77 | 0.94 | 0.88 | 0.88 |
| Random Forest | 0.93 ± 0.02 | 0.93 (0.83-1) | 0.003 | 1 | 0.86 | 0.88 | 1 | 0.98 (0.96-1) | 0.000 | 1 | 0.96 | 0.94 | 1 |
| Gradient Boosting | 0.9 ± 0.03 | 0.89 (0.77-1) | 0.007 | 1 | 0.82 | 0.85 | 1 | 0.98 (0.96-1) | 0.000 | 0.98 | 0.94 | 0.9 | 0.99 |
| Logistic Regression | 0.82 ± 0 | 0.85 (0.68-1) | 0.018 | 0.91 | 0.73 | 0.77 | 0.89 | 0.91 (0.86-1) | 0.000 | 0.83 | 0.81 | 0.71 | 0.9 |
ROC curves of the improved HallMark-32 random forest model and the predictive HCC probability for each sample. A) ROC curves for the test set (n=27) and the whole dataset (n=133). B) The HCC probability predicted by the improved HallMark-32 random forest model (n=133).
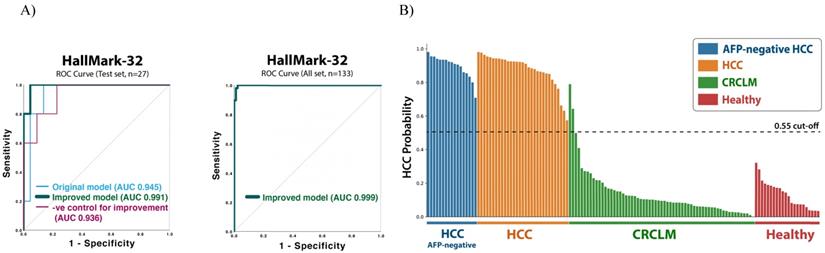
Comparison of performance before and after improvement of HallMark-32 model. A) Performance indexes of the sample quality-adjusted HallMark-32 Random Forest model. B) Accuracy and AUC after model improvement (i.e. sample quality-adjusted), before improvement (No adjustment), and negative control for improvement.
| A) | Sample quality-adjusted HAllMark-32 | |||||||||||
| Model | Test set (n = 27) | All data (n=133) | ||||||||||
| Mean accuracy | AUC (95% CI) | P value for AUC | Sensitivity | Specificity | PPV | NPV | AUC | Sensitivity | Specificity | PPV | NPV | |
| Random Forest | 0.95±0.01 | 0.991 (0.96-1) | 0.001 | 1 | 0.91 | 0.92 | 1 | 1 | 1 | 0.98 | 0.96 | 1 |
| B) | HallMark-32 (Test set, n = 27) | |||||||||||
| Sample quality-adjusted | No adjustment | Negative control for improvement | ||||||||||
| Accuracy | 0.95 | 0.91 | 0.84 | |||||||||
| AUC(95% CI) | 0.991 (0.96-1) | 0.945 (0.86-1) | 0.936 (0.836-1) | |||||||||
| P value for AUC | 0.001 | 0.002 | 0.003 | |||||||||
KM survival and Cox proportional hazards analyses of TCGA LIHC dataset (n=372).
Virtually all cancer progresses with ten cancer hallmarks proposed by Hanahan and Weinberg [16]. The development of HCC in hepatitis B virus (HBV)-infected patients, for example, involves dysregulation in p53, PI3K, TGF-β, IL-6, VEGF, and TERT pathways [17, 18], thereby acquiring oncogenic hallmarks for HCC development (Table S5). To develop an HCC-specific assay, we selected miRNA candidates that regulate ten hallmark properties in HCC based on the literature (Table S2). In the expression profiling comparing HCC and healthy individuals, six signature miRNAs (miR-221-3p, miR-223-3p, miR-26a-5p, miR-30c-5p, miR-365a-3p, and miR-423-3p) were identified. Intriguingly, the expression levels of miR-221-3p, miR-223-3p, and miR-26a-5p were significantly lower in HCC and higher in CRCLM than in healthy individuals (Figure 1). The differences of this expression pattern may in part be explained by the cancer stage in HCC and CRCLM patients. For the intermediate-stage of HCC, cancer cells tend to progress by activating angiogenesis and extravasation. Whereas in CRCLM, the extravasation, intravasation, and colonization signaling pathways are activated to develop distant organ metastasis. The differences of the activated signaling pathways may explain the distinct expression patterns. In fact, miR-221-3p, miR-223-3p, and miR-26a-5p are functionally linked to proliferation, invasion, and metastasis in both HCC [19-23] and CRC [24-26]. The miR-221 is known to regulate epithelial-mesenchymal transition (EMT) by regulating zinc finger E-box binding homeobox 2 (ZEB2) pathway in HCC. Both miR-223 and miR-26a are linked to invasion and metastasis in HCC. Whereas, in CRC, miR-223 and miR-26a promote proliferation and cell invasion. The miR-221 enhances the cancer stem cell property in CRC by targeting human Quaking (QKI) gene. The signaling interaction between these miRNAs warrants further investigation, but their differential expression pattern could provide an important fingerprint to distinguish primary liver cancer against secondary liver metastases, as demonstrated in this study.
When diagnosing HCC, tissue biopsy procedures are needed, especially when CT and MRI fail to show typical HCC features. However, tissue biopsy may lead to clinical complications and sampling bias. Liquid biopsy may serve as a non-invasive and complementary approach to improve the HCC diagnosis and surveillance. In this study we demonstrated the performance of Signature-Six and HallMark-32 miRNA panel to identify HCC and differentiate HCC from liver metastasis. We also illustrated the capability of HallMark-32 to complement AFP for HCC detection. Taken together, we demonstrated potential applications of Signature-Six and HallMark-32 in supporting HCC surveillance and diagnosis.
Future work with an increased sample size of HCC is needed to evaluate the performance of these models to predict HCC at different stages. Our group has previously conducted technical validation and established clinical utilities of circulating tumor cells in colorectal, lung, breast, gastric, liver, prostate, esophageal, and nasopharyngeal cancers [27-29]. The clinical application of combining miRNA, circulating tumor cell [27-29], and bioinformatics technology [30, 31] merits further exploration. To sum up, we discovered circulating microRNA signatures for identifying HCC and demonstrated the promising performance of the liquid biopsy assay for HCC identification and differential diagnosis.
Supplementary Material
Supplementary figure and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250-61
2. Llovet JM, Bustamante J, Castells A, Vilana R, Ayuso Mdel C, Sala M. et al. Natural history of untreated nonsurgical hepatocellular carcinoma: rationale for the design and evaluation of therapeutic trials. Hepatology. 1999;29:62-7
3. Padhya KT, Marrero JA, Singal AG. Recent advances in the treatment of hepatocellular carcinoma. Curr Opin Gastroenterol. 2013;29:285-92
4. Wu Q, Qin SK. Features and treatment options of Chinese hepatocellular carcinoma. Chin Clin Oncol. 2013;2:38
5. Patel N, Yopp AC, Singal AG. Diagnostic delays are common among patients with hepatocellular carcinoma. J Natl Compr Canc Netw. 2015;13:543-9
6. Singal AG, Waljee AK, Patel N, Chen EY, Tiro JA, Marrero JA. et al. Therapeutic delays lead to worse survival among patients with hepatocellular carcinoma. J Natl Compr Canc Netw. 2013;11:1101-8
7. Kim JK, Kim HD, Jun MJ, Yun SC, Shim JH, Lee HC. et al. Tumor Volume Doubling Time as a Dynamic Prognostic Marker for Patients with Hepatocellular Carcinoma. Dig Dis Sci. 2017;62:2923-31
8. Russo FP, Imondi A, Lynch EN, Farinati F. When and how should we perform a biopsy for HCC in patients with liver cirrhosis in 2018? A review. Dig Liver Dis. 2018;50:640-6
9. Li J, Han B, Li H, Deng H, Mendez-Sanchez N, Guo X. et al. Association of coagulopathy with the risk of bleeding after invasive procedures in liver cirrhosis. Saudi J Gastroenterol. 2018;24:220-7
10. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A. et al. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology. 2018;154:1706-18 e1
11. Esfeh JM, Hajifathalian K, Ansari-Gilani K. Sensitivity of ultrasound in detecting hepatocellular carcinoma in obese patients compared to explant pathology as the gold standard. Clin Mol Hepatol. 2020;26:54-9
12. Colli A, Fraquelli M, Casazza G, Massironi S, Colucci A, Conte D. et al. Accuracy of ultrasonography, spiral CT, magnetic resonance, and alpha-fetoprotein in diagnosing hepatocellular carcinoma: a systematic review. Am J Gastroenterol. 2006;101:513-23
13. Komaki Y, Komaki F, Micic D, Ido A, Sakuraba A. Risk of colorectal cancer in chronic liver diseases: a systematic review and meta-analysis. Gastrointest Endosc. 2017;86:93-104 e5
14. Zamcheck N, Pusztaszeri G. CEA, AFP and other potential tumor markers. CA Cancer J Clin. 1975;25:204-14
15. Ren F, Weng W, Zhang Q, Tan C, Xu M, Zhang M. et al. Clinicopathological features and prognosis of AFP-producing colorectal cancer: a single-center analysis of 20 cases. Cancer Manag Res. 2019;11:4557-67
16. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
17. Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-62
18. D'Souza S, Lau KC, Coffin CS, Patel TR. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J Gastroenterol. 2020;26:5759-83
19. Li Y, Ren M, Zhao Y, Lu X, Wang M, Hu J. et al. MicroRNA-26a inhibits proliferation and metastasis of human hepatocellular carcinoma by regulating DNMT3B-MEG3 axis. Oncol Rep. 2017;37:3527-35
20. Li ML, Zhang Y, Ma LT. LncRNA HCG11 accelerates the progression of hepatocellular carcinoma via miR-26a-5p/ATG12 axis. Eur Rev Med Pharmacol Sci. 2019;23:10708-20
21. Wan L, Yuan X, Liu M, Xue B. miRNA-223-3p regulates NLRP3 to promote apoptosis and inhibit proliferation of hep3B cells. Exp Ther Med. 2018;15:2429-35
22. Chen Z, Xiang B, Qi L, Zhu S, Li L. miR-221-3p promotes hepatocellular carcinogenesis by downregulating O6-methylguanine-DNA methyltransferase. Cancer Biol Ther. 2020;21:915-26
23. Yang J, Cui R, Liu Y. MicroRNA-212-3p inhibits paclitaxel resistance through regulating epithelial-mesenchymal transition, migration and invasion by targeting ZEB2 in human hepatocellular carcinoma. Oncol Lett. 2020;20:23
24. Mukohyama J, Isobe T, Hu Q, Hayashi T, Watanabe T, Maeda M. et al. miR-221 Targets QKI to Enhance the Tumorigenic Capacity of Human Colorectal Cancer Stem Cells. Cancer Res. 2019;79:5151-8
25. Chai B, Guo Y, Cui X, Liu J, Suo Y, Dou Z. et al. MiR-223-3p promotes the proliferation, invasion and migration of colon cancer cells by negative regulating PRDM1. Am J Transl Res. 2019;11:4516-23
26. Coronel-Hernandez J, Lopez-Urrutia E, Contreras-Romero C, Delgado-Waldo I, Figueroa-Gonzalez G, Campos-Parra AD. et al. Cell migration and proliferation are regulated by miR-26a in colorectal cancer via the PTEN-AKT axis. Cancer Cell Int. 2019;19:80
27. Wong VC, Ko JM, Lam CT, Lung ML. Succinct workflows for circulating tumor cells after enrichment: From systematic counting to mutational profiling. PLoS One. 2017;12:e0177276
28. Ko JM, Vardhanabhuti V, Ng WT, Lam KO, Ngan RK, Kwong DL. et al. Clinical utility of serial analysis of circulating tumour cells for detection of minimal residual disease of metastatic nasopharyngeal carcinoma. Br J Cancer. 2020;123:114-25
29. Ko JMY, Ng HY, Lam KO, Chiu KWH, Kwong DLW, Lo AWI. et al. Liquid Biopsy Serial Monitoring of Treatment Responses and Relapse in Advanced Esophageal Squamous Cell Carcinoma. Cancers (Basel). 2020 12
30. Wong VC. Nuclear EGFR and Integrator/Super Elongation Complex concurrently binds to Immediate Early Genes for gene transactivation. J Cancer. 2018;9:108-16
31. Dai W, Chung DL, Chow LK, Yu VZ, Lei LC, Leong MM. et al. Clinical Outcome-Related Mutational Signatures Identified by Integrative Genomic Analysis in Nasopharyngeal Carcinoma. Clin Cancer Res. 2020;26:6494-504
Author Biography
Dr. Victor Chun-Lam Wong received his Ph.D. in Clinical Oncology at the University of Hong Kong, pursued his postgraduate training at Brown University and Memorial Sloan Kettering Cancer Center in the USA, obtained his postgraduate diploma in Machine Learning Artificial Intelligence in Emeritus Institute in collaboration with Columbia University. His research integrates circulating tumor cells, DNA, microRNA, and machine learning technologies for supporting cancer surveillance and monitoring. In recognizing the excellence and innovation of the R&D work, he and his team have received many awards, such as the Hong Kong Science Park Incubator Program Award in 2014, the Technology Start-up Support Scheme Award in 2015 and 2016, the Best Pitching Award in 2017, and the Public Sector Trial Scheme Award in 2021. Dr. Wong's publications in cancer research have over 300 citations. He is currently an ad-hoc reviewer/editorial board member in over ten international journals such as Frontiers of Oncology, Aging, Oncotarget, Cancer Letters, Cancer Biology & Therapy, Journal of Experimental & Clinical Cancer Research, Plos One, Oncology Research and Review, and Trends in Cancer Updates.
Ming-In Wong received her Bachelor's degree of Science in Biochemistry and Cell Biology at The Hong Kong University of Science and Technology. Currently working as an R&D assistant scientist in OncoSeek Ltd, her R&D interests include machine learning, biomedical science, and genetics.
Dr. Chi-Tat Lam received his Ph.D. in the Department of Surgery at the University of Hong Kong, and he is a co-founder of OncoSeek Ltd. He has a long-term interest in studying the molecular mechanisms of esophageal and liver cancers by using essential state-of-the-art approaches. Dr. Lam's recent research has advanced the use of liquid biopsy in clinical settings.
Prof. Maria Li Lung received her Ph.D. from the Department of Medical Microbiology at Stanford University, Stanford, California, USA in the Cancer Biology Research Laboratory in Radiology at the Stanford Medical School. She was a postdoctoral fellow at the Center for Cancer Research at the Massachusetts Institute of Technology, Cambridge, Massachusetts, USA. She became a Lecturer in the Department of Microbiology at the University of Hong Kong. She subsequently became a founding member of the Department of Biology at the Hong Kong University of Science and Technology in Hong Kong, where she progressed from Lecturer to Chair Professor. She was the Founding Director of the Center for Cancer Research at HKUST. She is now a Chair Professor of the Department of Clinical Oncology at the University of Hong Kong. She is an elected Fellow of the American Academy of Microbiology. Her primary research interests involve deciphering the molecular genetic basis for nasopharyngeal carcinoma (NPC) and esophageal squamous cell carcinoma (ESCC), which are two strategically important cancers in Chinese. Using a functional chromosome transfer approach, she identified critical regions and candidate tumor suppressor genes vital for NPC and ESCC development. Recent approaches using whole genome next-generation sequencing (NGS) for targeted gene and genome sequencing were used to elucidate the genetic basis for inherited cancers, as well as determining the molecular landscape of tumor variants contributing to tumor pathogenesis. Together with global transcriptomic and methylome approaches, these integrative studies helped to identify key genes and pathways responsible for NPC and ESCC and led important biomarker studies for both these cancers. Her current interest in liquid biopsies includes studies on circulating tumor cells, as well as ctDNAs. She established a Hong Kong territory-wide Center for NPC Research including individuals from three universities and five public hospitals. Currently she leads a group on translational studies for elucidating the tumor heterogeneity and molecular evolution in metastatic gastrointestinal tract cancers for personalized medicine. She established OncoSeek with funding from HKU and the Hong Kong Innovation and Technology Commission and is a co-founder of this company.
Dr. Ka-On Lam is currently a Clinical Associate Professor in the Department of Clinical Oncology, The University of Hong Kong (HKU). He graduated with distinction in M.B.B.S. from HKU in 2005. He was awarded the Frank Doyle Medal in 2008 and the Rohan Williams Medal in 2010 for outstanding performance at Examinations of Fellowship of the Royal College of Radiologists (FRCR, UK). He then joined HKU as clinical assistant professor and obtained his specialist qualification in Clinical Oncology in 2013. He was selected Distinguished Young Fellow of Hong Kong Academy of Medicine in 2014. His research interests are novel treatment strategy and circulating tumor cells in gastrointestinal cancers and nasopharyngeal cancer. He was awarded the HKU Overseas Fellowship in 2013 which funded his elective on drug development at The Royal Marsden. Since then he participated as Principal Investigator (PI) or site PI in 19 clinical trials in gastrointestinal oncology, including 5 phase I/II trials He has published over 80 articles in international journals including BJC, Cell Stem Cell, PNAS, IJROBP and IJC, and presented over 60 conference abstracts at DDW, ESMO, ESMO World Congress on Gastrointestinal Cancer and other major conferences. He has been serving the Editorial Board of the Asia-Pacific Journal of Clinical Oncology (Hong Kong Supplement) since 2018, BMC Cancer since 2020, and guest editor for Frontiers in Oncology in 2019-21. He is a founding member of the HK SBRT Study Group. He has given over 50 invited lectures on gastrointestinal cancer in Hong Kong and in other Asia-Pacific countries.
Dr. Victor Ho-Fun Lee is currently Clinical Associate Professor of the Department of Clinical Oncology, Li Ka Shing Faculty of Medicine, The University of Hong Kong. He graduated in the University of Hong Kong in 2002. He joined the Department of Clinical Oncology, Li Ka Shing Faculty of Medicine, The University of Hong Kong as Clinical Assistant Professor in 2008. He obtained his fellowship in Royal College of Radiologists in Clinical Oncology in 2010. Afterwards, he received further specialist training in interstitial brachytherapy for head and neck cancers and sarcoma in Institut Gustave Roussy in Paris, France and novel radiation techniques like stereotactic radiosurgery and stereotactic ablative radiotherapy in Stanford University USA. In 2013, he received further training on stereotactic body radiation therapy for liver tumors at Princess Margaret Hospital, Toronto, Canada. More recently in 2015 he was awarded HKCR 15A Traveling Fellowship and pursued subspecialty training in image-guided brachytherapy for cervical cancer and pediatric oncology in Addenbrooke's Hospital in Cambridge. He has obtained a Doctor of Medicine (MD) in 2015 for his research on nasopharyngeal carcinoma. He was promoted to Clinical Associate Professor in 2017 and appointed as Assistant Dean (Assessment) of Li Ka Shing Faculty of Medicine, The University of Hong Kong since August 2018. He was awarded the Outstanding Young Researcher Award of The University of Hong Kong in December 2018. His current interests include clinical and genetic studies on nasopharyngeal cancer, head and neck cancers, lung cancers, liver cancers and gastrointestinal cancers and has published extensively in these respects. In addition, he has special interest in preclinical and clinical dosimetric studies on intensity-modulated radiation therapy, stereotactic body radiation therapy (SBRT), stereotactic radiosurgery (SRS) and selective internal radiation therapy (SIRT) with Yttrium-90 microspheres for liver tumors.
![]() Corresponding authors: Department of Clinical Oncology, The University of Hong Kong, 21 Sassoon Road, Pokfulam, Hong Kong Special Administrative Region, People's Republic of China; OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China. E-mail addresses: lamkaonhk (KL), vhfleehk (VL), victorcom (VW). Phone: (+852) 2255 4352 (KL, VL); (+852) 31889335 (VW).
Corresponding authors: Department of Clinical Oncology, The University of Hong Kong, 21 Sassoon Road, Pokfulam, Hong Kong Special Administrative Region, People's Republic of China; OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China. E-mail addresses: lamkaonhk (KL), vhfleehk (VL), victorcom (VW). Phone: (+852) 2255 4352 (KL, VL); (+852) 31889335 (VW).

 Global reach, higher impact
Global reach, higher impact