3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(16):5035-5045. doi:10.7150/jca.59391 This issue Cite
Review
Emerging Diagnostic Potential of Tumor-derived Exosomes
1. College of Pharmacy, School of Medicine, Hangzhou Normal University, Hangzhou, Zhejiang 311121, China.
2. Key Laboratory of Elemene Class Anti-Cancer Chinese Medicines; Engineering Laboratory of Development and Application of Traditional Chinese Medicines; Collaborative Innovation Center of Traditional Chinese Medicines of Zhejiang Province, Hangzhou Normal University, Hangzhou, Zhejiang 311121, China.
*Co-first authors with equal contributions to this work.
Received 2021-2-14; Accepted 2021-5-23; Published 2021-6-16
Abstract
Exosomes carry genetic information originating from their parental cells, raising their possibility as novel noninvasive biomarkers for cancer. Tumor-derived exosomes (TEXs) have a variety of endogenous cargos that reflect the pathophysiology status and information of tumor cells. TEXs are increasingly being recognized as potential biomarkers for cancer diagnosis prognosis, and monitoring. It is important to develop a variety of sensitive methods, including probes and biomaterials to isolate exosomes. A variety of approaches for detecting exosomes have been established. By combining exosome DNA and RNA sequencing tools, exosome proteomics analysis and immunoassay technology, it is expected that exosomes will gain widespread use in the diagnosis and treatment of cancer.
Keywords: exosomes, tumor-derived exosomes, biomarkers, cancer detection
Introduction
Exosomes are small extracellular vehicles (EVs) ranging from ~30 to ~200 nm diameter that were released by almost all types of cells, including tumor cells, presenting pathophysiological roles and clinical value. Exosomes participate in mediating cell-to-cell communication, or multiple processes of tumor development, presenting pathophysiological roles and clinical value. They are nanosized membrane-bound nanovesicles that carry genetic information originating from their parental cells, raising their possibility as novel noninvasive biomarkers for cancer [1]. Tumor-related exosomes have various contents that reflect pathophysiological changes in parental cells, whose information could be important in cancer diagnosis. Their presence in physiological fluids can be used to reveal important clinical information regarding these tissues [2]. Exosomes have been widely reported in tumor research. Tumor-related exosomes are involved in the establishment or transformation of tumor microenvironments, advancing tumor angiogenesis and invasion, as well as mediating immune inhibition and escape [3]. Exosomes with CD63 and caveolin-1 in plasma can be utilized as non-invasive markers of melanoma [4]. Circulating exosomes with overexpressed glypican 1 (GPC1) in the serum of patients closely correlate with pancreatic cancer, which could be used as a highly specific biomarker for pancreatic cancer [5]. Exosomes are involved in multiple processes of tumor genesis and development, including promoting angiogenesis [6], differentiation and infiltration [7], regulating immunity, and mediating drug resistance [8]. Therefore, exosomes can be used as a potential biomarker for clinical applications and but exosomes-measuring technologies need to be standardized.
Biological features of exosomes
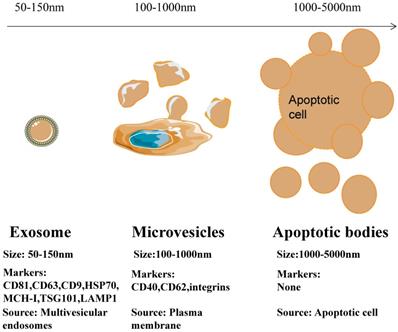
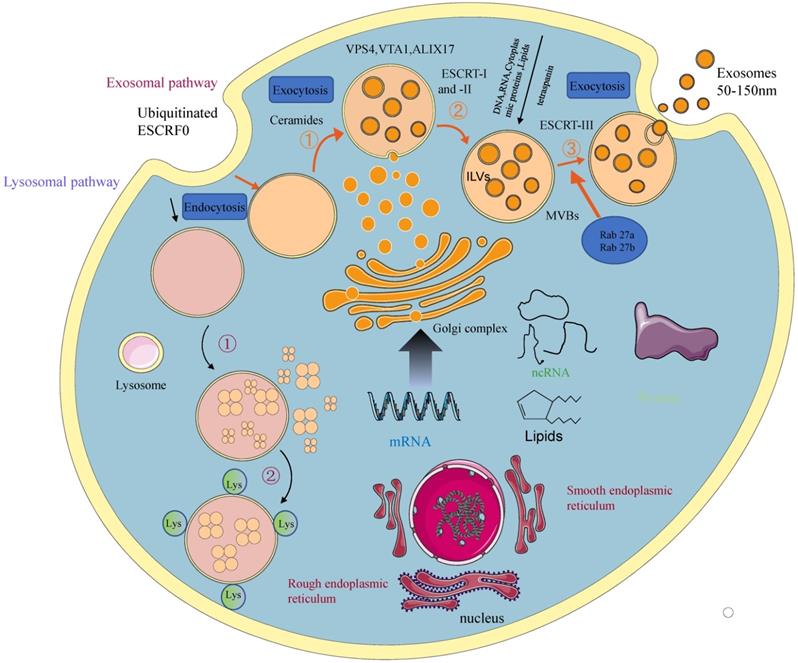
Depending on their biogenesis and generation pathways, EVs can be broadly classified into three subpopulations; exosomes, microvesicles, and apoptotic bodies [9] (Figure 1). Compared to microvesicles and apoptotic bodies, exosomes have a different biogenesis machinery [10]. Exosomes are generated through a complex process (Figure 2). They are a class of cellular secreted nanovesicles that form multivesicular bodies (MVBs), which contain numerous intraluminal vesicles (ILVs).
Exosomes, as messengers of intercellular information transmission, are involved in the transfer of materials and information between cells by combining fusion or endocytosis [11]. Its outer layer is the phospholipid bilayer similar to the cell membrane and related proteins such as membrane transport and fusion. Its interior contains functional biomolecules such as proteins and nucleic acids [12]. Non-coding small RNA molecules (ncRNAs) are important in cell proliferation, differentiation and apoptosis [13].
Differences in size, markers and origin of exosomes, microvesicles and apoptotic bodies.
A schematic presentation of exosomal biogenesis. An early endosome is formed from the plasma membrane through the endocytic pathway. With the involvement of the Golgi apparatus, they mature into late endosomes and MVBs. The invagination from MVBs forms the ILVs. The process of exosomal biogenesis is dependent on either the ESCRT or independent of ESCRT mechanisms. Eventually, depending on the function and content of MVB, MVB fuses with the cell membrane and is released into the extracellular space as exosomes or fuse with lysosomes for degradation.
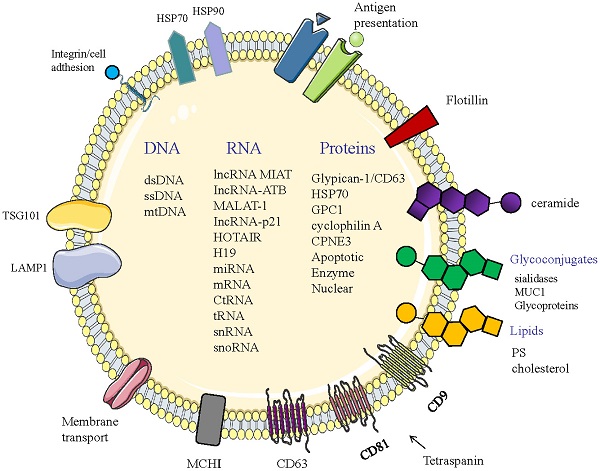
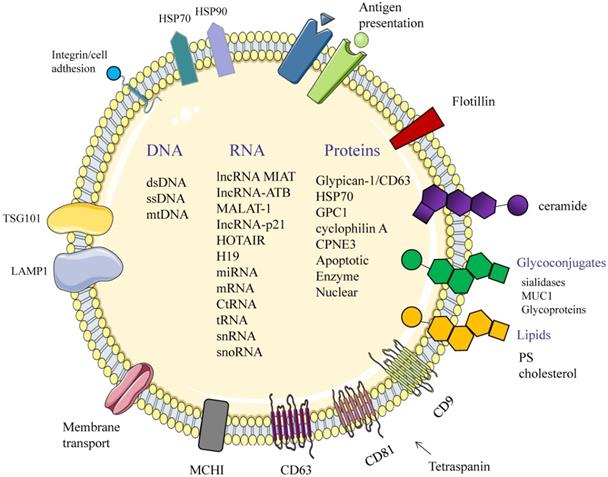
Structure and composition of exosomes. Exosomes consist of an aqueous core and a phospholipid bilayer. They carry important biomolecules such as proteins, DNA, and RNA, while the exosome membrane is rich in a variety of specific proteins, such as: Tetraspanins (CD9, CD63, CD81); major histocompatibility complexes I and II (MHC I, II); TSG101; HSP70, 90 (heat shock protein 70,90); LAMP1; Surface receptor; Ceramide; Glycoconjugates.
Potential of Exosomes in Tumor Diagnosis
Early diagnosis and efficient therapy for patients are the best options for enhancing their survival outcomes. However, efficient diagnosis and therapy is inhibited by the lack of biomarkers that are highly specific, stable and noninvasive [14]. As cancer biomarkers, the significance of exosomes in tumor diagnosis has been highlighted [15]. Studies have documented that exosomes are important in intercellular communication and molecular exchange of constitutive lipids [16], characteristic proteins [17], functional messenger RNAs (mRNAs) and microRNAs (miRNAs) (Figure 3, Table 1) [18]. TEXs carry a variety of endogenous cargos that reflect the pathophysiology status and information of tumor cells. Generally, cancer cells secrete more exosomes than their non-malignant counterparts. TEXs can be separated from body fluids to provide information about the tumor microenvironment. Accordingly, TEXs are increasingly being recognized as potential biomarkers for cancer diagnosis [19], prognosis, and monitoring [20].
RNA
In recent years, liquid biopsies have been widely used in the early diagnosis of tumors, especially in the analysis of tumor-related substances extracted from body fluids of patients, such as miRNAs [59], exosomes [60] and circulating tumor cells (CTCs) [61]. Exosomes contain various functional nucleic acids, some of which (such as miRNA, IncRNA and CircRNA) are promising options for cancer diagnosis. Studies have documented that exosomes play crucial roles in cancer onset, progression and metastasis regulating the stability of target mRNAs or by inhibiting translation [62]. Although miRNAs have been well characterized in exosomes, a lack of specific expression and their small amounts limit their use as biomarkers [63]. Recently, there are abundant data indicating that exosomal miRNAs play an vital role in cancer biology and may be potential diagnostic for cancer. A study indicated that there was a significant difference in the levels of serum exosomal microRNAs in patients with hepatocellular carcinoma (HCC) compared with patients with chronic hepatitis B (CHB) or liver cirrhosis (LC) [64]. However, studies have shown that exosomal length non-coding RNA (IncRNA) has a significant therapeutic potential, including disease diagnosis [65]. It has also been documented that TEXs IncRNA GAS5 is a potential diagnostic marker for early non-small cell carcinoma [66]. Moreover, breast cancer preoperatively diagnosed as ductal carcinoma in situ (DCIS) patients are likely to develop invasive ductal carcinoma (IDC). miR-223-3p enhances breast cancer cell invasion, and its exosomes may be used as minimally invasive biomarkers for the diagnosis of IDC patients by biopsy [67]. In addition, miR-375 [68], miR-21 [69] from urinary exosomes in both urine and blood have been identified to be specific exosomal miRNAs that can be used to distinguish between prostate cancer (PCa) patients from healthy individuals [70]. Interestingly, studies used miRNA sequencing to investigate the exosomal miRNA profiles of patients with Papillary thyroid cancer (PTC). This method combined small RNA sequencing from numerous samples and RT-qPCR validation. A total of 41 significantly upregulated miRNAs were found to be concurrently expressed in plasma. It was also found that miR-485-3p and miR-4433a-5p are potential markers for PTC in clinical diagnosis. Moreover, miR-204-3p and miR-4306 were confirmed as clinical biomarkers for PTC [71]. Serum exosomal long noncoding RNA lnc-GNAQ-6:1 is lowly expressed in gastric cancer patients. The difference in expression levels might be used as a new diagnostic marker for disease assessment in large-scale gastric cancer studies [72]. In addition, circular RNAs (circRNAs) are associated with cancer progression and development. In Cholangiocarcinoma (CCA), circ-0000284 (circHIPK3) was found to elevate lymphocyte antigen 6E (LY6E) expression by competitively binding miR-637Owing to circRNAs that are enriched and stable in exosomes. Exosome-mediated circ-0000284 has been reported to contribute to the development of cancer in surrounding normal cells [73]. Exosomal circRNAs are easy to be detected due to stable structure, extensive expression, conserved sequence, and abundant content, thus providing additional evidence for conventional diagnostic methods and holding great promise as diagnostic markers for cancer.
Exosomal cargos as biomarkers for various cancers
| Cargos | Cancer type | Source | Method | Detection | Reference |
|---|---|---|---|---|---|
| Nucleic acid | |||||
| RNA | |||||
| IncRNAs | |||||
| lncRNA MIAT | GC | Serum | qRT-PCR detected serum exosomal MIAT | Compared to healthy controls, pre-treatment and recurrence, MIAT levels were higher in GC patients | [21] |
| IncRNA-ATB | HCC | Serum/plasma | NcRNAs were isolated from exosomes using the miRNeasy serum/plasma micro kit, characterized and analyzed. | lncRNA-ATB were correlated with TNM stage. The more tumor volume and CRP are, the more level of IncRNA-ATB is. | [22] |
| MALAT-1 | NSCLC | Serum | To measure the expression levels of MALAT-1 contained in exosomes | Exosomal MALAT-1 was highly expressed in NSCLC patients | [23] |
| IncRNA-p21 | PCa | Urine | Quantification of IncRNA molecules was performed by real-time PCR | IncRNA-p21 levels were elevated in PCa patients than BPH | [24] |
| HOTAIR | GBM | Serum/tumors | HOTAIR expression was measured using qRT-PCR. | HOTAIR levels from GBM patients were significantly higher than in the controls | [25] |
| miR-21, HOTAIR | LSCC | Serum | RT-PCR was performed to detect the expression levels of miR-21 and HOTAIR | The expressions of miR-21 and HOTAIR were significantly higher in patients with LSCC than vocal cord polyps | [26] |
| HOTAIR/MALAT1/MEG3 | Cervical cancer | Cervicovaginal lavage specimens | Exosomal lncRNAs HOTAIR, MALAT1, and MEG3 were quantified by qRT-PCR. | Expression of HOTAIR, MALAT1 and MEG3 was predominantly observed in cervical cancer | [27] |
| H19 | BC | Serum | To measure the levels of lncRNA H19 by using quantitative real-time PCR | H19 expression levels were upregulated in BC patients | [28] |
| miRNAs | |||||
| miR-20a/miR-21 | cHL | Serum | Analysis of immunovirological data | miR-21 and miR-20a levels were highly associated with adverse clinical outcomes | [29] |
| miR-21/miR-27a/miR-375 | BC | Serum | Clinical samples were used to test exosomal miRNAs by using the biosensor | miR-21, miR-27a and miR-375 levels were highly expressed in breast cancer cells | [30] |
| miR-3940-5p/miR-8069 | PDAC | Urine | Novel biomarker candidates for PDCA were found using expression profiling, and validated in samples using 3D digital PCR | miR-3940-5p/miR-8069 ratio in urine exosomes was elevated in PDAC patients | [31] |
| miR-5189-3p | PTC | Plasma | Analysis of miRNA expression in PTC and NG | miR-5189-3p levels were significantly high in PTC patients | [32] |
| miR139/miR136/miR19/miR210 | Bladder cancer | Urine | Detection of miRNAs using Real-time PCR and ELISA | miR139/miR136/miR19/miR210 were upregulated in cancer patients | [33] |
| let-7b-5p/miR-122-5p/miR-146b-5p/miR-210-3p/miR-215-5p | BC | Plasma | Exiqon miRNA qPCR panel was used to select candidate miRNAs, which were further analyzed using qRT-PCR | miR-122-5p was upregulated in the plasma of BC patients. Let-7b-5p was contrarily down-regulated in BC tissue | [34] |
| miR-142-3p/miR-142-5p/miR-223-3p | PCa | Semen | An altered miRNA expression pattern was revealed by a high throughput profiling analysis in PCa ,and validated miRNA | Exosomal miRNA-based molecular biomarkers have the potential to improve PCa diagnosis | [35] |
| miR-141-3p/miR-375 | LARC | Plasma | Expression of exosomal miRNAs was profiled using a miRCURY LNA miRNA microarray | Exosomal miR-141-3p and miR-375 levels were higher in patients | [36] |
| miR-7977 | LUAD | Serum | Exosomal miRNAs expression levels in serum were further validated by the qRT-polymerase chain reaction. | Inhibition of miR-7977 enhanced the proliferation, invasion, and suppressed apoptosis in A549 cells | [37] |
| CtRNAs | |||||
| hsa-circ-0004771 | CRC | Plasma | The expression and diagnostic utility of circRNA were tested by qRT-PCR and ROC analysis | Expression of hsa-circ-0004771 in the serum of CRC patients was elevated | [38] |
| hsa-circ-0043603/hsa-circ-0062459 | ESCC | Plasma and cell culture | The expression of profile was identifiedmicroarray and displayed by qRT-PCR. | hsa-circ-0062459 was used as a diagnostic biomarker | [39] |
| Circ-IARS | PCa | Plasma | qRT-PCR was used to measure the expression levels of circ-IARS | circ-IARS expression was up-regulated in pancreatic cancer tissues and in plasma exosomes of patients with metastatic disease | [40] |
| F-circEA | NSCLC | Peripheral blood | The presence of the EML4-ALK fusion gene in H2228 cells was verified by sequencing RT-PCR | F-circEA is specifically present in the plasma of EML4-ALK positive NSCLC patients | [41] |
| Circ SATB2 | NSCLC | Serum | The expression of genes and proteins was detected by qPCR and western blot | circSATB2 was highly expressed in serum from lung cancer patients and was associated with lung cancer metastasis | [42] |
| DNA | |||||
| SOX2 DNA | GBM | Blood | To detect cancer-specific SNPs in exosomal SOX2 DNA sequence | SOX2 is stemness gene highly expressed in cancer stem cells | [43] |
| dsDNA | PCC/PGL | Serum | Compared the serum exosomal DNA and tumor tissue DNA from patients or mice with PCC or PGL | Serum-derived exosomal dsDNA in PCC and PGL was highly consistent with the paired tumor genome | [44] |
| mRNAs | |||||
| hnRNPH1 | HCC | Serum | hnRNPH1 mRNA expression levels were measured using the TaqMan real-time PCR | Serum exosomal hnRNPH1 mRNA levels in HCC patients were significantly higher than in the other groups | [45] |
| hTERT | 'Pan- cancer' | Serum | The expression of exosomal MT1-MMP mRNA in GC patients was detected using RT-PCR. | MT1-MMP mRNA expression levels were found to be higher in GC patients | [46] |
| MT1-MMP | GC | Serum | The expression of exosomal MT1-MMP mRNA in GC patients was detected using RT-PCR | MT1-MMP mRNA expression levels were found to be higher in GC patients | [47] |
| Proteins | |||||
| glypican-1/ CD63 | PDAC | Whole blood/plasma/serum | On-chip immunofluorescence analysis permits specific identification and quantification of target biomarkers | The elevated glypican-1 was observed in metastatic but not in non-metastatic disease | [48] |
| HSP70 | Solid cancers | Blood/urine | HSP70 concentration will be determine | HSP70-exosomes were detected and quantified in blood from patients with solid cancers. | [49] |
| GPC1 | PCa | Serum | Python-based standardized data processing method was established to analyze exosomal GPC1 expression. | The abundance of GPC-1 was elevated in patients with pancreatic cancer. | [50] |
| cyclophilin A | NPC | Serum | Protein expression levels were validated by reverse transcription -quantitative polymerase chain reaction, Western blotting, and ELISA | CYPA levels of NPC patients were significantly higher than in normal cases | [51] |
| CD82 | BC | Breast cancer tissue | Test CD82 expression in tissues and CD82 content in exosomes | CD82 expression levels in breast cancer tissues were significantly lower than in healthy and benign breast cancer tissues. | [52] |
| CPNE3 | CRC | Plasma | CPNE3 expression levels were determined by RT-PCR, western blot, and immunohistochemistry | CPNE3 expression was elevated in CRC tissues. | [53] |
| Glycoconjugates | |||||
| Sialidases | PCC/PCa | Plasma | Serum sialidase was assessed by a sandwich ELISA method using two anti-NEU3 antibodies. | NEU3 is markedly upregulated in cancers | [54] |
| MUC1 | Lung cancer | Plasma | LC-MS/MS by data-independent analysis mode identified proteins | MUC1 levels in NSCLC patients were 1.5-fold higher than in controls. | [55] |
| Glycoproteins | BC | Serum/plasma | Glycoproteins in extracellular vesicles (EVs) were identified through integrating quantitative glycoproteomics | 20 glycoproteins were verified to be significantly higher in individual breast cancer patients. | [56] |
| Lipids | |||||
| PS | BC | Plasma | Highly specific and sensitive ELISA for the capture of PS-expressing tumor exosomes in blood from tumor mice | PS-expressing tumor exosome levels were significantly high in blood | [57] |
| Cholesterol | PCa | Urine | A high-throughput mass spectrometry quantitative lipidomic analysis was used to reveal the lipid composition of urinary exosomes in PCa patients | The highest significance was shown for phosphatidylserine and lactosylceramide | [58] |
DNA
Exosomes are secreted by both viable and dying tumor cells and exosomal DNA (ExoDNA) is likely to be one of the most stable cargos due to protection by lipid bilayer [74]. The discovery that exosomes contain tumor specific DNA mutations reinvented the clinical diagnosis of tumor mutations and opened up the possibility to perform liquid biopsy on patients. Exosomal DNA represents the entire genome and reflects the status of mutations in parental tumor cells [75]. In non-small cell lung cancer (NSCLC), it has been shown that the mutant EGFR gene is enveloped in exosomes. Exosomal circulating mutations are present during early tumor development. About 50% of malignant melanomas have a BRAF (V600E) mutation, while ExoDNA mutant alleles have been reported in mutant cell lines. In pancreatic ductal adenocarcinoma (PDAC), KRAS mutations in ExoDNA were detected in 43.6% of early PDAC patients and in 20% of healthy controls [76]. The packaged amount of exoDNA varies depending on the cancer type.[77] The DNA content may be related to the size exosomal vesicles. High molecular weight tumor DNA fragments are specifically exported to exosomes. A comparative study by vagner et al found that larger vesicles (> 1 µm) contain more DNA and preferentially export genetic aberrations than smaller vesicles [78]. Mutations KRAS and P53 were detected in exogenous DNA from pancreatic cancer cell lines and in the genomic DNA from the serum of pancreatic cancer patients [79]. It has been demonstrated that the majority of DNA in exosomes derived from tumors is double stranded. Williams et al. definitively confirmed that dsDNA was present in tumor-associated exosomes [80]. Compared with serum exo-DNA and tumor tissue DNA in patients with pheochromocytoma (PCC) and paraganglioma (PGL), dsDNA fragments are highly concordant with paired oncogenome. The definitive evidence of the presence of exosomal double-stranded DNA (dsDNA) shows, for the first time, that it can be used as a non-invasive genetic marker for the diagnosis and preoperative evaluation of PCCs and PGLs [44]. Vaidya et al. performed a detailed analysis of the association between the NANOG gene family and exosomes [81]. Differential NANOGP8 sequences from other embryonic stemness genes (OCT3/4, SOX2, etc.) may establish a set of exosomal-based diagnostic markers. Exosomes contain large dsDNA fragments spanning multiple genomic regions and harbor EGFR (T790M and L858R) [82] and BRAF (V600E) [83] gene mutations, proposing the possibility of tracing these mutations in plasma exosomes of NSCLC and/or melanoma patients through liquid biopsy. Exo-DNA has a short half-life and can accurately reflect tumor status in real time. In addition, exoDNA detection has potential clinical application in minimal residual disease and early warning of recurrence, contributing to better management of cancer and benefiting more patients.
Proteins
Studies have shown the role for exosomal proteins in clinical diagnosis. Exosomes contain various tumor-associated proteins that reflect tumor statuses. In addition, they also contain proteins from cells. The membrane structure of exosomes protects proteins from external proteases and other enzymes [84,85]. Due to their small sizes, strong permeability and high sensitivity, exosomal proteins from cancer cells can be used as biomarkers for cancer monitoring and efficacy evaluation, thereby helping in cancer diagnosis and early detection [86]. Exosomal extracellular matrix protein 1 (ECM1) [87] and Alpha-2-HS-glycoprotein (AHSG) [88] respectively promote cancer progression and invasion in most tumors. A study using proteomic profiles confirmed that the combination of AHSG, ECM1, and carcinoembryonic antigen (CAE) improve the diagnostic potential of NSCLC [89]. Moreover, melanoma secreted exosomes were found to contain intact tumor antigens to activate CD8+ T cells, and showed anti-cancer activities after being absorbed by dendritic cells, suggesting that CD63 in plasma exosomes can be used as a protein marker for melanoma [90]. Exosomal membranes are homologous to cell membranes, and about 40% of cancer cell surface markers are found on the surface of exosomes. As biomarkers, exosomal proteins can provide a rich, stable, sensitive, and specific biological information. Kristine et al. [91] developed an EV array that coupled 37 antibodies targeting lung cancer-related proteins and CD9, CD63, and CD81 antibody groups to explore circulating exosomes from healthy subjects and patients with lung cancer. Proteomic characteristics of salivary and serum exosomes from lung cancer patients and healthy controls have been identified and comprehensively compared by liquid chromatography-tandem mass spectrometry (LC-MS/MS) [92]. Certain special proteins are only present on the exosomes derived from malignant cells [93]. Compared to healthy controls, plasma levels of epithelial cell adhesion molecule (EpCAM) -positive exosomes have been found to be significantly higher in breast cancer patients [94]. Protein phosphorylation is the fundamental, most prevalent mechanism controlling diverse cellular physiological functions, and few phosphoproteins have been identified as potential biomarkers to distinguish disease from health [95]. A recent study showed that phosphoproteins in plasma exosomes are highly feasible as disease biomarkers. Using label free quantitative Phosphoproteomics, researchers identified 144 phosphoproteins in plasma EVs that were significantly higher in patients diagnosed with breast cancer compared to healthy controls. Glypican-1 (GPC-1), the cell surface proteoglycan [96], is exclusively present on exosomes from malignant cells and is overexpressed in breast and pancreatic cancers. Application of GPC-1 as a diagnostic marker for colorectal cancer (CRC) has also been reported [97]. Gammaglutamyltransferase 1 (GGT1), a cell surface enzyme, and CD9 in exosomes are present in PCa patients [98]. In conclusion, even though there are associated difficulties and problems in the study of exosomal proteomics, it provided a novel method for cancer diagnosis [99].
Glycoconjugates
Studies on exosomes have addressed the role of glycosylation. The surface of exosomes consists of a polysaccharide canopy attached to surface proteins and certain outer leaflet lipids. Malignancy is best characterized by the feature of aberrant sialylation in glycoproteins and glycolipids, which have been implicated in cancer progression [100]. It has been established that plasma membrane‐associated ganglioside sialidase (NEU3) is up-regulated in a variety of cancers, including colon and prostate cancers. Keiko Hata et al. found different levels of ganglioside activity in cancer patients, and evaluated the sialidase NEU2 subtype as a potential biomarker for human cancer diagnosis by enzyme-linked immunosorbent assay (ELISA) [54]. TEXs are somewhat different because their glycan characteristics vary with the expression of various glycosidases and glycosyltransferases [101]. For example, hyaluronic acid synthase 3 levels are highly expressed in some cancers, leading to long polymers that synthesize hyaluronic acid, promote cancer growth and the biosynthesis of exosomes. Cancers can express heparanase to cleave the heparin sulfate proteoglycan chain. However, addition of exogenous heparinase was shown to alleviate the apparent spatial block of SDC budding, and increases the release of exosomes containing SDC [102]. In addition to special surface proteins and glycans, certain types of exosomal lipids have been studied. Glycolipids from exosomes have also been considered to be potential cancer biomarkers elsewhere. For example, glycosphingolipids such as hexosylceramides (HexCer) and lactosylceramides (LacCer) were abundantly enriched in PCa cells-derived exosomes [95]. Among them, phosphatidylserine (PS) is a type of phospholipid that is anchored within the inner leaflet of the normal cell membrane but is externalized in malignant and apoptotic cells. Y. T. Kang et al. reported that Tumor-related exosomes can be isolated by the PS targeting method [103]. PS is not only specific to cancer cells, but has also been shown to have a regulatory effect on immune cells in vivo. This method has been used to isolate exosomes from samples, and confirmed that exosomal PS expression is not only a characteristic of vesicle formation, but also a PS mediated immune response on the surface of exosomes from NK cells, which contribute to tumor diagnosis and treatment.
Exosomal detection and monitoring technology
Although TEXs characteristics enhance their potential as clinical biomarkers, effective methods for isolation and detection have not been established. Differential ultracentrifugation (UC) has been a classical method for EV separation, at least, until recently [104]. A variety of approaches for detecting exosomes have been established, which include the conventional nanoparticle tracking analysis (NTA) [105], ELISA [106], flow cytometry [107], digital detection [108], etc. The methods for exosomal quantification requires low sample bulk and high sensitivity. However, the step-by-step processing and analysis of exosomes is time and labor-consuming. Moreover, the samples are also vulnerable to external contamination [109]. Faster and more directional extraction procedures such as alternating current (AC) chip and microfluidic chip have been developed. An AC microarray chip that can isolate exosomes and other nanoscale substances (EVs and cf-DNA) from the whole blood of pancreatic cancer patients within 20 minutes and perform in situ immunofluorescence analysis has been designed [48]. This assay can distinguish between PDAC patients and healthy people, and can also detect patients with metastatic diseases. Xu and colleagues developed a two-stage microfluidic platform (ExoPCD-chip) that integrates on-chip separation and in situ electrochemical analysis of exosomes in serum [110]. Zhou et al. designed a hairpin-like structure aptasensor that integrates the highly specific MUC1 aptamer with a hemin/G-quadruplex to detect exosomes in breast cancer [111]. Compared to other methods, it was found to be a sensitive, simple and low-cost colorimetric method that could distinguish between breast cancer patients and healthy individuals. An aptasensor based on aptamer-capped Fe3O4 NPs was constructed for visual and label-free detection of PCa exosomes from plasma. Exosomes were directly isolated from clinical PCa plasma and detected by the aptasensor for the early diagnosis and staging of PCa. In addition to the new detection technology, there have been advances on the traditional detection methods, such as ELISA, to resolve reproducibility and sensitivity issues [112]. Moreover, UC has also been combined with polymer precipitation to separate small extracellular vesicles in the serum of breast cancer patients [113]. Lee et al. proposed a sandwich paper-based ELISA (p-ELISA) method [114] that is a portable diagnostic system with low cost, simple to operate and easy to use. It is efficient for the isolation/detection functions of targeted EVs/exosomes. The procedures for this method can be done within 2 h, and the SARBI p-ELISA assay provides strong binding due to the strong affinity between biotinylated Ab and streptavidin agarose. Exosome-containing microRNAs (exomiRs) are potential biomarkers for tumor diagnosis. A novel exosome-specific tumor diagnosis strategy was constructed by combining the rapid magnetic exosome-enrichment platform with the Ru(bpy)32+-polymer amplified electrochemiluminescence (ECL) strategy [115]. To realize the rapid and efficient capture of TEXs, the researchers constructed a non-destructive detection mode of tumor liquid biopsy through the biological affinity identification platform of EpCAM antibody. The glypican-1 and CD63 biomarkers reflect the presence of PDAC, which was quantified within as little as 30 min by subsequent on-chip immunofluorescence analysis [48]. Compared to healthy people, elevated glypican-1 in PDAC patients with 99% sensitivity and 82% specificity can improve early stage cancer diagnostics by ACE exosome capture and on-chip biomarker detection. Specificity and sensitivity are the two foremost concerns in biological detection [116]. Yang et al. [117] measured the expression of miR-21 microRNA and TTF-1 mRNA in the exosomes containing EGFR or PD-L1 in human plasma using an immune biochip. Compared to the traditional immunomagnetic separation, immuno-biochip enhanced lung cancer diagnostic platform uses a shorter time and consumes less than 30 μL of samples. However, it is important to develop a variety of sensitive methods, including probes and biomaterials to isolate exosomes.
Conclusion
The complexity of exosomes bring many challenges to isolate large pure and specific exosomes from mixture. Maybe, only a small subtype of exosomes in an abundance mixture has the feature of biomarkers. It is an important process to determine the specific or characteristic exosomes from the abundance mixture. The concentration of TEXs were an important parameter that indicate the tumor pathological circumstances. This means that the TEXs is more abundant in serum, which is helpful for cancer diagnosis.
Compared with conventional biopsy or liquid biopsy, exosome-based diagnosis has higher sensitivity and specificity. By combining exosome DNA and RNA sequencing tools, exosome proteomics analysis and immunoassay technology, it is expected that exosomes will gain widespread use in the diagnosis and treatment of cancer.
Abbreviations
TEXs: tumor-derived exosomes; EVs: extracellular vesicles; GPC1: glypican 1; MVBs: multivesicular bodies; ILVs: intraluminal vesicles; ncRNA: non-coding small RNA; mRNAs: messenger RNAs; miRNAs: microRNAs; MCH I and II: major histocompatibility complexes I and II; HSP70,90: heat shock protein; lncRNAs: long non-coding RNAs; GC: gastric cancer; qRT-PCR: Quantitative reverse transcription-polymerase chain reaction; lncRNA-ATB: lncRNA-activated by tumor growth factor-beta (TGF-β); HCC: hepatocellular carcinoma; MALAT-1: metastasis-associated lung adenocarcinoma transcript 1; PCa: prostate cancer; BPH: benign prostatic hyperplasia; GBM: Glioblastoma multiforme; LSCC: laryngeal squamous cell carcinoma; HOTAIR: Hox transcript antisense intergenic RNA; MEG3: maternally expressed gene 3; BC: breast cancer; cHL: classical Hodgkin lymphoma; NG: nodular goiter; LARC: locally advanced rectal cancer; miRNA: microRNA; LUAD: lung adenocarcinoma; CRC: colorectal cancer; circRNAs: circular RNAs; ESCC: Esophageal squamous cell cancer; EML4-ALK: Echinoderm Microtubule-associated protein-Like 4- Anaplastic Lymphoma Kinase; f-circM9: MLL/AF9 fusion gene; SOX2: Sex-determining region Y (SRY)-box 2; dsDNA: double-stranded DNA; PCC: Pheochromocytoma; PGL: paraganglioma; hnRNPH1: heterogeneous nuclear ribonucleoprotein H1; hTERT: human telomerase reverse transcriptase; MT1-MMP: Membrane Type 1-Matrix Metalloproteinase; NPC: nasopharyngeal carcinoma; CYPA: Cyclophilin A; CD82: cluster of differentiation 82; CPNE3: clinical significance of exosomal Copine III; CTCs: circulating tumor cells; CHB: chronic hepatitis B; LC: liver cirrhosis; DCIS: ductal carcinoma in situ; IDC: invasive ductal carcinoma; PTC: Papillary thyroid cancer; CCA: Cholangiocarcinoma; LY6E: lymphocyte antigen 6E; NSCLC: non-small cell lung cancer; PDAC: pancreatic ductal adenocarcinoma; ECM1: exosomal extracellular matrix protein 1; AHSG: Alpha-2-HS-glycoprotein; LC-MS/MS: liquid chromatography-tandem mass spectrometry; EpCAM: epithelial cell adhesion molecule; CAE: carcinoembryonic antigen; GPC1: glypican-1; CRC: colorectal cancer; GGT1: gammaglutamyltransferase 1; NEU3: plasma membrane-associated ganglioside sialidase; ELISA: enzyme-linked immunosorbent assay; HexCer: hexosylceramides; LacCar: lactosylceramides; PS: phosphatidylserine; UC: ultracentrifugation; NTA: nanoparticle tracking analysis; AC: alternating current; p-ELISA: paper-based ELISA; exomiRs: Exosome-containing microRNAs; ECL: electrochemiluminescence; MUC1: mucin 1.
Acknowledgements
This work was supported by Zhejiang Provincial Natural Science Foundation of China (LY20H160008), Department of Health of Zhejiang Province (2019329972), The National Natural Science Foundation of China (82074052), Hangzhou Social Development of Scientific Research projects (20191203B17), Key projects of National Natural Science Foundation of China (81730108).
Author Contributions
Qingchang Tian, Shuling Wang, Tian Xie drafted the work organized co-author to write this review, and give final approval of the version to be published. Ruhua Luo, Qian Yang, Mengmeng Liu, Huijuan Cheng, Huimin Yang, Minhui Li, Xue Bai, Yue Wang, Honghua Zhang accomplished the main text of manuscript. And Ruhua Luo, Mengmeng Liu, Qian Yang, Huijuan Cheng accomplished the tables and abbreviations in the manuscript. Ruhua Luo accomplished the figure in the manuscript. Ruhua Luo and Mengmeng Liu contributed equally to this work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Valadi H, Ekström K, Bossios A. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654-9
2. Simpson RJ, Jensen SS, Lim JWE. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083-99
3. Olejarz W, Dominiak A, Zolnierzak A. et al. Tumor-Derived Exosomes in Immunosuppression and Immunotherapy. J Immunol Res. 2020;2020:11
4. Logozzi M, De Milito A, Lugini L. et al. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS ONE. 2009;4:e5219
5. Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-82
6. Wang H, Wang L, Zhou XC. et al. OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. Biomed Res Int. 2020;2020:13
7. Baj-Krzyworzeka M, Mytar B, Szatanek R. et al. Colorectal cancer-derived microvesicles modulate differentiation of human monocytes to macrophages. J Transl Med. 2016;14:15
8. Sun ZQ, Shi K, Yang SX. et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:19
9. Soo CY, Song YQ, Zheng Y. et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192-7
10. Mannan A, Muhammad SA, Anwar F. et al. AN EMERGING PARADIGM EXOSOMES: COMPOSITION, BIOLOGICAL FUNCTIONS, AND DIAGNOSTIC AND THERAPEUTIC POTENTIALS. Indo Am J Pharm Sci. 2016;3:1297-308
11. Jiang WY, Wang M. New insights into the immunomodulatory role of exosomes in cardiovascular disease. Rev Cardiovasc Med. 2019;20:153-60
12. Wee I, Syn N, Sethi G. et al. Role of tumor-derived exosomes in cancer metastasis. Biochim Biophys Acta-Rev Cancer. 2019;1871:12-19
13. Nanbo A, Kawanishi E, Yoshida R. et al. Exosomes Derived from Epstein-Barr Virus-Infected Cells Are Internalized via Caveola-Dependent Endocytosis and Promote Phenotypic Modulation in Target Cells. Journal of Virology. 2013;87:10334-47
14. Naeli P, Yousefi F, Ghasemi Y. et al. The Role of MicroRNAs in Lung Cancer: Implications for Diagnosis and Therapy. Current Molecular Medicine. 2020;20:90-101
15. Kim JH, Kim E, Lee MY. Exosomes as diagnostic biomarkers in cancer. Mol Cell Toxicol. 2018;14:113-22
16. Pascual M, Ibanez F, Guerri C. Exosomes as mediators of neuron-glia communication in neuroinflammation. Neural regeneration research. 2020;15:796-801
17. Wang Q, Han C-L, Wang K-L. et al. Integrated analysis of exosomal lncRNA and mRNA expression profiles reveals the involvement of lnc-MKRN2-42:1 in the pathogenesis of Parkinson's disease. CNS neuroscience & therapeutics. 2020;26:527-37
18. Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in Tumour Microenvironment: Overview of the Crosstalk between Normal and Cancer Cells. Biomed Res Int. 2014;2014:10
19. Amrollahi P, Rodrigues M, Lyon CJ. et al. Ultra-Sensitive Automated Profiling of EpCAM Expression on Tumor-Derived Extracellular Vesicles. Front Genet. 2019;10:10
20. Leal AC, Mizurini DM, Gomes T. et al. Tumor-Derived Exosomes Induce the Formation of Neutrophil Extracellular Traps: Implications for the Establishment of Cancer-Associated Thrombosis. Sci Rep-Uk. 2017;7:12
21. Xu H, Zhou J, Tang J. et al. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. Journal of clinical laboratory analysis. 2020;34:7
22. Lee YR, Kim G, Tak WY. et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444-52
23. Zhang R, Xia YH, Wang ZX. et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Bioph Res Co. 2017;490:406-14
24. Isin M, Uysaler E, Ozgur E. et al. Exosomal IncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:5
25. Tan SK, Pastori C, Penas C. et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 2018;17:7
26. Wang JT, Zhou YD, Lu JG. et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. 2014;31:8
27. Zhang J, Liu SC, Luo XH. et al. Exosomal Long Noncoding RNAs are Differentially Expressed in the Cervicovaginal Lavage Samples of Cervical Cancer Patients. Journal of Clinical Laboratory Analysis. 2016;30:1116-21
28. Zhong GB, Wang KQ, Li JW. et al. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Breast Cancer Diagnosis. OncoTargets Ther. 2020;13:2563-71
29. Hernandez-Walias FJ, Vazquez E, Pacheco Y. et al. Risk, Diagnostic and Predictor Factors for Classical Hodgkin Lymphoma in HIV-1-Infected Individuals: Role of Plasma Exosome-Derived miR-20a and miR-21. J Clin Med. 2020;9:18
30. Wang H, He D, Wan K. et al. In situ multiplex detection of serum exosomal microRNAs using an all-in-one biosensor for breast cancer diagnosis. Analyst. 2020;145:3289-96
31. Yoshizawa N, Sugimoto K, Tameda M. et al. miR-3940-5p/miR-8069 ratio in urine exosomes is a novel diagnostic biomarker for pancreatic ductal adenocarcinoma. Oncol Lett. 2020;19:2677-84
32. Pan QX, Zhao JM, Li MZ. et al. Exosomal miRNAs are potential diagnostic biomarkers between malignant and benign thyroid nodules based on next-generation sequencing. Carcinogenesis. 2020;41:18-24
33. Amuran GG, Tinay I, Filinte D. et al. Urinary micro-RNA expressions and protein concentrations may differentiate bladder cancer patients from healthy controls. Int Urol Nephrol. 2020;52:461-8
34. Li MH, Zou X, Xia TS. et al. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019;8:7006-17
35. Barcelo M, Castells M, Bassas L. et al. Semen miRNAs Contained in Exosomes as Non-Invasive Biomarkers for Prostate Cancer Diagnosis. Sci Rep-Uk. 2019;9:16
36. Meltzer S, Bjornetro T, Lyckander LG. et al. Circulating Exosomal miR-141-3p and miR-375 in Metastatic Progression of Rectal Cancer. Transl Oncol. 2019;12:1038-44
37. Chen LY, Cao PJ, Huang CL. et al. Serum exosomal miR-7977 as a novel biomarker for lung adenocarcinoma. J Cell Biochem. 2020;121:3382-91
38. Pan B, Qin J, Liu XX. et al. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front Genet. 2019;10:10
39. Fan LY, Cao Q, Liu J. et al. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. 2019;18:6
40. Li J, Li ZH, Jiang P. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Canc Res. 2018;37:16
41. Tan SY, Gou QH, Pu WC. et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28:693-5
42. Zhang N, Nan AR, Chen LJ. et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:16
43. Vaidya M, Sugaya K. Differential sequences and single nucleotide polymorphism of exosomal SOX2 DNA in cancer. Plos One. 2020;15:19
44. Wang L, Li Y, Guan X. et al. Exosomal double-stranded DNA as a biomarker for the diagnosis and preoperative assessment of pheochromocytoma and paraganglioma. Mol Cancer. 2018;17:6
45. Xu H, Dong XY, Chen YM. et al. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56:479-84
46. Goldvaser H, Gutkin A, Beery E. et al. Characterisation of blood-derived exosomal hTERT mRNA secretion in cancer patients: a potential pan-cancer marker. Br J Cancer. 2017;117:353-7
47. Dong ZG, Sun XY, Xu JJ. et al. Serum Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) mRNA Protected by Exosomes as a Potential Biomarker for Gastric Cancer. Med Sci Monitor. 2019;25:7770-83
48. Lewis JM, Vyas AD, Qiu YQ. et al. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. Acs Nano. 2018;12:3311-20
49. Chanteloup G, Cordonnier M, Isambert N. et al. Membrane-bound exosomal HSP70 as a biomarker for detection and monitoring of malignant solid tumours: a pilot study. Pilot and feasibility studies. 2020;6:35
50. Xiao D, Dong Z, Zhen L. et al. Combined Exosomal GPC1, CD82, and Serum CA19-9 as Multiplex Targets: A Specific, Sensitive, and Reproducible Detection Panel for the Diagnosis of Pancreatic Cancer. Molecular Cancer Research. 2020;18:300-10
51. Liu LZ, Zuo LL, Yang J. et al. Exosomal cyclophilin A as a novel noninvasive biomarker for Epstein-Barr virus associated nasopharyngeal carcinoma. Cancer Med. 2019;8:3142-51
52. Wang XD, Zhong WL, Bu JY. et al. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol Carcinog. 2019;58:674-85
53. Sun B, Li YM, Zhou YM. et al. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234:1416-25
54. Hata K, Tochigi T, Sato I. et al. Increased sialidase activity in serum of cancer patients: Identification of sialidase and inhibitor activities in human serum. Cancer Sci. 2015;106:383-9
55. Pan D, Chen JX, Feng CC. et al. Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells. Int J Mol Sci. 2019;20:12
56. Chen IH, Aguilar HA, Paez JSP. et al. Analytical Pipeline for Discovery and Verification of Glycoproteins from Plasma-Derived Extracellular Vesicles as Breast Cancer Biomarkers. Anal Chem. 2018;90:6307-13
57. Sharma R, Huang XM, Brekken RA. et al. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br J Cancer. 2017;117:545-52
58. Skotland T, Ekroos K, Kauhanen D. et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer. 2017;70:122-32
59. Wu H, Wang Q, Zhong H. et al. Differentially expressed microRNAs in exosomes of patients with breast cancer revealed by next-generation sequencing. Oncol Rep. 2020;43:240-50
60. Reese M, Flammang I, Yang Z. et al. Potential of Exosomal microRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers. 2020;12:16
61. Cieslikowski WA, Budna-Tukan J, Swierczewska M. et al. Circulating Tumor Cells as a Marker of Disseminated Disease in Patients with Newly Diagnosed High-Risk Prostate Cancer. Cancers. 2020 12, 16
62. Duijvesz D, Luider T, Bangma CH. et al. Exosomes as Biomarker Treasure Chests for Prostate Cancer. European Urology. 2011;59:823-31
63. Ma CH, Shi XF, Zhu QQ. et al. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumor Biol. 2016;37:1437-44
64. Sohn W, Kim J, Kang SH. et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Experimental & Molecular Medicine. 2015;47:e184
65. Dong L, Lin WR, Qi P. et al. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:1158-66
66. Li CL, Lv YL, Shao CY. et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721-27
67. Yoshikawa M, Iinuma H, Umemoto Y. et al. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett. 2018;15:9584-92
68. Huang X, Yuan T, Liang M. et al. Exosomal miR-1290 and miR-375 as Prognostic Markers in Castration-resistant Prostate Cancer. European Urology. 2015;67:33-41
69. Samsonov R, Shtam T, Burdakov V. et al. Lectin-Induced Agglutination Method of Urinary Exosomes Isolation Followed by mi-RNA Analysis: Application for Prostate Cancer Diagnostic. Prostate. 2016;76:68-79
70. Rodriguez M, Bajo-Santos C, Hessvik NP. et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer. 2017;16:6
71. Dai DF, Tan YW, Guo LF, Tang AF, Zhao YS. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur J Endocrinol. 2020;182:111-21
72. Li S, Zhang M, Zhang H. et al. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clinica chimica acta; international journal of clinical chemistry. 2020;501:252-7
73. Louis C, Desoteux M, Coulouarn C. Exosomal circRNAs: new players in the field of cholangiocarcinoma. Clinical science (London, England: 1979). 2019;133:2239-44
74. Jin Y, Chen K, Wang Z. et al. DNA in serum extracellular vesicles is stable under different storage conditions. Bmc Cancer. 2016;16:9
75. Williams C, Rodriguez-Barrueco R, Silva JM. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766-9
76. Hilmi M, Tlemsani C. High Prevalence of Mutant KRAS in Circulating Exosome-Derived DNA from Early-Stage Pancreatic Cancer Patients. Oncologie. 2018;20:41-2
77. Mohammadi S, Yousefi F, Shabaninejad Z. et al. Exosomes and cancer: From oncogenic roles to therapeutic applications. Iubmb Life. 2020;72:724 48
78. Vagner T, Spinelli C, Minciacchi VR. et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7:16
79. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-15
80. Bernard V, Kim DU, San Lucas FA. et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology. 2019;156:108-18.e4
81. Vaidya M, Bacchus M, Sugaya K. Differential sequences of exosomal NANOG DNA as a potential diagnostic cancer marker. Plos One. 2018;13:13
82. Castellanos-Rizaldos E, Grimm DG, Tadigotla V. et al. Exosome-Based Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018;24:2944-50
83. Ahn J, Konno H, Barber GN. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene. 2015;34:5302-8
84. Penfornis P, Vallabhaneni KC, Whitt J. et al. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int J Cancer. 2016;138:14-21
85. Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom Clin Appl. 2015;9:358-67
86. Li WH, Li CY, Zhou T. et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:12
87. Gomez-Contreras P, Ramiro-Diaz JM, Sierra A. et al. Extracellular matrix 1 (ECM1) regulates the actin cytoskeletal architecture of aggressive breast cancer cells in part via S100A4 and Rho-family GTPases. Clinical & Experimental Metastasis. 2017;34:37-49
88. Guillory B, Sakwe AM, Saria M. et al. Lack of Fetuin-A (alpha(2)-HS-Glycoprotein) Reduces Mammary Tumor Incidence and Prolongs Tumor Latency via the Transforming Growth Factor-beta Signaling Pathway in a Mouse Model of Breast Cancer. Am J Pathol. 2010;177:2635-44
89. Niu LM, Song XG, Wang N. et al. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 2019;110:433-42
90. Logozzi M, De Milito A, Lugini L. et al. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. Plos One. 2009;4:10
91. Jakobsen KR, Paulsen BS, Baek R. et al. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:10
92. Sun Y, Liu S, Qiao Z. et al. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Analytica Chimica Acta. 2017;982:84-95
93. Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-U182
94. Fang S, Tian H, Li X. et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. Plos One. 2017;12:3
95. Chen IH, Xue L, Hsu CC. et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114:3175-80
96. Lai XY, Wang M, McElyea SD. et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86-93
97. Li J, Chen Y, Guo X. et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. Journal of Cellular and Molecular Medicine. 2017;21:838-47
98. Kawakami K, Fujita Y, Matsuda Y. et al. Gamma-glutamyltransferase activity in exosomes as a potential marker for prostate cancer. Bmc Cancer. 2017;17:12
99. Fontana S, Saieva L, Taverna S. et al. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: State of the art and new perspectives. Proteomics. 2013;13:1581-94
100. Freitas D, Balmana M, Pocas J. et al. Different isolation approaches lead to diverse glycosylated extracellular vesicle populations. J Extracell Vesicles. 2019;8:17
101. Rilla K, Pasonen-Sepptanen S, Deen AJ. et al. Hyaluronan production enhances shedding of plasma membrane-derived microvesicles. Exp Cell Res. 2013;319:2006-18
102. Thompson CA, Purushothaman A, Ramani VC. et al. Heparanase Regulates Secretion, Composition, and Function of Tumor Cell-derived Exosomes. J Biol Chem. 2013;288:10093-9
103. Kang YT, Purcell E, Palacios-Rolston C. et al. Isolation and Profiling of Circulating Tumor-Associated Exosomes Using Extracellular Vesicular Lipid-Protein Binding Affinity Based Microfluidic Device. Small. 2019;15:e1903600
104. Benmoussa A, Laugier J, Beauparlant CJ. et al. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J Dairy Sci. 2020;103:16-29
105. Vaswani K, Mitchell MD, Holland OJ. et al. A Method for the Isolation of Exosomes from Human and Bovine Milk. J Nutr Metab. 2019;2019:6
106. Zhang Y, Liu ZY, Li SY. et al. Upregulation of E-cadherin in bronchoalveolar lavage fluid-derived exosomes in patients with lung cancer. Thorac Cancer. 2020;11:41-7
107. Sawada S-I, Sato YT, Kawasaki R. et al. Nanogel hybrid assembly for exosome intracellular delivery: effects on endocytosis and fusion by exosome surface polymer engineering. Biomaterials science. 2020;8:619-30
108. Tian Q, He C, Liu G. et al. Nanoparticle Counting by Microscopic Digital Detection: Selective Quantitative Analysis of Exosomes via Surface-Anchored Nucleic Acid Amplification. Analytical chemistry. 2018;90:6556-62
109. Ferreira N, Marques A, Aguas H. et al. Label-Free Nanosensing Platform for Breast Cancer Exosome Profiling. Acs Sensors. 2019;4:2073-83
110. Xu HY, Liao C, Zuo P. et al. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal Chem. 2018;90:13451-58
111. Zhou Y, Xu HY, Wang H. et al. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor. Analyst. 2020;145:107-14
112. Wang SM, Ge L, Song XR. et al. Paper-based chemiluminescence ELISA: Lab-on-paper based on chitosan modified paper device and wax-screen-printing. Biosens Bioelectron. 2012;31:212-8
113. Ryu KJ, Lee JY, Park C. et al. Isolation of Small Extracellular Vesicles From Human Serum Using a Combination of Ultracentrifugation With Polymer-Based Precipitation. Ann Lab Med. 2020;40:253-8
114. Lee J, Kim H, Heo Y. et al. Enhanced paper-based ELISA for simultaneous EVs/exosome isolation and detection using streptavidin agarose-based immobilization. Analyst. 2020;145:157-64
115. Fan ZJ, Yu J, Lin JY. et al. Exosome-specific tumor diagnosis via biomedical analysis of exosome-containing microRNA biomarkers. Analyst. 2019;144:5856-65
116. Li W, Wang H, Zhao Z. et al. Emerging Nanotechnologies for Liquid Biopsy: The Detection of Circulating Tumor Cells and Extracellular Vesicles. Adv Mater. 2019;31:25
117. Yang Y, Kannisto E, Yu G. et al. An Immuno-Biochip Selectively Captures Tumor-Derived Exosomes and Detects Exosomal RNAs for Cancer Diagnosis. Acs Appl Mater Inter. 2018;10:43375-86
Author contact
![]() Corresponding authors: Shuling Wang (E-mail: wsling222com); Tian Xie (E-mail: xbsedu.cn); Qingchang Tian (E-mail: tianqcedu.cn).
Corresponding authors: Shuling Wang (E-mail: wsling222com); Tian Xie (E-mail: xbsedu.cn); Qingchang Tian (E-mail: tianqcedu.cn).

 Global reach, higher impact
Global reach, higher impact