Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(19):5838-5847. doi:10.7150/jca.57926 This issue Cite
Research Paper
18F-FDG PET/CT metabolic parameters correlate with EIF2S2 expression status in colorectal cancer
1. Postgraduate Training Basement of Jinzhou Medical University, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, China.
2. Department of Nuclear Medicine and Institute of Anesthesiology and Pain, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, China.
3. Department of Pathology, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, China.
4. Hubei Key Laboratory of Embryonic Stem Cell Research, Shiyan, Hubei, China.
5. Hubei Key Laboratory of WudangLocal Chinese Medicine Research, Shiyan, Hubei, China.
6. Department of Radiology, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, China.
*Contributed equally to this work.
Received 2021-1-7; Accepted 2021-7-9; Published 2021-8-3
Abstract
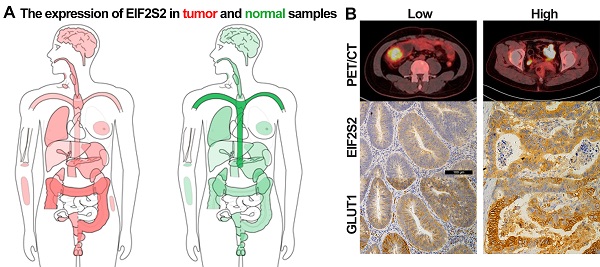
Background: We sought to investigate whether the expression of the gene EIF2S2 is related to 18F-FDG PET/CT metabolic parameters in patients with colorectal cancer (CRC).
Materials and methods: The expression of EIF2S2 in CRC and its relationship with clinicopathological features were obtained through the ONCOMINE, UALCAN and GEPIA databases. EIF2S2 and GLUT1 expression were examined by immunohistochemistry in 42 CRC patients undergoing preoperative PET-CT examination. Spearman correlation analysis was used to assess the relationship between EIF2S2 and GLUT1 levels and clinical parameters. Correlation analysis between EIF2S2 and Reactome-Glycolysis signatures was performed using GEPIA2. We describe the effect of EIF2S2 knockdown on lactate production and the mRNA levels of glycolysis-related genes in human colon cancer SW480 cells.
Results: Immunohistochemistry revealed an upregulation of EIF2S2 protein expression in tumor tissues of colorectal cancer patients, which is consistent with the significant upregulation of EIF2S2 transcript levels in the database. These colorectal cancer patients included 24 cases of colon cancer and 18 cases of rectal cancer, ranging in age from 31 to 78 years. The transcription was significantly related to histological subtypes and TP53 mutations (P <0.05). The value of SUVmax in CRC significantly correlated with the expression of EIF2S2 (rho = 0.462, P <0.01). Although SUVmax and SUVmean was not correlate with the expression of GLUT1 (P <0.05), a significant correlation was observed between the expression of GLUT1 and the volumetric PET parameters, such as MTV and TLG (P < 0.01). GLUT1 expression in CRC was positively correlated with EIF2S2 status (rho = 0.470, P <0.01). In SW480 cells, RNAi-mediated depletion of EIF2S2 inhibited lactic acid production (P <0.05) and SLC2A1, SLC2A3, SLC2A10, HK2, PKM2, LDHA mRNA level (P <0.01).
Conclusions: Primary CRC FDG uptake is strongly associated with the overexpression of EIF2S2, and EIF2S2 may promote glycolysis in CRC by mediating GLUT1.
Keywords: Colorectal cancer, EIF2S2, 18F-FDG, SUVmax, glucose transporter 1
Introduction
Colorectal cancer (CRC) is the fourth most common cancer [1]. Although the incidence and mortality of CRC have declined over the past two decades, unresectable locally advanced CRC has a poor prognosis. Early diagnosis and timely surgical treatment can significantly reduce the mortality of patients. There is a need for effective biomarkers of CRC for screening and early detection. Cancer cells mainly obtain energy by breaking down glucose into ATP using aerobic glycolysis (the Warburg effect) [2, 3]. 18F-FDG PET/CT has been widely used in the detection, treatment monitoring, and prognostic evaluation of various tumors [4-7]. 18F-FDG is glucose radiolabeled with 18F. Drawn in by glucose transporters (GLUTs) and phosphorylated by hexokinases (HK), it cannot be metabolized further, allowing its irreversible entry into cells. 18F-FDG is used to detect the amount of glucose uptake in tumors. The mechanism may be mediated by the increased number of GLUTs [8] and HK [9, 10] in malignant cells. The relationship between expression of GLUT-1 or HK2 and degree of 18F-FDG uptake has been extensively studied [11-16]. High expression of GLUT-1 is a crucial factor for 18F-FDG uptake in many malignant tumors. GLUT1 is encoded by the SLC2A1 gene and plays an important role in the development of many tumors, such as nonsmall cell lung cancer [17] and breast cancer [18]. In previous reports, it was found that the relationship between 18F-FDG uptake and GLUT-1 expression in different malignant tumors is not entirely consistent [19, 20]. It has been reported that 18F-FDG uptake correlates with GLUT1 expression in patients with liver metastasis from CRC [21]. In addition, it showed that SUVmax may be useful for predicting the NPM1 expression of lung adenocarcinoma [22] and METTL3 expression of esophageal cancer [23].
RNA binding protein (RBP) is widely recognized as a protein that plays an important role in regulating gene expression [24]. At present, more and more RBPs have been discovered, which play an important role in promoting the occurrence and development of tumors [25]. Some RBPs can also mediate energy metabolism [26]. EIF2 is a heterotrimer consisting of eIF2α, eIF2β and eIF2γ [27, 28]. EIF2S2 (Eukaryotic Translation.
Initiation Factor 2 Subunit β) showed a significant relationship between DNA copy number levels and mRNA expression in luminal breast tumors [29]. The eIF2β gene is highly expressed in lung cancer and can indicate the prognosis of patients with lung adenocarcinoma [30]. EIF2S2 may play a role in promoting tumors by regulating the WNT signaling pathway [31]. But whether EIF2S2 mediates aerobic glycolysis has not been reported in CRC.
Transcriptional data for EIF2S2 in CRC and its clinically relevant data were obtained from The Cancer Genome Atlas (TCGA) database. We confirmed the high expression of EIF2S2 in CRC through immunohistochemistry. The correlation between EIF2S2 and 18F-FDG uptake in colorectal cancer and the effect of EIF2S2 knockdown on the levels of genes related to lactate production and partial glycolysis in SW480 cells were investigated.
Methods
ONCOMINE database analysis
The ONCOMINE database (www.oncomine.org) is an integrated online cancer microarray database for DNA or RNA sequence analysis, designed to facilitate discoveries from whole gene expression analysis[32]. In this study, using the ONCOMINE database, we obtained data on the transcriptional expression differences of EIF2 between CRC and normal colorectal tissues. The cutoff values were as follows: P value: 1E-4; fold change: 2; gene rank: 10%; data type: mRNA.
UALCAN
UALCAN (http://ualcan.path.uab.edu) is a comprehensive, user-friendly, and interactive web resource for analyzing cancer omics data from the TCGA database [33]. Differences in EIF2S2 transcript levels were identified through the UALCAN website, including 286 colon cancer tissues and 41 normal colon tissues; 166 rectal cancer tissues and normal rectal tissues in the TCGA database. At the same time, we analyzed the relative expression of EIF2S2 in different groups with variations in sex, age, tumor histology, tumor grade, tumor stage, and lymphnode status). P < 0.05 was considered statistically significant.
GEPIA
Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/index.html) is a tool to analyze RNA sequence expression data of 9736 tumors and 8587 normal tissues [34]. We found 83 Reactome-Glycolysis signaling molecules from the Reactome database [35]. The correlation between EIF2S2 and glycolysis and glucose transport-related proteins was analyzed.
Study population
The study included 42 patients with CRC undergoing 18F-FDG PET/CT examination and surgical resection from February 2015 to October 2019. Inclusion criteria were as follows: (a) CRC confirmed by pathology; (b) no biopsy, radiotherapy or chemotherapy before PET/CT; (c) surgery was performed within 4 weeks after the scan; (d) tissue samples were available for immunohistochemistry (IHC) staining; and (e) case records were complete. The age range of the 28 male patients was 31-78 with a mean age of 57.5 years. 14 female patients had an age range of 40-80 years with a mean age of 57.7 years. They were pathologically diagnosed with CRC and did not receive radiotherapy or chemotherapy before surgical resection and PET/CT imaging.
18F-FDG PET/CT
PET/CT images (Biograph MCT; Siemens) were acquired 1 h after intravenous injection of 18F-FDG and fasting for at least 6 h. Regions of interest (ROIs) were drawn around the contours of the primary tumors on the PET images. SUVmax, SUVmean, total lesion glycolysis (TLG), and metabolic tumor volume (MTV) of each primary tumor were automatically calculated and recorded.
Immunohistochemistry and analysis
After the CRCs were resected, the tumor tissues and adjacent tissues were subjected to immunohistochemical analysis of the expression of EIF2S2 and GLUT1. The sections were deparaffinized, rehydrated, and incubated with citrate buffer under high pressure for 3 min. After cooling to room temperature, the slices were placed in 3% hydrogen peroxide and incubated for 12 min. After blocking the sections with the serum for 20 min, they were incubated with anti-EIF2S2 antibody (1:400, Abcam) and anti-GLUT1 antibody (1:200, Abcam) overnight at 4 °C. The slices were cleaned with tris-buffered saline Tween-20. Donkey anti-rabbit (1:300, Abcam) and goat anti-mouse (1:400, Abcam) secondary antibodies were dropped onto the slice to cover the tissues and then incubated at room temperature for 50 minutes. After the sections were cleaned with TBST, diaminobenzaldehyde (DAB) reagent was added dropwise to develop the color. After rinsing, they were stained with hematoxylin. Next, the slices were dehydrated and sealed. Two experienced physicians evaluated all immunohistochemical staining results and reached agreement. The intensity of membrane/cytoplasmic staining was scored as follows: strong staining of >50% of cancer cells, 3+; moderate staining of ≥10%-50% of cancer cells, 2+; weak staining of <10% of cancer cells, 1+; and no staining, 0. Cases with scores of 2+ and 3+ were rated as highly positive.
Cell culture and cell transfection
Colon cancer cell line SW480 was purchased from the cell bank of the Shanghai Institute of Life Sciences. SW480 was cultured in H-DMEM (GIBCO) supplemented with 10% fetal bovine serum and maintained at 37 °C and 5% CO2 in humid air. EIF2S2 siRNA and a negative control siRNA were designed and synthesized by the Shanghai Gene Pharmaceutical Company. siRNA sequences were listed in Table A .
On the day before transfection, approximately 2 × 106 cells per well were inoculated into 6-well plates. According to the manufacturer's instructions, siRNA was transfected with Lipofectamine™ Reagent (Invitrogen).
List of siRNA sequences
| Name | Sequence |
|---|---|
| siEIF2S2 | Forward 5'- GUCGUCCGAGUAGGAACCATT-3' |
| Reverse 5'- UGGUUCCUACUCGGACGACUU-3' | |
| NC siRNA | Forward 5'- UUCUCCGAACGUACGUTT-3' |
| Reverse 5'- ACGUGACACGUUCGGAGAATT-3' |
Quantitative real-time PCR assay
SW480 cells in the logarithmic growth phase were inoculated with 1 × 105 cells in 24-well plates and infected with siEIF2S2 or siCtrl. Total RNA was extracted from cells and tissues with Trizol reagent (Invitrogen) and a real-time polymerase chain reaction (RT-PCR) was performed with PrimeScript™ RT Master Mix (TakaRa). The expression level of the target gene was analyzed by qRT-PCR in SYBR Green qPCR main mixed reagent system (TakaRa). The PCR primers used are listed in Table B.
List of PCR primers
| Gene name | Primer sequence |
|---|---|
| EIF2S2 | Forward 5'- ACACATACGAGGAGCTGCTGAATC-3' |
| Reverse 5'- AG CTTGGTTCCTACTCGGACGACTTG-3' | |
| SLC2A1 | Forward 5'- TGTCTGGCATCAACGCTGTCTTC-3' |
| Reverse 5'- TC CCTGCTCGCTCCACCACAAAC-3' | |
| SLC2A3 | Forward 5'- TCAATGTGCAGTGTAGCCCA-3' |
| Reverse 5'- AG CTGCCTTACTGCCAACCTAC-3' | |
| SLC2A10 | Forward 5'- TCATTGGCACCATCGGCTTGTC-3' |
| Reverse 5'- CC GGTGAACCGTCTCTTCTGGAACTG-3' | |
| HK2 | Forward 5'- CGACAGCATCATTGTTAAGGAG-3' |
| Reverse 5'- CA GCAGGAAAGACACATCACATTT-3' | |
| PKM2 | Forward 5'- TGCCGCCTGGACATTGATTCAC-3' |
| Reverse 5'- GA GTTCAGACGAGCCACATTCATTCC-3' | |
| LDHA | Forward 5'- TGGCAACTCTAAAGGATCAGC-3' |
| Reverse 5'- TA CCAACCCCAACAACTGTAATCT-3' | |
| β-actin | Forward 5'- GGAGA TTACTGCCCTGGCTCCTA-3' |
| Reverse 5'- GACTCA TCGTACTCCTGCTTG-CTG-3' |
Lactate production assays
An L-Lactic Acid Colorimetric Assay Kit (Elabscience) was used to measure the lactate production according to the manufacturer's protocols. The transfected cells were seeded into 96-well cell culture plate and incubated at 37 °C overnight. After starvation for 2 h, the supernatant was collected to determine lactic acid production. Lactate production was measured at 530 nm with a microplate reader.
Statistical analyses
The expression of EIF2S2 and GLUT1 in tumor tissues and adjacent normal tissues was analyzed by an independent sample t-test. The chi-squared test was used to analyze the correlation between EIF2S2 expression and clinical parameters in patients with CRC. Spearman's correlation coefficient was used for correlation analysis among SUVmax, EIF2S2, and GLUT1 expression. The ROC curve was used to analyze the accuracy of SUVmax to predict the expression of EIF2S2. Multivariate analysis was used to analyze the factors related to the expression of EIF2S2. P <0.05 was considered significant. SPSS software (SPSS, version 26.0) was used for statistical analysis.
Results
Overexpression of EIF2S2 gene in CRC
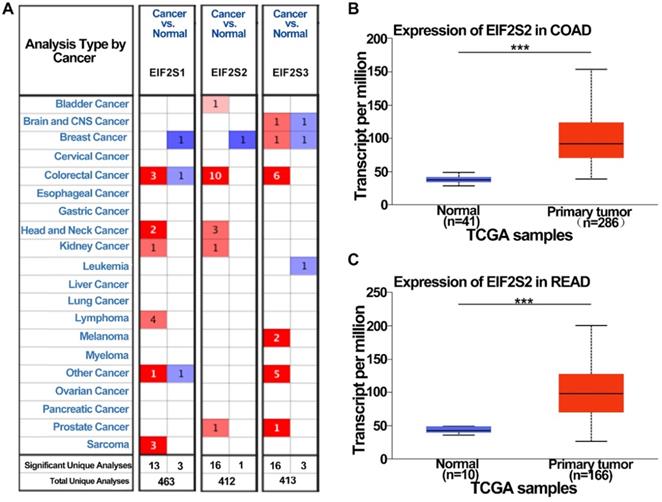
To examine the potential role of EIF2s in CRC development, we first analyzed the expression of EIF2s in normal samples and CRC tissues with the Oncomine database. We found that the expression of EIF2S1, EIF2S2, and EIF2S3 in CRC was higher than that in normal colorectal tissues in several databases (Fig. 1A, P < 0.001). The transcription rate of EIF2S2 in CRC tissues was significantly higher than that in normal colorectal tissues by the UALCAN database (Fig. 1B, C; P < 0.001). These results suggest that EIF2S2 may promote tumorigenesis and progression of CRC.
Relationship between transcriptional rate of EIF2S2 and clinical data of CRC patients
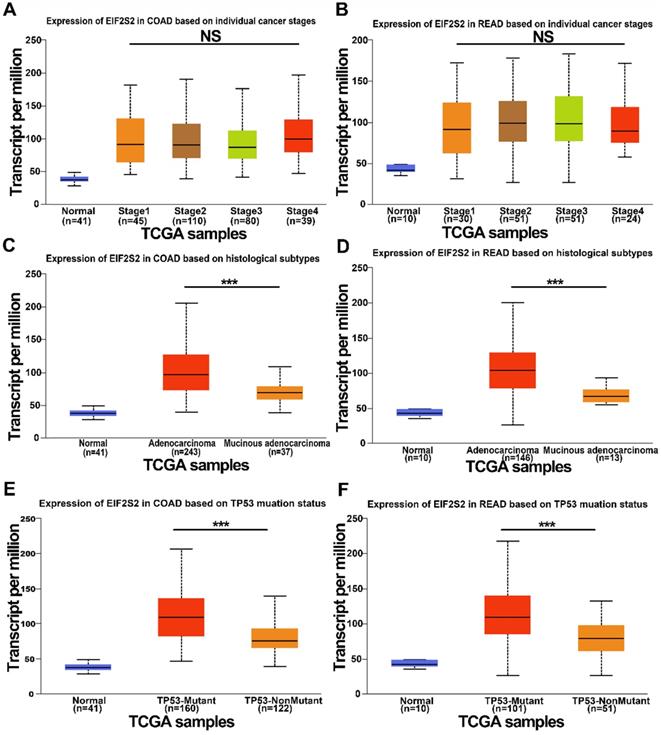
We further examined the expression of EIF2S2 expression in human CRC in the TCGA data using the UALCAN database. It revealed that EIF2S2 remained the same regardless of sex, age, or node metastasis status in CRC patients. Surprisingly, the expression of EIF2S2 had no significant correlation with tumor stage (Fig. 2A, B; P >0.05). However, EIF2S2 expression in colorectal cancer was significantly correlated with TP53 mutation status and tumor histological subtype (Fig. 2C-F [P < 0.001]).
Patient characteristics
The clinical data of the 42 patients with CRC was shown in Table 1. There were 28 males and 14 females. There were 21 patients aged ≥ 60 y and 21 patients < 60 y. A total of 24 cases occurred in the colon, and 18 cases occurred in the rectum. There were 15 tumors ≤ 3 cm and 27 > 3 cm. The degree of differentiation of tumors was divided into 4 types: 3 patients with poorly differentiated, 11 patients with poorly-moderately differentiated, 25 patients with moderately differentiated, and 3 patients with well-differentiated tumors. The tumor stages were 4 patients with Stage 1, 15 with Stage 2, 22 with Stage 3, and 1 with Stage 4. Further analysis of clinical data revealed that the mean SUVmax of the CRCs was 22.4, ranging from 6.88-56.66 (Table 2).
The expression level of EIF2S2 in CRC and adjacent normal tissues and from the TCGA. A, EIF2S2-EIF2S3 mRNA expression (cancer tissue VS normal tissue) was analyzed using the Oncomine database. The numbers in colored cells show the quantities of datasets with statistically significant mRNA overexpression (red) or underexpression (blue) of target genes. Cell color was determined by the best gene rank percentile for the analysis within the cells. The number in each cell represents the number of analyses that satisfied the threshold, such as gene rank percentile (10%), P-value (1E-4), and fold change (2). B, Comparison of EIF2S2 expression between COAD and normal colon tissue. C, Comparison of EIF2S2 expression between READ and normal colon tissue. ***P<0.001.
The expression level of EIF2S2 in CRC and adjacent normal tissues from the TCGA. A, The expression of EIF2S2 in COAD based on different individual cancer stages VS normal tissues by RNA-Seq derived expression data. B, The expression of EIF2S2 in READ, based on different individual cancer stages VS normal tissues by RNA-Seq derived expression data. C, The expression of EIF2S2 in COAD based on different histological subtypes VS normal tissues by RNA-Seq derived expression data. D, The expression of EIF2S2 in READ based on different histological subtypes VS normal tissues by RNA-Seq derived expression data. E, The expression of EIF2S2 in COAD based on different TP53 mutation status VS normal tissues by RNA-Seq derived expression data. F, The expression of EIF2S2 in READ based on different TP53 mutation status VS normal tissues by RNA-Seq derived expression data. ***P<0.001.
Expression of EIF2S2 in CRC patients and its relationship to clinical features
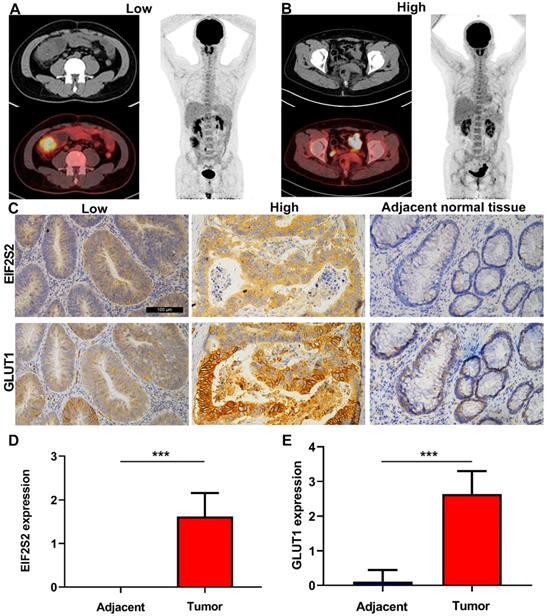
The SUVmax values of tumors in CRC patients were higher than those in normal tissues, but varied (Fig. 3A). Through immunohistochemical staining of the tissues of 42 patients with CRC, we found that EIF2S2 and GLUT1 were more highly expressed in tumor tissues than in adjacent normal tissues. In addition, we found that EIF2S2 is mainly expressed in the cytoplasm, while GLUT1 is expressed in both the cytoplasm and the cell membrane (Fig. 3B). The positive rate of EIF2S2 expression in tumor tissues is 97.6% (41/42) and 0% (0/10) in adjacent normal tissues. Positive GLUT1 expression was observed in 100% (42/42) of the tumors and 10% (1/10) of adjacent normal tissues. The average staining intensity scores of EIF2S2 and GLUT1 in tumor tissues were significantly higher than those in adjacent normal tissues (Fig. 3C, D; P <0.001). Multivariate analysis demonstrated there were no significant differences between EIF2S2 expression and sex, age, tumor size, histological differentiation, tumor staging and microsatellite stability immunohistochemical scores (P > 0.05). However, there was a significant correlation between SUVmax and EIF2S2 expression (Tables 3 & 4).
Representative images of PET/CT and immunohistochemistry. A, PET/CT imaging for CRC patient with high SUVmax (left) and low SUVmax (right). B, Immunohistochemical staining for EIF2S2 and GLUT1 in CRC tissue with different SUVmax and adjacent normal lung tissues (magnification, ×100). C, The mean immunohistochemical staining levels of EIF2S2 (1.96 ± 0.84) in CRC tissue was significantly higher than that of matched adjacent normal lung tissue (0.13 ± 0.34). D, The mean immunohistochemical staining levels of GLUT1 (1.65 ± 0.90) in CRC tissue was significantly higher than that of matched adjacent normal Colorectal tissue (0.83 ± 0.38). ***P<0.001.
Clinicopathological characteristics of 42 patients
| Variables | N | EIF2S2 expression | GLUT1 expression | ||||
|---|---|---|---|---|---|---|---|
| Low | High | P-value | Low | High | P-value | ||
| Sex | |||||||
| Male | 28 | 11 | 17 | 0.506 | 3 | 25 | 0.718 |
| Female | 14 | 4 | 10 | 1 | 13 | ||
| Age | |||||||
| <60 years | 21 | 10 | 11 | 0.113 | 2 | 19 | 1 |
| ≥60 years | 21 | 5 | 16 | 2 | 19 | ||
| Size | |||||||
| ≤3 cm | 15 | 8 | 7 | 0.079 | 2 | 13 | 0.542 |
| >3 cm | 27 | 7 | 20 | 2 | 25 | ||
| Primary lesion | |||||||
| Colon | 24 | 8 | 16 | 0.718 | 3 | 21 | 0.460 |
| Rectum | 18 | 7 | 11 | 1 | 17 | ||
| Histological differentiation | |||||||
| Poorly | 3 | 2 | 1 | 0.826 | 0 | 3 | 0.079 |
| Poorly-Moderately | 11 | 3 | 8 | 0 | 11 | ||
| Moderately | 25 | 9 | 16 | 3 | 22 | ||
| Well | 3 | 1 | 2 | 1 | 2 | ||
| Tumor staging | |||||||
| 1 | 4 | 1 | 3 | 0.455 | 1 | 3 | 0.708 |
| 2 | 15 | 5 | 10 | 0 | 15 | ||
| 3 | 22 | 8 | 14 | 3 | 19 | ||
| 4 | 1 | 1 | 0 | 0 | 1 | ||
Median values of PET parameters (min-max)
| PET metabolic parameter | Value |
|---|---|
| SUVmax | 22.4 (6.88-56.66) |
| SUVmean | 11.9 (3.96-25.81) |
| TLG | 194.7 (9.47-1025.46) |
| MTV | 17.5 (1.56-101.4) |
SUVmax positively correlated with the expression of EIF2S2
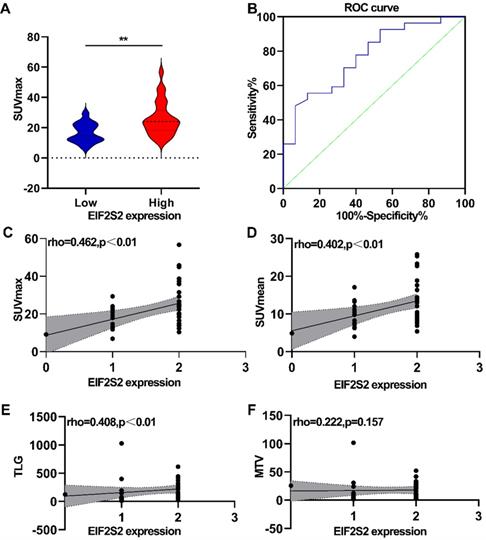
The correlation between PET/CT metabolic parameters and EIF2S2 expression in CRC was analyzed. In CRC, tumor tissues with high expression of EIF2S2 had a significantly higher SUVmax than tissues with low expression (25.6 ± 11.29 vs 16.56 ± 6.39) (Fig. 4A, P <0.05). The ROC curve analysis showed that the area under the curve was 0.769 ± 0.090 (95% CI = 0.623-0.915, P = 0.0042) (Fig. 4B). The expression rate of EIF2S2 in CRC patients was significantly positively correlated with SUVmax, SUVmean, and TLG (Figure 4C-E; P < 0.01), but not with MTV (Fig. 4F, P >0.05). These results suggest that the SUVmax of the tumor in patients with CRC has a certain predictive value for the expression of EIF2S2.
Correlation analysis between PET/CT metabolic parameters and IHC score. A, The SUVmax was higher in high expression of EIF2S2.B, Determination of the cutoff value of SUVmax by the receiver operating characteristics (ROC) curve. The ROC curve was used to determine the optimal cutoff value of SUVmax for predicting EIF2S2 high positive CRC. Area under the curve: 0.769; 95% CI: 0.623 to 0.915. C, D The SUVmax, SUVmean TLG showed a linear correlation with the EIF2S2 IHC score with a correlation coefficient of 0.462, 0.402, 0.408respectively (P< 0.001). E, F, There was no significant correlation between EIF2S2 IHC score and MTV. **P<0.01.
Multivariate analysis of EIF2S2 expression in 42 CRC patients
| Parameter | Univariate analysis | P | |
|---|---|---|---|
| OR | 95%CI | ||
| Sex | 1.997 | 0.409-9.740 | 0.393 |
| Age (years) | 2.472 | 0.610-10.025 | 0.205 |
| Tumor size (cm) | 3.110 | 0.760-12.722 | 0.115 |
| Histological differentiation | 1.111 | 0.437-2.825 | 0.825 |
| Tumor staging | 0.599 | 0.212-1.689 | 0.333 |
| SUVmax | 5.000 | 1.143-21.864 | 0.033 |
OR: Odds ratio. 95% CI: 95% Confidence interval.
Correlation between the parameters of 18F-FDG uptake and GLUT1 expression
By analyzing the relationship between PET/CT parameters and GLUT1 expression, no significant correlation was detected between SUVmean and SUVmax in 18F-FDG and GLUT1 expression (P > 0.05). In contrast, TLG and MTV were significantly correlated with GLUT1 expression (P < 0.01) (Table 5).
Comparison of STR IHC score with EIF2S2 expression
| Variables | N | EIF2S2 expression | ||
|---|---|---|---|---|
| Low | High | P-value | ||
| MSH1 | ||||
| Low | 2 | 0 | 2 | 0.203 |
| High | 29 | 10 | 19 | |
| MSH2 | ||||
| Low | 0 | 0 | 0 | |
| High | 31 | 10 | 21 | |
| MSH6 | ||||
| Low | 0 | 0 | 0 | |
| High | 31 | 10 | 21 | |
| PMS2 | ||||
| Low | 8 | 2 | 6 | 0.605 |
| High | 23 | 8 | 15 | |
Comparison of PET parameter according to GLUT1 expression
| PET metabolic parameter | GLUT1-Low (n = 11) | GLUT1-High (n = 31) | P-value |
|---|---|---|---|
| SUVmax (mean ± SD) | 25.02 ±14.06 | 22.14 ±10.48 | 0.714 |
| SUVmean (mean ± SD) | 13.99± 8.18 | 11.71 ±5.09 | 0.620 |
| TLG (mean ± SD) | 73.64±51.14 | 207.47± 202.89 | 0.005 |
| MTV (mean ± SD) | 6.22± 4.37 | 18.70± 18.36 | 0.003 |
EIF2S2 may mediate glycolytic metabolism in CRC patients
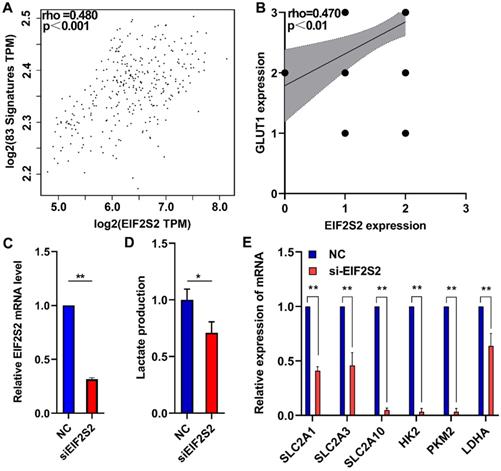
A total of 83 glycolysis and glucose transporters were found from the Reactome database. A significant correlation was found between glycolysis signal molecules and EIF2S2 by using the GEPIA database (Fig. 5A). From the analysis of the previous immunohistochemistry results, there was a significant positive correlation between the expression of EIF2S2 and GLUT1 (Fig. 5B). Further lactate production assays showed that lactic acid production (a key metabolite of glycolysis) was significantly decreased after EIF2S2 knockdown in SW480 cells (Fig. 5C, D). SLC2A1, SLC2A3, SLC2A10, HK2, PKM2, and LDHA were significantly decreased after EIF2S2 knockdown in SW480 cells (Fig. 5E). These data indicate that EIF2S2 may mediate glycolysis in CRC, thereby promoting the occurrence and development of CRC.
Discussion
In this paper, to study the expression of EIF2S2 in CRC comprehensively, the overexpression of EIF2S2 and its relationship with clinicopathological features were identified using the TCGA database. The results showed that the expression of EIF2S2 was significantly higher in CRC than adjacent normal tissues, the higher expression of EIF2S2 is associated with worse overall survival of patients with CRC [31]. Therefore, EIF2S2 is expected to be a prognostic marker and potential therapeutic target for CRC.
EIF2S2 is closely correlated with glycolytic metabolism in CRC. A, The expression of EIF2S2 showed a linear correlation with related to Reactome-Glycolysis and Reactome-Glucose-Transport gene sets with a correlation coefficient of 0.480, respectively (P<0.001) . B, The expression of EIF2S2 showed a linear correlation with the expression of GLUT1 with a correlation coefficient of 0.470 in CRC, respectively (P<0.01). C, Real-time PCR assays were performed to analyze the relative EIF2S2 levels in SW480 cells after knockdown of EIF2S2, n=3. D, Lactate production were assessed in SW480 cells after knockdown of EIF2S2, n= 3 E, Real-time PCR assays were performed to analyze the relative SLC2A1, SLC2A3, SLC2A10,HK2,PKM2,LDHA levels and in SW480 cells after knockdown of EIF2S2, n=3. * P<0.05, ** P<0.01.
It was found that EIF2S2 expression is associated with tissue subtype and TP53 mutation, but not related to age, sex, tumor differentiation, tumor stage, or lymphnodal status in CRC patients. We performed immunohistochemistry on the surgically removed tumor tissues and adjacent normal tissues in 42 patients with CRC. That EIF2S2 expression is not related to age, sex, tumor differentiation, or tumor stage, is consistent with the data from the UALCAN database. When we analyzed the correlation between EIF2S2 and Mismatch Repair (MMR) protein expression, it was found that there was no significant correlation between the expression of EIF2S2 and the expression of MSH2 and PMS2. But there are no patients with microsatellite instability in 31 patients, the relationship between EIF2S2 and microsatellite stability could not be evaluated. By analyzing the correlation between the expression level of EIF2S2 in CRC patients and the related parameters of PET/CT (SUVmax, SUVmean, MTV, and TLG), we found that CRC with high EIF2S2 expression is accompanied by high uptake of 18F-FDG. ROC analysis showed that 18F-FDG uptake could predict the expression of EIF2S2 in CRC patients. In addition, multivariate analysis showed that there was a correlation between SUVmax and EIF2S2 expression. This result suggests that we can predict the expression of EIF2S2 through SUVmax, and also suggests that EIF2S2 may affect the glucose uptake process of aerobic glycolysis in CRC.
Immunohistochemical staining confirmed the correlation between EIF2S2 expression and GLUT1 expression. We confirmed that some glycolysis-related mRNA levels and lactate production and some glycolysis-related mRNA levels (SLC2A1, SLC2A3, SLC2A10, HK2, PKM2, LDHA) were reduced after EIF2S2 knockdown. Therefore, these results suggest that EIF2S2 may be involved in the glycolysis by regulating GLUT1 in CRC. In previous reports, there have been different opinions about SUVmax and GLUT1 in CRC [36, 37]. Moreover, the values of MTV and TLG have been linked with the prognosis of CRC [38, 39]. While TLG and MTV significantly correlated with GLUT1, the correlation between SUVmax, SUVmean and the expression of GLUT1 had no significant correlation in the study. Although EIF2S2 was significantly correlated with SUVmax and GLUT1 in CRC, it is not known whether EIF2S2 affects glucose uptake by acting directly on GLUT1.
The TCGA database shows that the expression of EIF2S2 is related to the mutation level of TP53 in CRC. The p53 pathway plays a vital role in the aerobic glycolysis pathway of malignant tumor [40, 41]. Jiwei Zhang et al. reported that EIF2S2 could interact with c-MYC [31]. In tumor cells, c-MYC can promote the expression of glucose transporter 1 (GLUT1) [42]. We speculate that EIF2S2 may regulate the metabolism of CRC by inhibiting the p53 signaling pathway or promoting the c-MYC-GLUT1 signaling pathway. In the future, we plan to analyze how EIF2S2 acts on the anaerobic glycolysis mechanism of CRC.
The present study is not without limitations. First of all, due to geographical limitations and the absence of multicenter sampling, the samples in this study are not representative of the whole population, which increases the possibility of selection bias. Second, our study sample size was relatively small. Third, our study was retrospective in nature. Therefore, a larger sample of cases, multicenter sampling, and prospective randomized studies are needed to verify our findings.
We tentatively put forward that the common PET/CT parameter SUVmax has a certain predictive value for the expression of EIF2S2 in CRC. Meanwhile, the comprehensive analysis of the TCGA database and clinicopathological features including PET parameters not only provides us with new ideas for finding new diagnostic and therapeutic targets for tumors, but also provides a theoretical basis for us to study further the pathways of differential gene expression affecting tumor aerobic glycolysis analysis.
Abbreviations
CRC: Colorectal carcinoma; COAD: colon adenocarcinoma; READ: rectal adenocarcinoma; EIF2S2: Eukaryotic Translation Initiation Factor 2 Subunit Beta; PET/CT: Positron emission tomography/computed tomography; 18F-FDG: 18F-fluorodeoxyglucose; SUV: standard uptake value; GLUT1: Glucose transporter 1; HK2: hexokinase 2; TCGA: The Cancer Genome Atlas; ROI: region of interest; TBST: Tris-Buffered Saline Tween-20; SD: standard deviation; ROC: receiver operating characteristic.
Acknowledgements
This work was supported by the Hubei Province's Outstanding Medical Academic Leader program, the Foundation for Innovative Research Team of Hubei Provincial Department of Education T2020025, the HuBei Provincial Department of Science and Technology Innovation Group Programme (grant no. 2019CFA034), Science and Technology Development Foundation of Shiyan City (grant no. 19Y33), Innovative Research Program for Graduates of Hubei University of Medicine (grant no. YC2020011) and the Key Discipline Project of Hubei University of Medicine.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70:7-30
2. Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927-30
3. Warburg O. On the origin of cancer cells. Science. 1956;123:309-14
4. Cerfolio RJ, Bryant AS, Ohja B, Bartolucci AA. The maximum standardized uptake values on positron emission tomography of a non-small cell lung cancer predict stage, recurrence, and survival. J Thorac Cardiovasc Surg. 2005;130:151-9
5. Cornelis FH, Petre EN, Vakiani E, Klimstra D, Durack JC, Gonen M. et al. Immediate Postablation F-FDG Injection and Corresponding SUV Are Surrogate Biomarkers of Local Tumor Progression After Thermal Ablation of Colorectal Carcinoma Liver Metastases. J Nucl Med. 2018;59:1360-5
6. Shi D, Cai G, Peng J, Li D, Li X, Xu Y. et al. The preoperative SUVmax for (18)F-FDG uptake predicts survival in patients with colorectal cancer. BMC Cancer. 2015;15:991
7. Gao Y, Li F, Zhou H, Yang Y, Wu R, Chen Y. et al. Down-regulation of MRPS23 inhibits rat breast cancer proliferation and metastasis. Oncotarget. 2017;8:71772-81
8. Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654-62
9. Pastorino JG, Hoek JB. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr Med Chem. 2003;10:1535-51
10. Golshani-Hebroni SG, Bessman SP. Hexokinase binding to mitochondria: a basis for proliferative energy metabolism. J Bioenerg Biomembr. 1997;29:331-8
11. Yang H-J, Xu W-J, Guan Y-H, Zhang H-W, Ding W-Q, Rong L. et al. Expression of Glut-1 and HK-II in Pancreatic Cancer and Their Impact on Prognosis and FDG Accumulation. Transl Oncol. 2016;9:583-91
12. Kitasato Y, Yasunaga M, Okuda K, Kinoshita H, Tanaka H, Okabe Y. et al. Maximum standardized uptake value on 18F-fluoro-2-deoxy-glucose positron emission tomography/computed tomography and glucose transporter-1 expression correlates with survival in invasive ductal carcinoma of the pancreas. Pancreas. 2014;43:1060-5
13. Bos R, van Der Hoeven JJM, van Der Wall E, van Der Groep P, van Diest PJ, Comans EFI. et al. Biologic correlates of (18)fluorodeoxyglucose uptake in human breast cancer measured by positron emission tomography. J Clin Oncol. 2002;20:379-87
14. Kaida H, Azuma K, Kawahara A, Sadashima E, Hattori S, Takamori S. et al. The assessment of correlation and prognosis among F-FDG uptake parameters, Glut1, pStat1 and pStat3 in surgically resected non-small cell lung cancer patients. Oncotarget. 2018;9:31971-84
15. Lu K, Yang J, Li DEC, He S-B, Zhu D-M, Zhang L-F. et al. Expression and clinical significance of glucose transporter-1 in pancreatic cancer. Oncol Lett. 2016;12:243-9
16. Kaira K, Okumura T, Ohde Y, Takahashi T, Murakami H, Oriuchi N. et al. Correlation between 18F-FDG uptake on PET and molecular biology in metastatic pulmonary tumors. J Nucl Med. 2011;52:705-11
17. Ancey P-B, Contat C, Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS J. 2018;285:2926-43
18. Martel F, Guedes M, Keating E. Effect of polyphenols on glucose and lactate transport by breast cancer cells. Breast Cancer Res Treat. 2016;157:1-11
19. Alvarez JV, Belka GK, Pan T-C, Chen C-C, Blankemeyer E, Alavi A. et al. Oncogene pathway activation in mammary tumors dictates FDG-PET uptake. Cancer research. 2014;74:7583-98
20. Baschnagel AM, Wobb JL, Dilworth JT, Williams L, Eskandari M, Wu D. et al. The association of (18)F-FDG PET and glucose metabolism biomarkers GLUT1 and HK2 in p16 positive and negative head and neck squamous cell carcinomas. Radiother Oncol. 2015;117:118-24
21. Riedl CC, Akhurst T, Larson S, Stanziale SF, Tuorto S, Bhargava A. et al. 18F-FDG PET scanning correlates with tissue markers of poor prognosis and predicts mortality for patients after liver resection for colorectal metastases. J Nucl Med. 2007;48:771-5
22. Zhou L-M, Yuan L-L, Gao Y, Liu X-S, Dai Q, Yang J-W. et al. Nucleophosmin 1 overexpression correlates with F-FDG PET/CT metabolic parameters and improves diagnostic accuracy in patients with lung adenocarcinoma. Eur J Nucl Med Mol Imaging. 2021;48:904-12
23. Liu X-S, Yuan L-L, Gao Y, Zhou L-M, Yang J-W, Pei Z-J. Overexpression of METTL3 associated with the metabolic status on F-FDG PET/CT in patients with Esophageal Carcinoma. J Cancer. 2020;11:4851-60
24. Turner M, Diaz-Munoz MD. RNA-binding proteins control gene expression and cell fate in the immune system. Nat Immunol. 2018;19:120-9
25. Saini Y, Chen J, Patial S. The Tristetraprolin Family of RNA-Binding Proteins in Cancer: Progress and Future Prospects. Cancers (Basel). 2020;12:1539
26. Shah M, Su D, Scheliga JS, Pluskal T, Boronat S, Motamedchaboki K. et al. A Transcript-Specific eIF3 Complex Mediates Global Translational Control of Energy Metabolism. Cell Rep. 2016;16:1891-902
27. Koromilas AE. Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim Biophys Acta. 2015;1849:871-80
28. Kashiwagi K, Takahashi M, Nishimoto M, Hiyama TB, Higo T, Umehara T. et al. Crystal structure of eukaryotic translation initiation factor 2B. Nature. 2016;531:122-5
29. Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nature genetics. 2014;46:1051-9
30. Tanaka I, Sato M, Kato T, Goto D, Kakumu T, Miyazawa A. et al. eIF2β, a subunit of translation-initiation factor EIF2, is a potential therapeutic target for non-small cell lung cancer. Cancer Sci. 2018;109:1843-52
31. Zhang J, Li S, Zhang L, Xu J, Song M, Shao T. et al. RBP EIF2S2 Promotes Tumorigenesis and Progression by Regulating MYC-Mediated Inhibition via FHIT-Related Enhancers. Mol Ther. 2020;28:1105-18
32. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1-6
33. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-58
34. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
35. Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P. et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649-D55
36. Gu J, Yamamoto H, Fukunaga H, Danno K, Takemasa I, Ikeda M. et al. Correlation of GLUT-1 overexpression, tumor size, and depth of invasion with 18F-2-fluoro-2-deoxy-D-glucose uptake by positron emission tomography in colorectal cancer. Dig Dis Sci. 2006;51:2198-205
37. Hong R, Lim S-C. ¹⁸F-fluoro-2-deoxyglucose uptake on PET CT and glucose transporter 1 expression in colorectal adenocarcinoma. World J Gastroenterol. 2012;18:168-74
38. Jiang H, Zhang R, Jiang H, Zhang M, Guo W, Zhang J. et al. Retrospective analysis of the prognostic value of PD-L1 expression and F-FDG PET/CT metabolic parameters in colorectal cancer. J Cancer. 2020;11:2864-73
39. Xu J, Li Y, Hu S, Lu L, Gao Z, Yuan H. The significant value of predicting prognosis in patients with colorectal cancer using F-FDG PET metabolic parameters of primary tumors and hematological parameters. Ann Nucl Med. 2019;33:32-8
40. Assaily W, Benchimol S. Differential utilization of two ATP-generating pathways is regulated by p53. Cancer Cell. 2006;10:4-6
41. Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer research. 2004;64:2627-33
42. Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M. et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797-800
Author contact
![]() Corresponding authors: Zhijun Pei, E-mail: pzjun1980net; Changbin Ke, E-mail: changbinke-iapcom.
Corresponding authors: Zhijun Pei, E-mail: pzjun1980net; Changbin Ke, E-mail: changbinke-iapcom.

 Global reach, higher impact
Global reach, higher impact