Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(20):6242-6253. doi:10.7150/jca.58004 This issue Cite
Research Paper
Pan-cancer Analysis of NEDD4L and Its Tumor Suppressor Effects in Clear Cell Renal Cell Carcinoma
1. Fujian Provincial Key Laboratory of Transplant Biology, Fuzong Clinical College, Fujian Medical University, Fuzhou 350025, China
2. Laboratory of Basic Medicine, Dongfang Hospital (900 Hospital of the Joint Logistics Team), Xiamen University, Fuzhou 350025, China
3. Department of Urology, 900 Hospital of the Joint Logistics Team, Fuzhou 350025, Fujian, China
4. Nephrology and Urology Department, The Second Affiliated Hospital of Wannan Medical College, Wuhu, China.
# These authors have contributed equally to this work.
Received 2021-1-9; Accepted 2021-8-9; Published 2021-8-28
Abstract
The expression level of NEDD4L, an E3 ubiquitin ligase, has changed significantly in human cancers. In this study, we aimed to study the expression of NEDD4L in pan-carcinoma and its function in malignant tumors. We analyzed the gene expression level of NEDD4L in pan-cancer from The Cancer Genome Atlas (TCGA) microarray data set, the correlation between gene expression and overall survival, disease-specific survival, and tumor immune microenvironment changes. NEDD4L expression changes in half of the cancer types. Low expression of NEDD4L gene predicts poor overall survival and disease-specific survival (DSS) in renal clear cell carcinoma (KIRC) and renal chromophobe cell carcinoma (KIRP). NEDD4L is negatively related to interstitial cell infiltration and immune cell infiltration in most common cancers. Furthermore, the low expression of NEDD4L was verified in our clear cell renal cell carcinoma (ccRCC) clinical tissues. In ccRCC cells, NEDD4L overexpression significantly reduced cell proliferation and migration. In the functional analysis, we proved that NEDD4L could inhibit ERBB3 and MAPK signaling pathways. When cells are deficient in nutrition, NEDD4L promoted the degradation of the autophagy regulatory protein ULK1. Our study provides novel insights into the role of NEDD4L in pan-cancer. NEDD4L may play a tumor suppressor effect in ccRCC, through tumor immune regulation and ubiquitination of key intracellular kinases.
Keywords: NEDD4L, pan-cancer, TCGA, immune cell infiltration, ubiquitination
Introduction
Ubiquitination mediated by the ubiquitin-proteasome system is an important process of post-translational modification of biological proteins, and plays a very important role in protein localization, metabolism, regulation and degradation [1]. Studies have found that the ubiquitin-proteasome system can regulate the biological functions of tumor cell proliferation and metastasis by mediating the degradation of a variety of tumor-related proteins [2]. The key enzymes involved in this system include ubiquitin activating enzyme E1, ubiquitin conjugating enzyme E2 and ubiquitin ligase E3 [3]. Different ubiquitin ligase E3 mediates the transfer of activated ubiquitin from the conjugate enzyme E2 to the substrate, and acts on different substrate proteins, which determines the specificity of ubiquitination modification. The substrate specificity of ubiquitin ligase E3 makes it considered a good target for cancer treatment.
NEDD4L (Neural Precursor Cell Expressed Developmentally Down-Regulated 4-Like), also known as NEDD4-2, is a member of the neural precursor cell expression down-regulated factor 4 (NEDD4) protein family. The expression level of NEDD4L has also changed significantly in human cancers. In gallbladder cancer cells, the expression level of NEDD4L was significantly up-regulated compared with normal epithelial cells [4]. At the same time, NEDD4L is generally highly expressed in melanoma cells [5]. However, in prostate cancer specimens, the expression of NEDD4L was significantly reduced compared with neighboring normal tissues [6]. It is reported that NEDD4L expression level is significantly down-regulated in non-small cell lung cancer [7]. Down-regulation of NEDD4L expression enhances the aggressiveness of NSCLC tumors [8]. At the same time, NEDD4L low expression group has a shorter survival time than non-low expression group. Similar observations have also been reported in colorectal cancer [9]. NEDD4L is related to the regulation of many central pathways in cancer, including TGF-β, Wnt and EGFR signaling pathways [9, 10].
The Cancer Genome Atlas (TCGA) contains data on 33 cancers from more than 10,000 patients [11]. The expression of NEDD4L in the data of TCGA pan-cancer species, its correlation with patient survival, the main function of genes, and whether it will affect the tumor immune microenvironment and other issues are in this paper through TCGA data analysis to draw conclusions. Select a tumor type whose NEDD4L gene expression significantly affects the prognosis of patients, and focus on how NEDD4L affects the malignant phenotype of tumor cells.
Materials and Methods
Data obtained from UCSC Xena Databases
We downloaded gene expression RNASeq (HTSeq-Fragments Per Kilobase per Million (FPKM)) data, somatic mutation data (varscan2_snv.tsv.gz), survival data and an integrated TCGA pan-cancer clinical resource (TCGA-CDR) from UCSC Xena Databases (http://xena.ucsc.edu/).
Pan-Cancer NEDD4L Expression Profile Analysis
We used a Perl script to generate a txt file of NEDD4L gene expression. Gene expression summary was performed by a R package “ggpubr”. NEDD4L protein expression summary among cancer types was obtain from The Human Protein Atlas (www.proteinatlas.org).
Survival Analysis of NEDD4L Expression
According to the median expression value of NEDD4L, we divided the patients in each cohort into "high" and "low" groups. The Kaplan-Meier method is used to estimate the survival function, and the R software package is used for overall survival analysis and the corresponding visualization "survival" and "survminer". Disease-related survival analysis and forest plots of all tumors are visualized using R packages "survival", "survminer", and "forestplot".
Gene Set Enrichment Analysis (GSEA) analysis of KIRC
The c2.cp.kegg.v7.1.symbols.gmt and c5.all.v7.1.symbols.gmt files are downloaded from the molecular signature database (MSigDB, http://software.broadinstitute.org/gsea/index.jsp). These two files were used for GSEA analysis. According to the median expression of NEDD4L gene in renal clear cell carcinoma samples, we divided them into high expression group and low expression group. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were completed by using the clusterprofiler R package. According to the default weighted enrichment statistical method, this process is repeated 1,000 times for each analysis. According to p value < 0.05, false discover rate (FDR) < 0.25 and p value < 0.05, FDR < 0.05 were used to screen statistically significant pathways and biological processes.
The relationship between NEDD4L gene expression and tumor microenvironment
The content of stromal cells and total immune cells in tumor tissues is realized by Pearson correlation analysis, which is realized by R package "estimate" analysis [12]. Only the tumor sample data in TCGA-kidney renal clear cell carcinoma (KIRC) is retained. CIBERSORT's (cell type identification by estimating relative subsets of known RNA transcripts) algorithm has been used to evaluate the composition of immune cells in many cancer types [13]. The content of different types of immune cells in KIRC tumor tissues is achieved by "CIBERSORT.R", p Value < 0.001. Relevant pictures are visualized by R package "ggplot2" "ggpubr" "ggExtra".
Clinical specimen
From November 2013 to November 2015, 50 cases of renal clear cell carcinoma and matched adjacent tissues (> 2 cm from the edge of the cancerous tissue) were selected from the renal cancer patients in Fuzhou General Hospital. The patients were 28-77 years old, with an average age of 55.5 years. All cancer tissue specimens were confirmed to be clear cell renal cell carcinoma (ccRCC) by pathology. The isolation and use of human tissues were approved by the Human Research Ethics Review Committee of Fuzhou General Hospital (approval number 2013-017). All patients provided written informed consent. The pathological classification was verified according to Furhman classification. The details were described in our previous study [14, 15].
Cell culture
ACHN and 786-O cells were provided by Shanghai Jikai Gene Technology Co., Ltd. (Shanghai, China). Cells are routinely checked for mycoplasma contamination every 2-3 months. ACHN and 786-O cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Inc.). All the cells were cultured at 37 °C in a humidified incubator under 5% CO2.
Quantitative polymerase chain reaction
Total RNA was extracted from tissues using Total RNA Kit II (Omega Bio-Tek, Inc., Norcross, GA, USA). After RNA quantification, 3 μg of total RNA was reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis kit. PowerUp SYBR Green mix (Applied Biosystems, Thermo Fisher Scientific, Inc.) is used for quantitative polymerase chain reaction (qPCR). Use NEDD4L and β-actin primers to perform qPCR on cDNA: NEDD4L forward primer, 5'-CAGGGTCCAGAAGCAGATGA-3' and reverse primer, 5'-GAGTAGCACAGCCTTCCAGA-3'; α-actin forward primer 5'-TGACGTGGACATCCGCAAAG-3' and reverse primer 5'-CTGGAAGGTGGACAGCGAGG-3'. PCR was carried out for 40 cycles under the following conditions: 30 seconds at 94 °C and 60 seconds at 58 °C. By using the formula 2-∆∆Ct to normalize β-actin mRNA, the relative fold change of mRNA expression is calculated [16].
Lentiviral and siRNA transfection
The lentivirus pLVX-Puro-NEDD4L and the control virus pLVX-Puro were purchased from a public protein/plasmid library (PPL, Nanjing, China). For virus infection, the virus is diluted with serum-containing cell culture medium containing 8 μg/mL polyethylene. Change to fresh medium after 12 hours. Then puromycin (1 μg/mL, Sigma) was added to select stable cells (10 days).
NEDD4L silencing was performed using one custom made siRNAs targeting NEDD4L mRNA region (si-NEDD4L sense: CCUCUGUAAUGAGGAUCAUUUTT, si-NEDD4L antisense: AAAUGAUCCUCAUUACAGAGGTT) and one scramble control siRNA (HippoBio, China) and transfected by Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.). Transfection Reagent followed the manufacturer's instructions.
Water-soluble tetrazole (WST)-1 detects cell proliferation
Take the cells of the experimental group and the control group, adjust the cell number to 2×104 cells/ml, and inoculate 100 μL of cells into a 96-well cell culture plate. After 24, 48, and 72 hours of adherence to the cells, 10 μL of water-soluble tetrazole (WST-1) reagent (Roche, Mannheim, Germany) was added, and incubated for 45 minutes. Then, a spectrophotometer (Multiskan GO, Thermo Scientific) was used to measure the optical density (D450) of each hole at a wavelength of 450 nm. Four repeat holes are set for each group.
NEDD4L integrated network analysis
In order to gain more insights into NEDD4L functionality. We used UbiBrowser (http://ubibrowser.ncpsb.org/ubibrowser/home/index) to predict the ubiquitination substrate of E3 ubiquitinase, NEDD4L, and obtained data from BioGRID (https://thebiogrid.org/) for molecules that interact with NEDD4L. Pathway Commons (https://www.pathwaycommons.org) is used for multi-gene interaction analysis. STRING (https://string-db.org/) is used for multi-gene function prediction.
Immunohistochemical staining
A tissue microarray (HKidCRC060CS01; Shanghai Outdo Biotech Co., China) was constructed with formalin-fixed paraffin-embedded 22 paired ccRCC and adjacent non-tumor renal tissues. Five of these pairs have distal normal renal tissues. The paraffin-embedded sections were stained with antibodies against NEDD4L (dilution at 1:500; HPA024618) at 4 °C overnight and incubated with secondary antibody. Negative controls were stained with isotype control IgG. Then, the sections were counterstained with hematoxylin and eosin. Immunohistochemical staining intensity was based on the proportion of cell staining and scored from 0 to 3 following criteria described previously [17]. The slides were analyzed by standard light microscopy.
Western Blot
For each group of proteins quantified by bicinchoninic acid (BCA), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed at 50 μg/well. After electrophoresis, the cells were transferred to the polyvinylidene fluoride (PVDF) membrane under a constant pressure of 60 V for 180 minutes. The blocking solution was blocked for 1 hour and reacted with the primary antibody overnight at 4 °C. Tris-Buffered Saline Tween-20 (TBST) was washed three times; horseradish peroxidase (HRP)-labeled goat anti-rabbit antibody was reacted at room temperature for 1 h, and TBST was washed 4 times; electrogenerated chemiluminescence (ECL) was added, exposed on X-ray film, film development, fixing, washing with water, drying, storage and photographing. NEDD4L, MEK, ERK, phospho-MEK, phospho-ERK, ERBB3, p62, LC3, ULK1, ACTB (β-actin) antibodies were purchased from Cell Signaling Technology (Denver, Massachusetts, USA). Horseradish peroxidase (HRP) labeled secondary antibody and ECL kit were purchased from Pierce (Thermo Fisher Scientific, Inc.).
Statistical processing
Use SPSS13.0 software for statistical analysis. The data obtained are expressed as mean ± SD. The single-factor repeated measures analysis of variance found a statistically significant difference when p value < 0.05.
Results
Pan-cancer expression landscape of NEDD4L
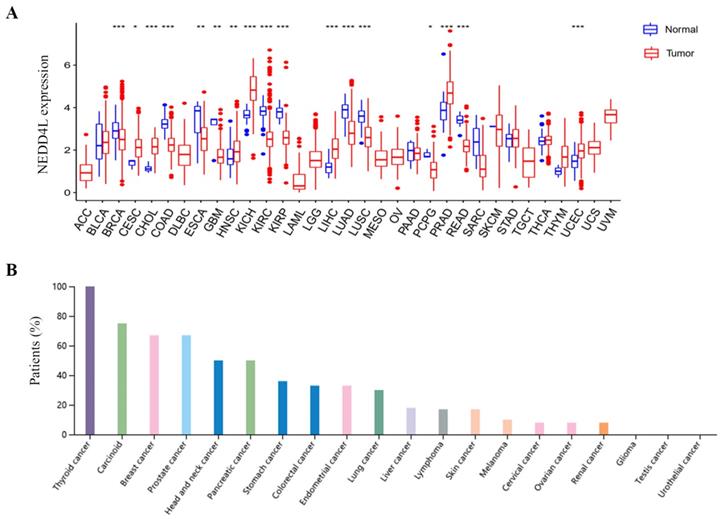
Among all 33 cancer types in the TCGA database, NEDD4L expression was significantly changed in 17 tumors, accounting for 51.52% of all cancer types. NEDD4L expression in cancer was significantly increased (according to cancer samples and normal samples) in cholangiocarcinoma (CHOL), kidney chromophobe (KICH), liver hepatocellular carcinoma (LIHC), prostate adenocarcinoma (PRAD) and uterine corpus endometrial carcinoma (UCEC), whereas, NEDD4L expression significantly reduced in breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), KIRC, kidney renal papillary cell carcinoma (KIRP), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and rectum adenocarcinoma (READ) (Figure 1A, ***, p-value < 0.001). Figure 1B shows the protein expression rates of NEDD4L in patients of various cancers. Weak to moderate cytoplasmic positivity was observed in most cancers, such as thyroid, carcinoid, breast, and prostate cancers. Urothelial, testis, glioma and renal cancers were almost negative.
Analysis of NEDD4L expression. (A) Box plots of NEDD4L mRNA levels in neoplastic tissues. Blue represents non-tumor tissues and red represents tumor tissues. * p-value < 0.05, ** p-value < 0.01, ***p-value < 0.001. (B) Bar charts of NEDD4L expression analysis (immunohistochemical staining) in various cancers. The results were download from The Human Protein Atlas.
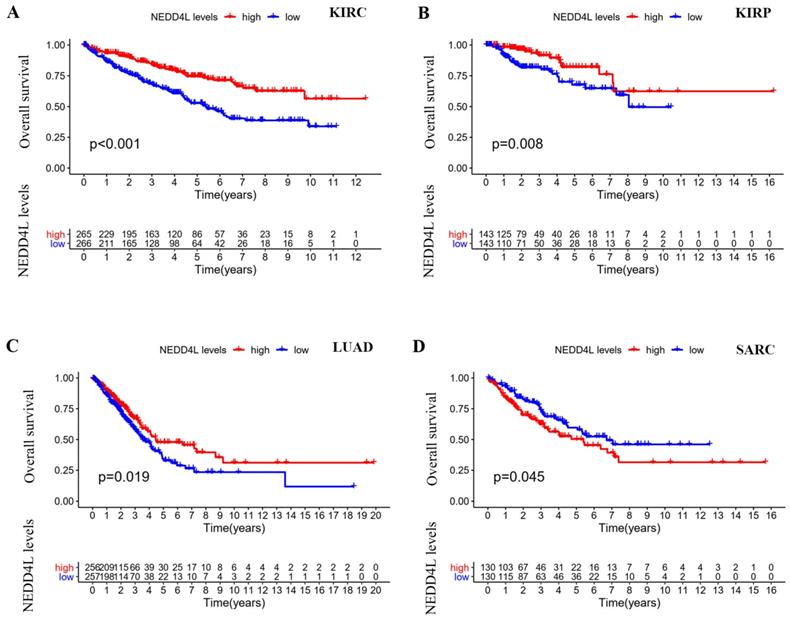
Overall survival analysis in TCGA cancers. (A-D) Kaplan-Meier curves represent NEDD4L association with patient overall survival in KIRC, KIRP, LUAD and SARC. p-value < 0.05.
NEDD4L expression and prognostic association
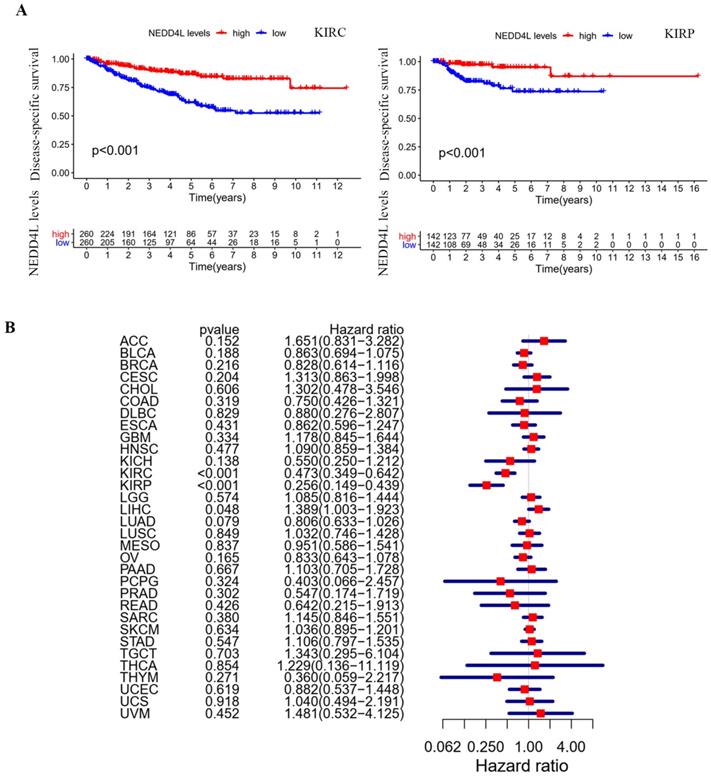
Kaplan-Meier plots [18] were used to evaluate overall survival (OS) and disease-specific survival (DSS). The DSS forest diagram is used to illustrate the correlation between NEDD4L gene expression and DSS in various cancer types in the TCGA database. Among the 33 cancer types in the TCGA database, there were four cancer types whose expression of NEDD4L protein can affect the overall survival of patients: KIRC, KIRP, LUAD, and sarcoma (SARC). NEDD4L was low in KIRC, KIRP, and LUAD, and lower gene expression predicts poor patient survival (Figure 2A). Low expression of NEDD4L gene in KIRC and KIRP was correlated with worse DSS (KIRC: hazard ratio: 0.473, 95% confidence interval: 0.349-0.642; p-value< 0.001; KIRP: hazard ratio: 0.256, 95% confidence interval: 0.149 -0.439; p-value< 0.001) (Figure 3B). These results indicated that NEDD4L gene expression may be one of the prognostic factors of KIRC and KIRP.
Pan-cancer analysis of NEDD4L expression and tumor microenvironment
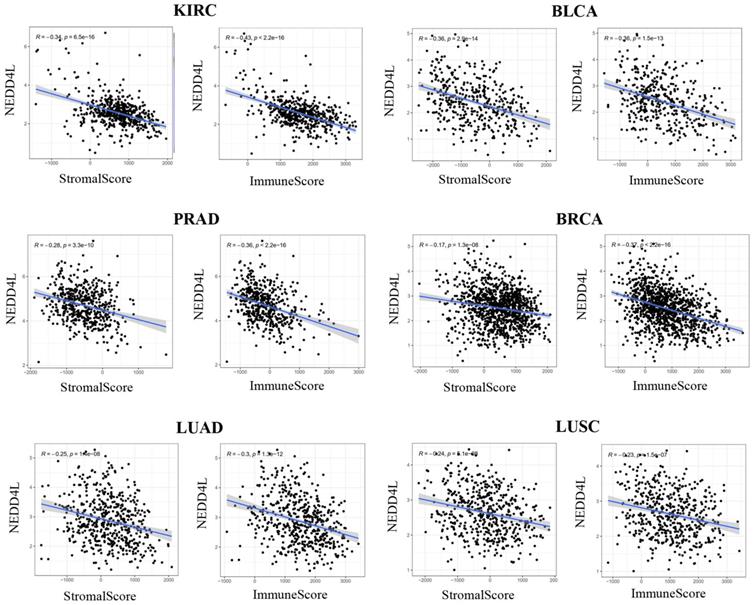
The tumor immune microenvironment plays a vital role in the development of many cancers [19, 20]. ESTIMATE algorithm can infer the StromalScore (stromal cell infiltration level) and ImmuneScore (immune cell infiltration level) in tumor tissue. The correlation between NEDD4L gene expression and stromal cell infiltration levels or immune cell infiltration level in various types of tumor tissues has not yet been reported. In most cancer types (20 in 34 types, p-value <0.05), the expression of NEDD4L is negatively correlated with the infiltration value of interstitial cells and immune cells in tumor tissues (Table S1). Figure 4 shows that the top six cancer types with negative correlation are: KIRC, Bladder urothelial carcinoma (BLCA), PRAD, BRCA, LUAD and LUSC.
Disease-specific survival (DSS) analysis in TCGA cancers. (A) Kaplan-Meier curves represent NEDD4L association with DSS in KIRC and KIRP. p-value < 0.05. (B) Forest plot of the association of NEDD4L gene with DSS in each of cancers.
The relationship between NEDD4L expression and immune cell infiltration in the TCGA-KIRC dataset
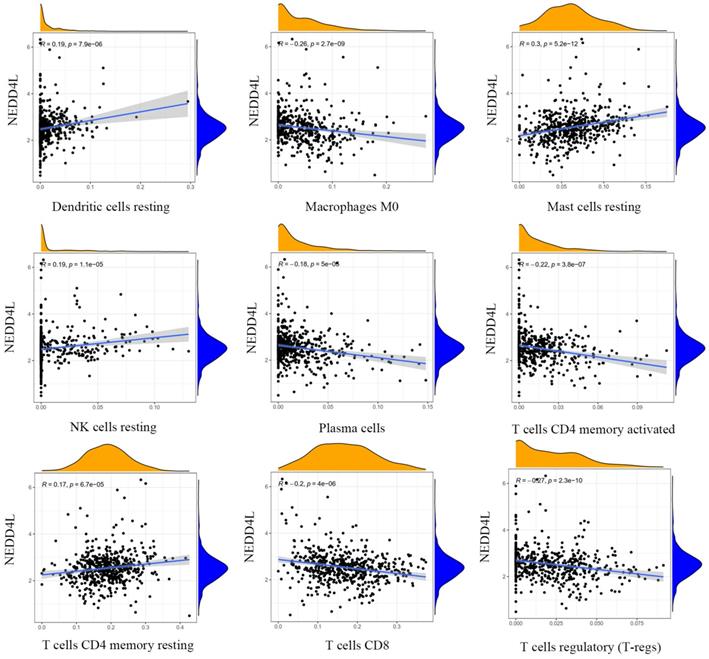
The results of gene expression and tumor microenvironment analysis indicate that NEDD4L may be closely related to immune function. There are a large number of lymphocyte infiltrations in tumor tissues, to further explore the influence of NEDD4L on the infiltrated patterns of specific lymphocyte types, using CIBERSORT, we investigated the relationship between the expression level of NEDD4L and the infiltration level of 22 immune cells in ccRCC (Figure 5). We found that NEDD4L expression is significantly negatively correlated with the ratio of three T cells, including cluster of differentiation 4 (CD4) memory activated T cells, CD8 positive T cells, regulatory T cells (T-regs), and positively correlated with CD4 positive T cells that have tumor-killing effects. T-regs are a type of CD4 + T cells that have powerful inhibitory functions. T-regs have been found to have an increased infiltration rate in many types of cancers, and a higher T-reg tumor infiltration rate indicates a poor clinical prognosis. In addition, most T cell are dysfunction in tumor tissues, such as only 10% of CD8-positive T cells have tumor recognition ability. These results suggest that NEDD4L may inhibit tumorigenesis and development by inhibiting the formation of tumor immune microenvironment.
Correlations between expression level of NEDD4L and stromal cell or immune cell infiltration levels in KIRC, BLCA, PRAD, BRCA, LUAD and LUSC.
Gene function analysis of NEDD4L in KIRC dataset
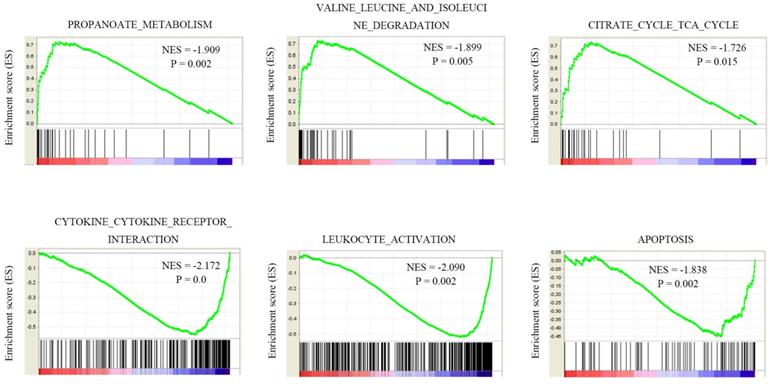
As we shown NEDD4L is significantly lower in KIRC, and the low expression of NEDD4L can be used as a predictor of poor prognosis in KIRC. The function of NEDD4L in the occurrence of ccRCC is not clear, so we chose to study the role of NEDD4L abnormal expression in tumorigenesis in KIRC. TCGA-KIRC data is used to analyze NEDD4L gene set enrichment analysis (GSEA). The analysis results found that acetone metabolism, valine, leucine and isoleucine degradation pathways, and citrate cycle (TCA cycle) are positively correlated with NEDD4L expression levels. These metabolic pathways are down-regulated in ccRCC. Cytokine-cytokine-receptor interaction, leukocyte-activation, apoptosis pathway, etc. are negatively related to NEDD4L. These results indicate that NEDD4L may participate in tumor immune regulation and affect tumor cell growth and apoptosis.
Overexpression of NEDD4L inhibits the growth and migration of renal clear cell carcinoma cell lines
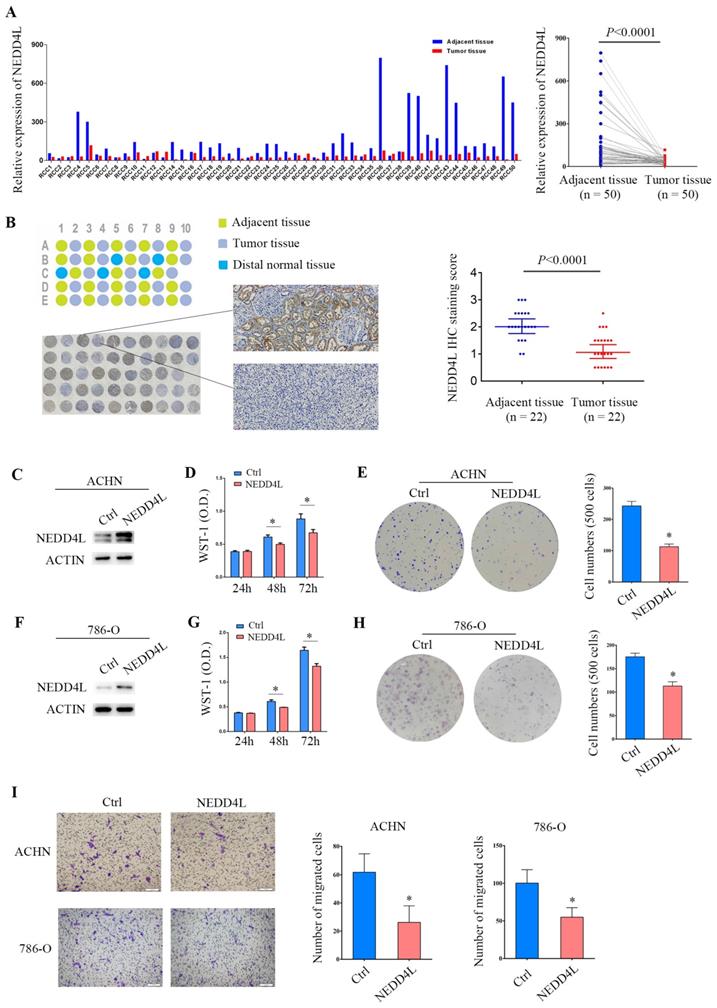
From 2013 to 2015, our center collected 50 pairs of ccRCC tumor tissues and their adjacent tissues that were pathologically identified and surgically removed [14, 15]. Quantitative PCR analysis showed that NEDD4L expression decreased in 82% of tumor tissues. Paired analysis showed that the level of NEDD4L mRNA in renal clear cell carcinoma tissue was significantly reduced (p <0.0001) (Figure 7A). A ccRCC paraffin tissue microarray was used for immunohistochemical detection. Results showed the protein levels of NEDD4L is also decreased in ccRCC tissues (p <0.0001) (Figure 7B).
Next, we tried to check the function of NEDD4L in ccRCC cells using a lentivirus-mediated overexpression system. It was confirmed that NEDD4L was highly expressed in ACHN cells (Figure 7C). The results of WST-1 determination showed that the proliferation of NEDD4L overexpression group was significantly lower than that of the control group at 48 and 72 hours after the cells grew adherently (p<0.05, Figure 7D). Compared with the control group, the ability of ACHN cells with stable NEDD4L expression to form colonies was significantly reduced (Figure 7E). These data were verified in another ccRCC cell line, 786-O cell line (Figure 7F-7H). In addition, in cells overexpressing NEDD4L, cell migration was significantly reduced (Figure 7I). These data collectively indicate that NEDD4L is a tumor suppressor of ccRCC.
NEDD4L deregulates the stability of several protein kinases via ubiquitination activity in ccRCC
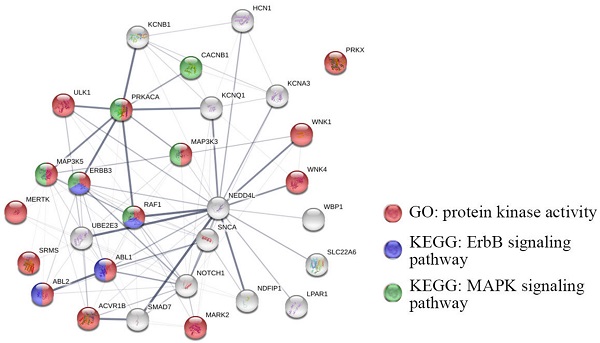
NEDD4L is an E3 ubiquitinase [21] and there are many proteins regulated by it [22, 23]. We used UbiBrowser to predict the ubiquitination substrate of E3 ubiquitinase, NEDD4L, and 743 molecules with the confidence score greater than 0.8. The data of molecules interacting with NEDD4L are obtained from BioGRID. There are 207 interacting molecules with evidence of ≥ 2 in BioGRID. Figure 8A shows 28 ubiquitinated substrates that interact with NEDD4L. STRING analysis results show that half of these genes have protein kinase activity (Figure 8B). Several genes are members of the ERBB3 or MAPK signaling pathway. In ACHN cells, NEDD4L is stably and highly expressed, and the phosphorylation levels of MEK and ERK in the cell are significantly reduced (Figure 8C), indicating that NEDD4L may degrade MEK upstream kinases through ubiquitination to inhibit the MAPK signaling pathway. ERBB3 is the direct ubiquitination substrate of NEDD4L [10, 24]. In ACHN cells, high expression of NEDD4L could result in a decrease in MAPK pathway and ERBB3 protein, whereas down-regulated NEDD4L caused increased levels of MAPK pathway and ERBB3 protein (Figure 8C).
Correlations between expression level of NEDD4L and the infiltration levels of dendritic cells resting, macrophage M0, mast cells resting, NK cells resting, plasma cells, T cells CD4 memory activated, T cells CD4 memory resting, T cells CD8, and T-regs.
GSEA analysis of NEDD4L co-expression genes in TCGA-KIRC samples.
Protein kinase ULK1 has been reported to regulate cell autophagy and energy metabolism. When cell nutrition is deficient, ULK1 ubiquitination and degradation participate in autophagy regulation. NEDD4L is an E3 ubiquitinase involved in the regulation of ULK1 degradation during autophagy [25, 26]. If NEDD4L is knockdown, the autophagy activity will be increased. In our ccRCC cell line, cells cultured by Earle's Balanced Salt Solution (EBSS) are deficient in nutrients and autophagy occurs. In cells with high NEDD4L expression, ULK1 is more easily degraded (Figure 8D). The results of protein interaction analysis in Pathway Common also showed that MAPK upstream kinases (MAP3K3, MAP3K5), ERBB3 and ULK1, which are important for tumor cell growth and proliferation, can bind to NEDD4L (Figure 8E). Our data indicate that in renal cancer cells, NEDD4L may inhibit tumor cell growth by ubiquitinating multiple kinase molecules.
Discussion
In this study, we first analyzed the overall expression of NEDD4L in 33 cancer types in TCGA data and found that NEDD4L expression was abnormally changed in a variety of tumors. Subsequent overall survival analysis and disease-specific survival analysis found that low NEDD4L expression in KIRC and KIRP patients was associated with poor prognosis. Tumor immune microenvironment analysis found that NEDD4L gene expression is negatively correlated with stromal cell infiltration level and immune cell infiltration level in most cancer types. KIRC negative correlation is the most significant. Therefore, we focus on KIRC.
In the analysis of various immune cell ratios in the TCGA-KIRC data set, it was found that the low expression of NEDD4L may indicate an increase in the ratio of immunosuppressive T cells. GSEA gene function enrichment analysis results suggest that NEDD4L participates in tumor immune regulation in KIRC, and affects tumor cell growth and apoptosis. In our samples, we verified that NEDD4L expression decreased in ccRCC tissues. Two ccRCC cell lines ACHN and 786-O were infected with NEDD4L gene expression lentivirus. In cells stably overexpressing NEDD4L, cell proliferation slows down and the ability to form cell clones decreases. Transwell experiments found that the cell migration ability of NEDD4L overexpressing cells was inhibited.
NEDD4L is E3 ubiquitinase, which can regulate multiple signaling pathways in cells through the ubiquitination degradation pathway [22, 23]. In previous studies, it has been shown that both NEDD4 and NEDD4L can bind to ERBB3 (HER3) to promote ubiquitination and degradation of ERBB3 [10, 24]. ERBB3 is an EGFR family protein, which is closely related to the occurrence and development and poor prognosis of breast cancer, ovarian cancer, prostate cancer, lung cancer and other tumors [27, 28]. Uncoordinated 51-like kinase 1 (ULK1) plays a vital role in autophagy, initiating autophagy when starved [29, 30]. ULK1 can also phosphorylate key glycolytic enzymes and maintain redox homeostasis [31]. In the progress of autophagy, NEDD4L is involved in ULK1 ubiquitination and degradation [24]. Lee et al. show that NEDD4L binds to ULK1, ULK1 expression was stable in NEDD4L knockdown in pancreatic cancer cells [25]. In recent years, ULK1 disorders have been found in several human cancers [32]. Our previous research results show that ULK1 is highly expressed in ccRCC, and ULK1 inhibitors can effectively inhibit the proliferation of ccRCC cells [33]. Here, our results indicate that NEDD4L can also degrade ERBB3 in kidney cancer cells. NEDD4L can promote the degradation of ULK1 when cells in starvation condition.
Effects of overexpression of NEDD4L on ccRCC cell growth. (A) 50 pairs of ccRCC samples were used to detect NEDD4L mRNA levels by quantitative-PCR. (B) Immunohistochemical detection of NEDD4L. There are 22 pairs of tissue samples in the tissue chip, 5 of which contain distal normal tissue controls. (C) ACHN cells stably transfected with NEDD4L lentivirus and performed by Western blot analysis. (D) The growth inhibition rates were measured by WST-1 assay kit at 24, 48, and 72 hours in ACHN cells that stably expressed NEDD4L. (E) Representative images of clonogenic assays of ACHN cells that stably expressed NEDD4L. (F) Western blot assay was performed in NEDD4L overexpressed 786-O cells. (G) WST-1 assay in 786-O cells that stably expressed NEDD4L. (H) Colony formation assay in 786-O cells that stably expressed NEDD4L. (I) Representative images of migration assays of ACHN and 786-O cells stably expressing NEDD4L and quantification of the relative migration cell number (right). Scale bar, 100 µm. All experiments were repeated three times. The p-value was measured using Student's t-test; * p < 0.05, compared with the control cells. Ctrl: cells transfected with empty control lentivirus.
NEDD4L overexpression degrades the protein kinases. (A) Overlap of genes showing those both with predicted the ubiquitination substrates and confirmed interacting molecules of NEDD4L. (B) Biological network enrichment of 28 intersection genes predicted by STRING analysis. (C) Representative western blot analysis of NEDD4L overexpressed (NEDD4L), down-regulated (si-NEDD4L) or control cells. The levels of NEDD4L, phospho (p)-MEK, MEK, p-ERK and ERK, ERBB3 and β-actin (ACTIN) in ACHN cells were measured. The result is representative of three separate experiments. (D) NEDD4L overexpressed or control cells were plated overnight and subsequently treated with EBSS for 6 h and resolved in an SDS-PAGE gel. Samples were immunoblotted against NEDD4L, LC3, p62, ULK1 and ACTIN. (E) Protein-protein interactions network of 28 intersection genes predicted by Pathway Common.
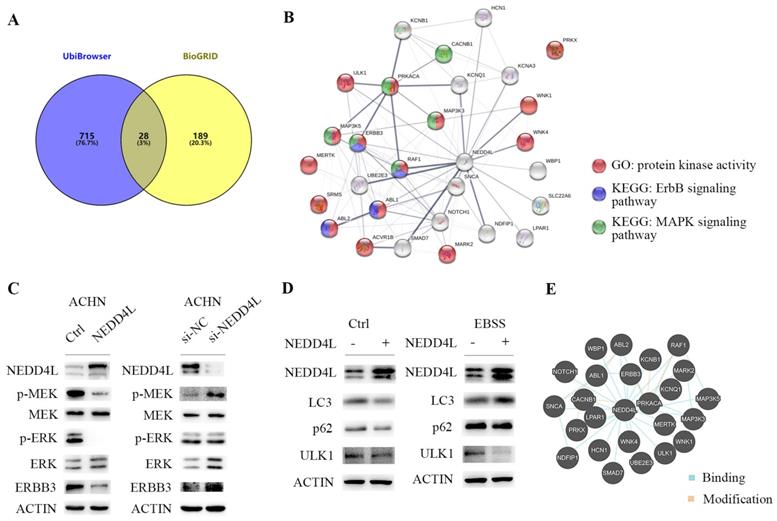
Among the predicted NEDD4L ubiquitination substrates, there are several genes in the MAPK signal pathway, especially MAP3K3 are important kinases upstream of MAPK. In our study, it was found that the MAPK signal pathway was reduced in NEDD4L overexpressed renal carcinoma cells but activated in NEDD4L knockdown cells. This result preliminarily indicates that NEDD4L may inhibit the activation of MAPK pathway by ubiquitinating MAPK upstream kinase. MAP3K3 has been shown to participate in the growth and proliferation of a variety of tumor cells [34, 35]. In future research, we will thoroughly explore the ubiquitination regulation effect of NEDD4L on MAP3K3.
Taken together, NEDD4L expression is abnormal in most common cancers. The low expression of NEDD4L predicts poor overall survival and disease-specific survival of KIRC and KIRP. NEDD4L may be involved in regulating the tumor immune microenvironment, and ubiquitinating and degrading key cell growth kinases to suppress the growth of renal cancer.
Abbreviations
BCA: bicinchoninic acid; ccRCC: clear cell renal cell carcinoma; CD4: cluster of differentiation 4; GSEA: gene set enrichment analysis; GO: gene ontology; HRP: horseradish peroxidase; KEGG: kyoto encyclopedia of genes and genomes; PVDF: polyvinylidene fluoride; SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis; TCGA: the cancer genome atlas; WST: water-soluble tetrazolium salt; ACC: adrenocortical carcinoma; BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: cholangiocarcinoma; COAD: colon adenocarcinoma; DLBC: diffuse large B-cell lymphoma; ESCA: esophageal carcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LAML: acute myeloid leukemia; LGG: lower grade glioma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MESO: mesothelioma; OV: ovarian serous cystadenocarcinoma; PADD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; SARC: sarcoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; UCEC: uterine corpus endometrial carcinoma; UCS: uterine carcinosarcoma; UVM: uveal melanoma.
Supplementary Material
Supplementary table S1.
Acknowledgements
The authors are grateful to the TCGA database for providing high-quality data.
Funding
This work was supported by the Joint Funds for the Innovation of Science and Technology of Fujian province (grant numbers: 2018Y9115), Natural Science Foundation of Fujian Province (grant numbers: 2018J01345 and 2019J01528), and Outstanding Youth Funds of the 900 Hospital of the Joint Logistics Team (grant numbers: 2017Q07).
Ethic Approval and Consent to Participate
The use of human tissues was approved by the Fuzhou General Hospital IRB, (Fuzhou, China), with written consent (2013-017).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jarome TJ, Devulapalli RK. The Ubiquitin-Proteasome system and memory: Moving beyond protein degradation. Neuroscientist. 2018;24(6):639-51
2. Yamaguchi H, Hsu JL, Hung MC. Regulation of ubiquitination-mediated protein degradation by survival kinases in cancer. Front Oncol. 2012;2:15
3. Varshavsky A. The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev Biochem. 2017;86:123-28
4. Takeuchi T, Adachi Y, Nagayama T. et al. Nedd4L modulates the transcription of metalloproteinase-1 and -13 genes to increase the invasive activity of gallbladder cancer. Int J Exp Pathol. 2011;92(2):79-86
5. Kito Y, Bai J, Goto N. et al. Pathobiological properties of the ubiquitin ligase Nedd4L in melanoma. Int J Exp Pathol. 2014;95(1):24-28
6. Gao C, Pang L, Ren C. et al. Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Med Oncol. 2012;29(3):1733-38
7. Sakashita H, Inoue H, Akamine S. et al. Identification of the NEDD4L gene as a prognostic marker by integrated microarray analysis of copy number and gene expression profiling in non-small cell lung cancer. Ann Surg Oncol. 2013;20(Suppl 3):S590-98
8. Qu MH, Han C, Srivastava AK. et al. MiR-93 promotes TGF-beta-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37(4):5645-51
9. Tanksley JP, Chen X, Coffey RJ. NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. Plos One. 2013;8(11):e81514
10. Verma N, Muller AK, Kothari C. et al. Targeting of PYK2 synergizes with EGFR antagonists in basal-like TNBC and circumvents HER3-Associated resistance via the NEDD4-NDRG1 axis. Cancer Res. 2017;77(1):86-99
11. Weinstein JN, Collisson EA, Mills GB. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113-20
12. Yoshihara K, Shahmoradgoli M, Martinez E. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612
13. Newman AM, Liu CL, Green MR. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-57
14. Lu J, Chen Z, Zhao H. et al. ABAT and ALDH6A1, regulated by transcription factor HNF4A, suppress tumorigenic capability in clear cell renal cell carcinoma. J Transl Med. 2020;18(1):101
15. Shen Z, Zhu L, Zhang C. et al. Overexpression of BHLHE41, correlated with DNA hypomethylation in 3'UTR region, promotes the growth of human clear cell renal cell carcinoma. Oncol Rep. 2019;41(4):2137-47
16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-08
17. Tang Y, Geng Y, Luo J. et al. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci Rep. 2015;5:9476
18. Jager KJ, van Dijk PC, Zoccali C. et al. The analysis of survival data: The Kaplan-Meier method. Kidney Int. 2008;74(5):560-65
19. Lei X, Lei Y, Li JK. et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126-33
20. Cassim S, Pouyssegur J. Tumor microenvironment: A metabolic player that shapes the immune response. Int J Mol Sci. 2019 21(1)
21. Ding Y, Zhang Y, Xu C. et al. HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J Biol Chem. 2013;288(12):8289-98
22. Donovan P, Poronnik P. Nedd4 and Nedd4-2: Ubiquitin ligases at work in the neuron. Int J Biochem Cell Biol. 2013;45(3):706-10
23. Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): The ubiquitin ligase for multiple membrane proteins. Gene. 2015;557(1):1-10
24. Huang Z, Choi BK, Mujoo K. et al. The E3 ubiquitin ligase NEDD4 negatively regulates HER3/ErbB3 level and signaling. Oncogene. 2015;34(9):1105-15
25. Nazio F, Carinci M, Valacca C. et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J Cell Biol. 2016;215(6):841-56
26. Lee DE, Yoo JE, Kim J. et al. NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. Cell Death Dis. 2020;11(1):38
27. Ma J, Lyu H, Huang J. et al. Targeting of erbB3 receptor to overcome resistance in cancer treatment. Mol Cancer. 2014;13:105
28. Kiavue N, Cabel L, Melaabi S. et al. ERBB3 mutations in cancer: Biological aspects, prevalence and therapeutics. Oncogene. 2020;39(3):487-502
29. Shang L, Wang X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy. 2011;7(8):924-26
30. Zachari M, Ganley IG. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61(6):585-96
31. Li TY, Sun Y, Liang Y. et al. ULK1/2 constitute a bifurcate node controlling glucose metabolic fluxes in addition to autophagy. Mol Cell. 2016;62(3):359-70
32. Liu L, Yan L, Liao N. et al. A review of ULK1-Mediated autophagy in drug resistance of cancer. Cancers (Basel). 2020 12(2)
33. Lu J, Zhu L, Zheng LP. et al. Overexpression of ULK1 represents a potential diagnostic marker for clear cell renal carcinoma and the antitumor effects of SBI-0206965. Ebiomedicine. 2018;34:85-93
34. Fan Y, Ge N, Wang X. et al. Amplification and over-expression of MAP3K3 gene in human breast cancer promotes formation and survival of breast cancer cells. J Pathol. 2014;232(1):75-86
35. Zhang Y, Wang SS, Tao L. et al. Overexpression of MAP3K3 promotes tumour growth through activation of the NF-kappaB signalling pathway in ovarian carcinoma. Sci Rep. 2019;9(1):8401
Author contact
![]() Corresponding author: Jun Lu, Xierhuan Road No. 156, Fuzhou General Clinical College / Dongfang Hospital / 900th Hospital of the Joint Logistics Team, Fuzhou, 350025, China. E-mail addresses: junlu.heatheredu.cn.
Corresponding author: Jun Lu, Xierhuan Road No. 156, Fuzhou General Clinical College / Dongfang Hospital / 900th Hospital of the Joint Logistics Team, Fuzhou, 350025, China. E-mail addresses: junlu.heatheredu.cn.

 Global reach, higher impact
Global reach, higher impact