Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(22):6600-6609. doi:10.7150/jca.60724 This issue Cite
Research Paper
MIR-4507 Targets TP53 to Facilitate the Malignant Progression of Non‐small‐cell Lung Cancer
1. Department of Oncology, Henan Provincial People's Hospital, People's Hospital of Zhengzhou University, Zhengzhou, Henan, 450003, China.
2. Shenzhen People's Hospital, The Second Clinical Medical College of Jinan University, Shenzhen 518020, China.
3. Cancer Center, Integrated Hospital of Traditional Chinese Medicine, Southern Medical University, Guangzhou, 510315, China.
4. Department of Medical Imaging, Henan Provincial People's Hospital, People's Hospital of Zhengzhou University, Zhengzhou, Henan, 450003, China.
#These authors contributed equally to this work.
Received 2021-3-22; Accepted 2021-8-15; Published 2021-9-13
Abstract
Lung cancer is a serious threat to human health due to its high morbidity and mortality. microRNAs (miRNAs) are involved in the tumorigenesis and progression of lung cancer. In this study, we elucidated the role of miRNA-4507 (miR-4507) in the pathogenesis of non‐small‐cell lung cancer (NSCLC). miR-4507 is found to be upregulated in NSCLC cells (A549, H460). MTT, 5-ethynyl-2'-deoxyuridine (EdU), wound healing, and transwell assays were performed to evaluate NSCLC cell proliferation and migration. The results demonstrated that miR-4507 inhibition significantly decrease the proliferation and migration of NSCLC cells. Subsequently, a luciferase activity assay was conducted to verify the regulation of the predicted gene target of miR-4507, namely, TP53. Mechanism experiments show that miR-4507 activates the PI3K/AKT signal. Further, we co-transfected miR-4507 mimics and TP53 plasmids and found that TP53 overexpression could recover the effects of miR-4507 mimics on proliferation, migration, and the PI3K/AKT signal activation. These results suggested that miR-4507 targets TP53 to facilitate the proliferation and migration of lung cancer cells through PI3K/AKT signal and that miR-4507 could serve as a potential target for NSCLC treatment.
Keywords: miR-4507, migration, non‐small‐cell lung cancer, proliferation, TP53
Introduction
Lung cancer is the most common cancer, with the highest tumor incidence and mortality in the world [1-3]. In the latest guidelines published by the World Health Organization (WHO) [4], lung cancer is divided into two subtypes according to the histological classification characteristics: small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC). NSCLC accounts for about 85% of the total lung cancer incidence [5, 6]. There are three main subtypes of NSCLC: adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma. Although great advances have been made in cancer treatments, including surgery, radiation, chemotherapy, and targeted therapies [7], lung cancer prognosis remains poor due to the presence of locally advanced or widely metastatic lesions in many patients by the time of diagnosis. Thus, exploring the underlying mechanisms of the tumorigenesis of lung cancer is especially important for lung cancer treatment.
MicroRNAs (miRNAs) are highly conserved in different organisms (including invertebrates, vertebrates, and plants), and they participate in many biological processes, such as cell cycle regulation, differentiation, and development [8]. miRNAs are also widely involved in the tumorigenesis of cancers, affecting progression in terms of proliferation, migration, and invasion [9-12]. In 2014, miRNA-4507 (miR-4507) was first found to affect the three prime untranslated regions (3'UTRs) of pulmonary surfactant-associated protein A1 (SFTPA1), which plays a vital role in maintaining normal lung function and in-host defense [13, 14]. In this study, miR-4507 was found to be highly expressed in NSCLC tissues, and further experiments demonstrated its role in tumor proliferation and migration. The target genes of miR-4507 were predicted using bioinformatics websites. Our results revealed that miR-4507 is overexpressed in NSCLC and promotes proliferation and migration of NSCLC cells via TP53.
The TP53 gene, which encodes the p53 protein (which has a molecular weight of 53 kDa), is a famous tumor suppressor gene [15, 16]. TP53 mutations occur in over 50% of malignant tumors [17, 18]. However, the mutation frequency of TP53 is different in different tumor types, with the highest frequency in uterine carcinosarcoma [19]. In lung adenocarcinoma, the frequency reaches more than 60%. TP53 is altered in most SCLC cases and in about 50% of NSCLC cases [20]. Researchers have found that lung cancer cells experience apoptosis when wild-type TP53 is re-expressed after TP53 mutation or deletion, and clinical trials have confirmed the anti-tumor activity, feasibility, and safety of TP53 gene therapy [21]. TP53 mutations may also affect the effectiveness of radiotherapy [22]. Introducing wild-type TP53 into tumor cells to recover the function of the p53 protein can help rebuild the tumor's sensitivity to radiotherapy and chemotherapy. Correlations between miRNAs and TP53 in NSCLC have already been reported, yet the relationships between miR-4507 and TP53 were first found in the present study. Our work further clarifies the mechanism of TP53 regulation in NSCLC.
Materials and Methods
Cell Cultures
Lung cancer cell lines including H1299, H460, SPC-A1, A549 and normal human lung epithelial cell (BEAS-2B) were obtained from the Cancer Research Institute of Zhengzhou University (Zhengzhou, China) and grown in 1640 supplemented with 10% fetal bovine serum (FBS) (Gibco,USA).
Cell Transfections
The lung cancer cells were transfected with miR-4507 mimics (cuggguugggcugggcuggg) or NC mimics, and miR-4507 inhibitor (cccagcccagcccaacccag) or NC inhibitor, the transfection efficiencies were evaluated by RT-qPCR. Plasmids were purchased from Vigene Biosciences (Shandong, China). Mimics and inhibitors were designed and synthesized by Guangzhou RiboBio (Guangzhou, China). Before transfection, exponentially growing cells were seeded in a cell culture plate or dish (NEST Biotech, China). Plasmids, mimics, and inhibitors were then transfected into cells using Lipofectamine TM 2000 (Invitrogen Biotechnology, Shanghai, China) according to the manufacturer's protocol. Cells were collected 48-72 h after transfection for further experimentation.
MTT and EdU assays
Cell proliferation capacity was determined by MTT assay. Cells were seeded in 96-well plates at a density of 3,000 cells/well. After incubation, each well was added using MTT (5 mg/mL) (Sigma-Aldrich, MO, USA), and incubated for 4 h. At the end of incubation, supernatants were removed, and dimethyl sulfoxide (Sigma-Aldrich, MO, USA) was added to each well. The absorbance value (OD) of each well was measured at 490 nm. The calculated rates were then used for curve fitting. All assays were independently performed three times. For EdU assay, proliferating NSCLC cells were examined using the Cell-Light EdU Apollo 488 or 567 In vitro Imaging Kit (RiboBio).
Transwell Assay
The transwell (BD Biosciences, NJ, USA) assay was used to test cell migration abilities. Cells were suspended in 100 mL 1640 without serum and seeded into the top chamber of the transwells coated with Matrigel (BD Biosciences, NJ, USA) or left uncoated, and the bottom chambers were filled with 500 mL 1640 supplemented with 10% FBS. The migrated cells were stained with crystal violet and then photographed and quantified by counting the cell numbers in five random fields. All assays were independently performed in triplicate.
Wound Healing Assay
Cells were seeded and grew in 6-well plates until a confluent monolayer was reached, and scratches (wounding) were created using a pipette tip. Progression of migration was photographed at initiation and 0 and 48h after wounding. All experiments were repeated at least three times.
In vivo tumorigenesis in nude mice
In vivo experiments were approved by the Animal Care and Use Committee of Zhengzhou University and were performed in accordance with the National Institute of Health guide for the care and use of laboratory animals. A total of 1×106 logarithmically growing NSCLC cells transfected with miR-4507 or the control (N=5 per group) in 0.1 ml Hank's solution were subcutaneously injected into the mice (BALB/C, nu/nu, 4 weeks-old, male). After 20 days, the mice were sacrificed and tumor tissues were excised and weighed.
Quantitative Real-Time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, USA), and cDNA was synthesized using Hiscript reverse transcriptase (Vazyme Biotech, USA) according to manufacturer's instructions. To measure the miRNA expression, real-time PCR was performed using One Step PrimeScript miRNA cDNA synthesis kit and SYBR premix Ex Taq II kit. Glyceraldehyde-3- phosphate dehydrogenase (GAPDH) or U6 small nuclear RNA (snRNA) was used as an endogenous normalization control. The primers are listed below: miR-4507 (cuggguugggcugggcuggg); TP53 (Forward: CCTCAGCATCTTATCCGAGTGG; Reverse: TGGATGGTGGTACAGTCAGAGC).
Western blot analysis
Western blotting analyses were performed with standard methods. Proteins were separated by 10% SDS-PAGE gels, and blotted onto polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membrane was probed with the specific antibodies (Cell Signaling Technology, USA; p53: #2524; GAPDH: # 5174S), and then with peroxidase-conjugated secondary antibodies. GAPDH were used as a protein loading control. The bands were visualized by eECL Western Blot Kit (CWBIO Technology, Beijing, China). The images were captured with ChemiDocTM CRS+ Molecular Imager (Bio-Rad, Hercules, CA, USA).
In situ hybridization
Lung cancer tissues were obtained from Henan Provincial People's Hospital, People's Hospital of Zhengzhou University. In situ hybridization was carried out by Bioscience Ltd. (Guangzhou, China) to examine miR-4507 expression level in 85 pairs of paraffin-embedded lung cancer specimens. DAB substrate kit (Axy bio, Guangzhou, China) was used to detect positive staining. The extent of staining, defined as the percentage of positively staining tumor cells in relation to the whole tissue area, was scored on a scale of 0-4 as follows: 0, <10%; 1, 10-25%; 2, 26-50%; 3, 50-75%; and 4, >75%. The staining intensity was scored as 0-3 (Negative: 0; Weak expression: 1; Positive expression: 2; Strong expression: 3. The sum of the staining intensity and staining extent scores was used as the final staining score for miR-4507. Inclusion criteria: patients with stage 1-4 NSCLC; patients between 18-85 years old; EGFR and ALK mutation status were not considered; measurable lesions. Cases without clinical staging or lost to follow-up were excluded (10 cases), and 75 cases remained for statistical analysis. This study was approved by the Medical Ethics Committee of Zhengzhou University. Written informed consent was signed by each patient prior to surgery.
Luciferase Reporter Assay
The day before transfection, the cells are trypsinized and plated, and the confluence of the cells on the second day is controlled to be about 70~80%. In the next day, balance the Hieff TransTM liposomal nucleic acid transfection reagent, vector, miR-4507 mimic, miR-4507 inhibitor, and serum-free MEM to 15~25 °C, dilute an appropriate amount of the vector with serum-free MEM medium, leave it at room temperature for 5 minutes, and dilute with the same medium Hieff TransTM liposomal nucleic acid transfection reagent, placed at room temperature for 5 minutes. Vortex the carrier solution gently, and add the diluted Hieff TransTM Liposome Nucleic Acid Transfection Reagent reagent. Leave it at room temperature for 10-25 minutes to form a DNA-Hieff TransTM complex. Add the compound dropwise to the culture plate, shaking while adding, detect the luciferase activity after 48 hours.
Put the room temperature Dual-Glo® Luciferase Reagent working solution into the sample wells, 70ul per well (at the same time set the culture medium to the empty well), put the 96-well plate on the shaker and shake for 10 minutes to ensure the cells are fully lysed. Move all 140 ul total sample volume to the whiteboard (blackboard) to detect the fluorescence value of firefly. Then add the same volume of 70ul Dual-Glo® Stop & Glo Reagent working solution to the sample well, leave it for 10 minutes to complete the fusion reaction (quench the firefly luciferase and start the Renilla luciferase), and proceed Renal fluorescence value measurement. (Detection ratio = Firefly luciferase detection value/Renilla luciferase detection value).
Statistical Analysis
All the data were analyzed by SPSS 24.0 (SPSS, Chicago, IL, USA). The data are expressed as the means ± SD from at least three independent experiments. Statistical significance was determined using the Student's two-tailed t test for two groups and a parametric generalized linear model with random effects for the MTT assay. Spearman correlation analysis was conducted to detect the correlation between miR-4507 and P53. All statistical tests were two-sided, and a p value of < 0.05 was considered statistically significant. * P < 0.05, ** P < 0.01, and *** P < 0.001.
Results
miR-4507 is elevated in lung cancer and is relates to poor prognosis
In situ hybridization assay was used to detect the expression of miR-4507 in lung cancer patients. Furthermore, survival analysis showed that miR-4507 high expression patients have a poor prognosis compared to low expression patients (P=0.0274, Fig. 1A,B), which suggested that miR-4507 serves as an unfavorable factor in lung cancer. Clinical features associated are presented (Table 1). However, no significant correlations were found between miR-4507 expression and patients' age, gender, pathological type or TNM stage.
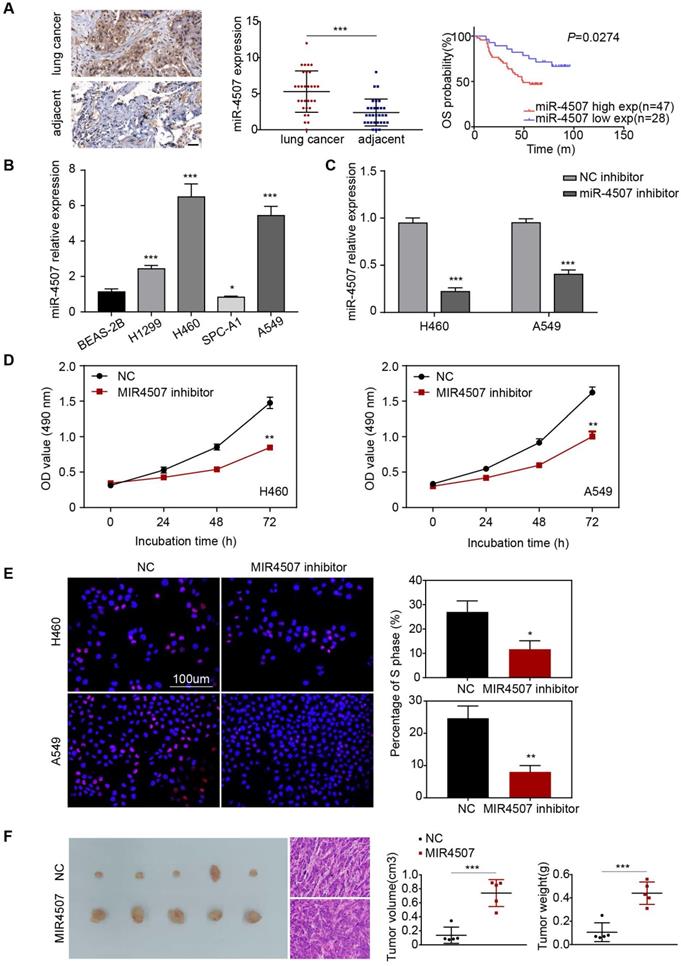
miR-4507 is elevated in lung cancer and miR-4507 inhibitor suppresses H460 and A549 cell proliferation. (A) In situ hybridization assay was used to detect the miR-4507 expression in lung cancer tissues and adjacent tissues. Scale bar: 50 µm. Kaplan-Meier survival analysis of overall survival of 75 pairs of lung cancer patients based on miR-4507 expression levels (P=0.0274). Log-rank test was used to calculate P values. (B) The expression fold of miR-4507 in BEAS-2B cell and in 4 NSCLC cells (H1299, H460, SPC-A1, A549). The data were shown as the mean ± SD (*P <0.05, *** P <0.001). (C) miR-4507 expression levels after transfection with miR-4507 inhibitor. Student's t-test. mean± SD (***P <0.001). (D) MTT assays, (E) Edu incorporation assays were conducted after transfection with miR-4507 inhibitor or NC inhibitor. Scale bar: 100 µm. (F) Xenograft tumors collected on day 20 post subcutaneous implantation of A549-NC, A549-miR-4507 cells on nude mice. Tumor volume and tumor weight were measured on day 20 (n=5), Student's t-test. mean± SD (*P <0.05, **P <0.01, ***P <0.001).
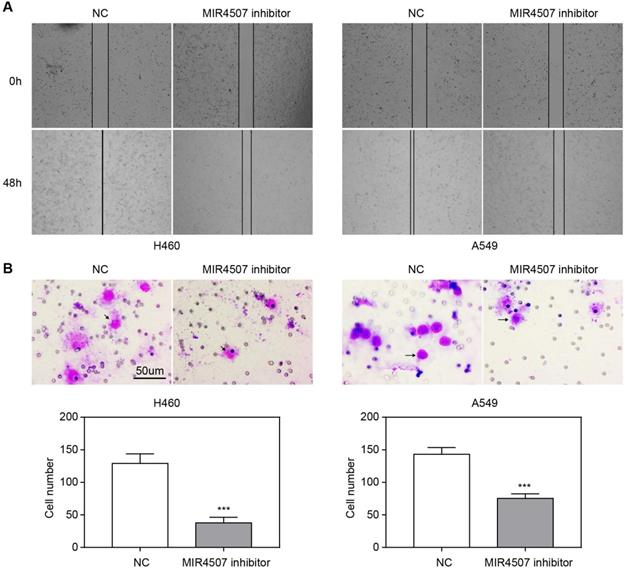
miR-4507 downregulation inhibits H460 and A549 cell migration. The effects of miR-4507 inhibitors on NSCLC cell migration capacity were measured by wound healing (A) and transwell assays (B) in H460 and A459 cells. NC inhibitors were used as negative control. Scale bar: 50 µm. Student's t-test. mean± SD (***P <0.001).
miR-4507 downregulation inhibits H460 and A549 cell proliferation
First, we detected the expression level of miR-4507 in different NSCLC cells (H1299, H460, SPC-A1, A549) and in normal human lung epithelial cells (BEAS-2B). miR-4507 was upregulated in NSCLC cells and was especially highly expressed in H460 and A549 cells (Fig. 1B). Next, miR-4507 inhibitors and NC (negative control) inhibitors were separately transfected into H460 and A549 cells, and the transfection efficacy was detected by RT‑qPCR (Reverse Transcription-Polymerase Chain Reaction) (Fig. 1C).
Then, we performed an MTT assay to investigate the effects of miR-4507 on NSCLC cell viability. The results showed that miR-4507 inhibition significantly decreased the viability of NSCLC cells compared to the NC group (Fig. 1D). We further conducted a 5-ethynyl-2'-deoxyuridine (EdU) incorporation assay to detect the effects of miR-4507 inhibitors on NSCLC cell proliferation capacity. The results showed that miR-4507 inhibitors significantly decreased the proliferation capacity compared to the NC group (Fig. 1E).
Further, we performed an in vivo tumor formation experiment by subcutaneously injected A549-NC or A549-miR-4507 cells into nude mice. After 20 days, mice injected with A549-miR-4507 had higher tumor burdens than the negative control (Fig. 1F). Statistical analysis showed that the tumor volume and weight of A549-miR-4507 group are bigger than A549-NC group, and is statistically significant (P<0.001).
These results demonstrated that miR-4507 downregulation inhibits the proliferation of H460 and A549 cells, and miR-4507 promotes lung cancer growth in vivo.
miR-4507 downregulation inhibits H460 and A549 cell migration
We next explored the effects of miR-4507 inhibitors on NSCLC cell migration capacity by conducting wound healing and transwell assays. The wound area was almost completely healed in the NC group after 48 hours, while it was only slightly reduced in the miR-4507 inhibitor group (Fig. 2A). Further, there were fewer traversing cells in the miR-4507 inhibitor group than in the NC group (Fig. 2B).
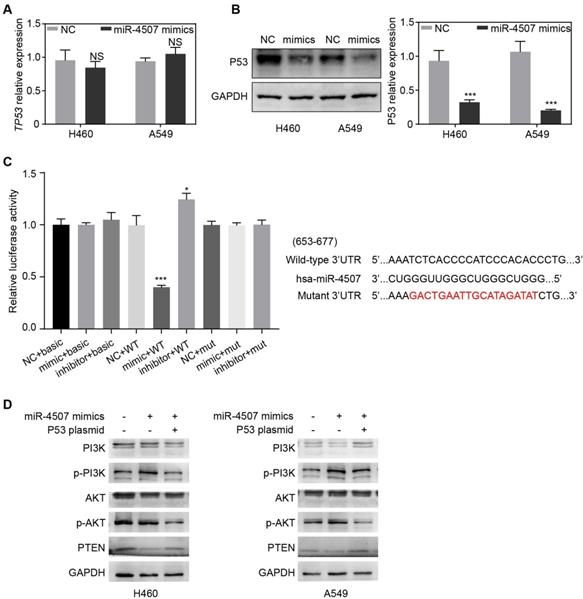
TP53 is a direct binding target of miR-4507. TP53 expression after miR-4507 overexpression in NSCLC cells were detected by RT-qPCR (A) and western blot (B), GAPDH was used as a loading control. Student's t-test. mean± SD (NS: no significance, ***P <0.001). (C) The luciferase reporter assay was conducted to detect the combination between miR-4507 and 3'UTR region of TP53 (left lane). Bioinformatics predictions of binding site by miR-4507 in TP53 3'-UTR region (right lane). (D) Western blot was conducted to detect the expression of p-PI3K/PI3K, p-AKT/AKT and PTEN. Student's t-test. mean± SD (*P <0.05, *** P <0.001).
Correlation between the clinicopathologic characteristics and expression of miR-4507 in lung cancer patients
| Characteristics | n | miR-4507 expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Age, years | ||||
| <60 | 39 | 23 | 16 | 0.9112 |
| ≥60 | 36 | 21 | 15 | |
| Gender | ||||
| Male | 39 | 24 | 15 | 0.7814 |
| Female | 36 | 20 | 16 | |
| T stage | ||||
| T1-T2a | 58 | 33 | 25 | 0.3792 |
| T2b-T4 | 17 | 11 | 6 | |
| N stage | ||||
| N0 | 40 | 21 | 19 | 0.7298 |
| N1-N3 | 35 | 23 | 12 | |
| M stage | ||||
| M0 | 71 | 42 | 29 | 0.5485 |
| M1 | 4 | 2 | 2 | |
| Pathological type | ||||
| Adenocarcinoma | 63 | 40 | 23 | 0.1502 |
| Other | 12 | 4 | 8 | |
TNM: tumor, node, metastasis.
Pathological type: other includes Bronchioloalveolar carcinoma and Mucoepidermoid carcinoma.
P values by t test.
These results demonstrated that miR-4507 downregulation inhibits the migration of H460 and A549 cells.
TP53 is a direct binding target of miR-4507
To investigate the underlying mechanism of miR-4507 in NSCLC cell proliferation and migration, we used the bioinformatic websites miRWalk and miRDB to predict the potential target genes of miR-4507. The screening results indicated that TP53 was a potential target of miR‑4507. To further clarified the regulatory relations between miR-4507 and TP53, RT-qPCR and western blot were used in both NSCLC cells transfected with NC mimics or miR-4507 mimics separately. RT-qPCR results showed no significant difference of TP53 expression between miR-4507 mimics and NC group while western blot analysis results showed that miR-4507 mimics downregulated P53 expression (Fig. 3A,B). A dual luciferase reporter assay was conducted to confirm the relationship between miR-4507 and TP53. Luciferase activity was reduced in cells co‑transfected with miR-4507 mimics and TP53‑WT (P<0.001), and increased in cells co‑transfected with miR-4507 inhibitor and TP53‑WT (P<0.05) compared with the NC group. However, the miR-4507 mimics did not change the luciferase activity in cells co-transfected with miR-4507 mimics and TP53‑MT or the cells co-transfected with miR-4507 inhibitor and TP53‑MT (Fig. 3C). These data confirmed that TP53 is a direct target of miR‑4507.
miR-4507 activates PI3K/AKT signal through TP53
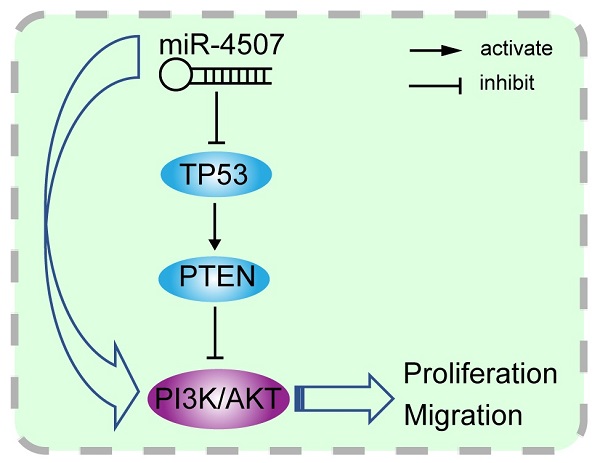
TP53 and PTEN are tumor suppressor which widely involved in the regulation of tumor-related signal pathways. It is well-known that PTEN is the negative regulator of the PI3K/AKT signal while it is transcriptionally regulated by TP53. Here, we generated the miR-4507 overexpressing cells with/without TP53 overexpression using miR-4507 mimics and TP53 plasmid. Western blot results showed that the expression of PTEN is downregulated while the PI3K/AKT signal is activated after miR-4507 overexpression in both NSCLC cells. And these effects were rescued with TP53 overexpression on that basis (Fig. 3D). These results demonstrated that miR-4507 targets TP53 to regulate the PTEN /PI3K/AKT signal axis.
miR‑4507 acts as a onco-miRNA in A549 and H460 cells via the negative regulation of TP53. Edu incorporation assays (A) and transwell assays (B) were conducted after transfection with miR-4507 mimics with/without TP53 plasmid. Scale bar: 100µm (Edu), 50 µm (transwell). mean± SD (*P <0.05; ** P <0.01, *** P <0.001).
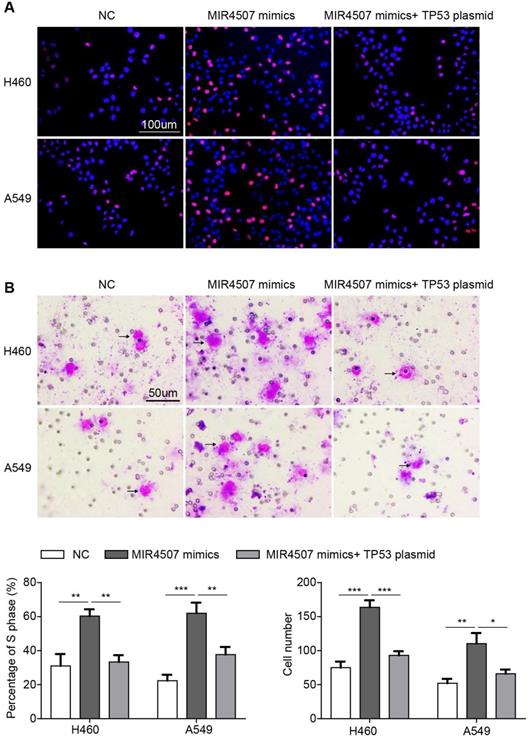
miR‑4507 acts as an onco-miRNA in A549 and H460 cells via the negative regulation of TP53
To further validate the role of TP53 in miR-4507 regulation in NSCLC progression, we conducted an EdU assay in three groups separately transfected with NC mimics, miR-4507 mimics, miR-4507 mimics and TP53 plasmids. The results showed that miR-4507 upregulation promotes proliferation, while this promotion is impeded by TP53 upregulation (Fig. 4A). Then, a transwell assay was performed to assess the migration capacity in the three aforementioned groups, and the same results were observed; miR-4507 upregulation promotes migration, while this promotion is impeded by TP53 upregulation (Fig. 4B).
These data indicated that miR-4507 acts as an onco-miRNA in H460 and A549 cells via the negative regulation of TP53.
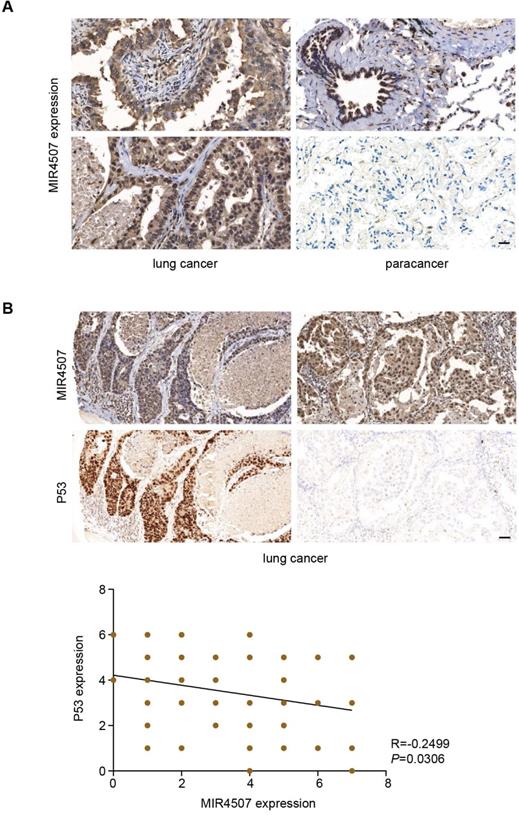
miR‑4507 expression is negatively correlated with P53 in lung cancer
Our study revealed that miR-4507 targets TP53 to promote the malignant progression of NSCLC, to further demonstrate this regulation on histological level, we used In situ hybridization and Immunohistochemistry assay to detect miR-4507 and P53 expression in 75 lung cancer tissues. Our results showed that miR-4507 is highly expressed in lung cancer tissues compared with para cancer tissues (Fig. 5A). Correlation analysis showed that miR‑4507 expression is negatively correlated with P53 expression in lung cancer (R=-0.2499, P=0.0306), which proved that our finding has also been confirmed at the histological level (Fig. 5B).
miR‑4507 is highly expressed in lung cancer tissues and is negatively correlated with P53. In situ hybridization and Immunohistochemistry assays were used to detect miR-4507 (A-B) and P53 (B) expression in 75 lung cancer tissues, scale bar: 200 µm. Spearman correlation analysis was conducted to detect the correlation between miR-4507 and P53. Two tailed Spearman correlation analysis. mean± SD (R=-0.2499, P =0.0306).
Discussion
Lung cancer, as one of the most malignant and lethal tumors, is a serious danger to human health. It is characterized by excessive cell proliferation, inevitable distant metastases, and recurrence [23]. Abnormal miRNA expression can seriously affect lung cancer progression [24-27]. Thus, the exploration of miRNA functions in the pathogenesis of lung cancer is of great significance. miRNAs are involved in every step of tumorigenesis and tumor progression through target genes [28-31]. In this study, we found an elevated expression of miR-4507 in NSCLC cells. Subsequently, the NSCLC cell proliferation capacity was determined by MTT and EdU assays in H460 and A549 cells transfected with miR-4507 inhibitors, with the control group transfected with NC inhibitors. The results showed that miR-4507 overexpression significantly promotes cell proliferation. In vivo experiments further demonstrated that miR-4507 promotes lung cancer growth. In addition, the NSCLC cell migration capacity was determined by wound healing and transwell assays in H460 and A549 cells transfected with miR-4507 inhibitors, with the control group transfected with NC inhibitors. The results showed that miR-4507 overexpression significantly promotes cell migration. These results indicated that miR-4507 could regulate cell proliferation and migration in NSCLC. However, the related mechanisms need to be studied further.
The miRWalk and miRDB databases were used to predict the target genes of miR-4507. We found that TP53 is the target gene of miR-4507. Luciferase reporter assay results further demonstrated the hypothesis that TP53 may be a direct target of miR-4507. Subsequently, TP53 expression was found to be downregulated in cells with miR-4507 overexpression. These results remind us that miR-4507 may serve as an onco-miRNA in NSCLC through the suppression of TP53. PTEN was reported to be the downstream effector of TP53 [32], and it is also the negative regulator of PI3K/AKT signal pathway [33, 34]. To fully understand the molecular mechanism of miR-4507 regulation, we overexpressed miR-4507 with/without TP53 overexpression in NSCLC cells, results showed that miR-4507 activates PI3K/AKT signal by inhibiting TP53/PTEN expression. Thus, we made a scientific hypothesis that miR-4507 targets TP53 to activate PI3K/AKT signal and promotes the proliferation and migration of lung cancer cells.
Illustration of the hypothesised signal mechanism of miR-4507. miR-4507 targets TP53 to facilitate the proliferation and migration of lung cancer cells through PI3K/AKT signal.
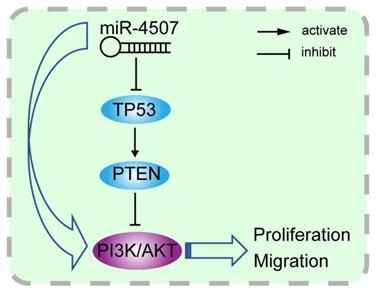
To confirm our inference, we co-transfected miR-4507 mimics with TP53 plasmids into H460 and A549 cells. Functional analysis results suggested that re-expression of TP53 in the cells with miR-4507 overexpression recovers the proliferation and migration capacity in both the H460 and A549 cells. Finally, we detected miR-4507 and P53 expression separately in 75 lung cancer tissues, results showed that miR-4507 is highly expressed in lung cancer tissues and is negatively correlated with P53 expression. These results demonstrated our deduction that miR-4507 promotes proliferation and migration through TP53 in NSCLC cells.
Abnormalities in the P53 signaling pathway are found in most malignant tumors [35-37], and TP53 mutations are found in over 50% of NSCLC cases[38, 39]. Studies have shown that the prognoses of patients with lung cancer with TP53 mutations are worse than those of patients with wild-type TP53, illustrating the importance of TP53 in lung cancer progression [40]. Thus, for patients with NSCLC without TP53 mutations, it is essential to maintain the physiological function of P53 in their treatment. In this study, two NSCLC cell lines without TP53 mutations were selected. Additional experiments have revealed the possibility that TP53 suppression by miR-4507 is an unfavorable factor for patients with NSCLC. This study found for the first time that miR-4507 regulates NSCLC cell proliferation and migration by targeting TP53. It may therefore be a potential therapeutic target for patients with NSCLC in the future.
Acknowledgements
Funding
This work was supported by the Youth Program of National Natural Science Foundation of China (NSFC) (No. 81702672).
Ethics statement
Our study was approved by Zhengzhou University Ethics Committee (approval NO. KY2018-015-03). All patients provided written informed consent prior to enrollment in the study.
Author contributions
M.Y.Z designed the study and interpreted the experimental data, and was the major contributors to the development of the first draft of the present manuscript. Z.B.T wrote the manuscript and prepared figures and tables, Y.J.W, J.J.D analyzed the data, Y.G, N.Z and T.H.G reviewed and approved the final draft of the manuscript prior to submission. All authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B. et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19:9-31
2. Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA. 2019;322:764-74
3. Rubin R. Declines in Lung Cancer Deaths Linked to Treatment Advances. JAMA. 2020;324:1128
4. Turner MC, Andersen ZJ, Baccarelli A, Diver WR, Gapstur SM, Pope CA 3rd. et al. Outdoor air pollution and cancer: An overview of the current evidence and public health recommendations. CA Cancer J Clin. 2020;70:460-479
5. Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M. et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med. 2020;383(18):1711-1723
6. Yao C, Su L, Zhang F, Zhu X, Zhu Y, Wei L. et al. Thevebioside, the active ingredient of traditional Chinese medicine, promotes ubiquitin-mediated SRC-3 degradation to induce NSCLC cells apoptosis. Cancer Lett. 2020;493:167-77
7. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA. et al. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N Engl J Med. 2020;383:640-9
8. Vychytilova-Faltejskova P, Slaby O. MicroRNA-215: From biology to theranostic applications. Mol Aspects Med. 2019;70:72-89
9. Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C. et al. miRomics and Proteomics Reveal a miR-296-3p/PRKCA/FAK/Ras/c-Myc Feedback Loop Modulated by HDGF/DDX5/beta-catenin Complex in Lung Adenocarcinoma. Clin Cancer Res. 2017;23:6336-50
10. Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q. et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 2016;7:11309
11. Tang Z, Chen W, Xu Y, Lin X, Liu X, Li Y. et al. miR-4721, Induced by EBV-miR-BART22, Targets GSK3β to Enhance the Tumorigenic Capacity of NPC through the WNT/β-catenin Pathway. Mol Ther Nucleic Acids. 2020;22:557-571
12. Ma X, Liang AL, Liu YJ. Research progress on the relationship between lung cancer drug-resistance and microRNAs. Journal of Cancer. 2019;10:6865-75
13. Kim S, Bae WJ, Ahn JM, Heo JH, Kim KM, Choi KW. et al. MicroRNA signatures associated with lymph node metastasis in intramucosal gastric cancer. Mod Pathol. 2020;34(3):672-683
14. Silveyra P, DiAngelo SL, Floros J. An 11-nt sequence polymorphism at the 3'UTR of human SFTPA1 and SFTPA2 gene variants differentially affect gene expression levels and miRNA regulation in cell culture. 2014; 307: L106-19.
15. Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471-80
16. Napoli M, Flores ER. Loss of p53 protein strikes a nerve to aid tumour growth. Nature. 2020;578:367-9
17. East MP, Laitinen T, Asquith CRM. PIP5K1A: a potential target for cancers with KRAS or TP53 mutations. Nat Rev Drug Discov. 2020;19:436
18. Miller JJ, Gaiddon C, Storr T. A balancing act: using small molecules for therapeutic intervention of the p53 pathway in cancer. Chem Soc Rev. 2020;49(19):6995-7014
19. Bell DW, Ellenson LH. Molecular Genetics of Endometrial Carcinoma. Annu Rev Pathol. 2019;14:339-67
20. Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ. et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52:177-86
21. Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89-102
22. Pitter KL, Casey DL, Lu YC, Hannum M, Zhang Z, Song X. et al. Pathogenic ATM Mutations in Cancer and a Genetic Basis for Radiotherapeutic Efficacy. J Natl Cancer Inst. 2020;113(3):266-273
23. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
24. Cen B, Lang JD, Du Y, Wei J, Xiong Y, Bradley N. et al. Prostaglandin E2 Induces miR675-5p to Promote Colorectal Tumor Metastasis via Modulation of p53 Expression. Gastroenterology. 2020;158:971-84 e10
25. Lampignano R, Kloten V, Krahn T, Schlange T. Integrating circulating miRNA analysis in the clinical management of lung cancer: Present or future? Mol Aspects Med. 2020;72:100844
26. Valencia K, Erice O, Kostyrko K, Hausmann S, Guruceaga E, Tathireddy A. et al. The Mir181ab1 cluster promotes KRAS-driven oncogenesis and progression in lung and pancreas. J Clin Invest. 2020;130:1879-95
27. Zhang JX, Yang W, Wu JZ, Zhou C, Liu S, Shi HB. et al. MicroRNA-32-5p inhibits epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting SMAD family 3. Journal of Cancer. 2021;12:2258-67
28. Sahraei M, Chaube B, Liu Y, Sun J, Kaplan A, Price NL. et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J Clin Invest. 2019;129:5518-36
29. Wei X, Zhao L, Ren R, Ji F, Xue S, Zhang J. et al. MiR-125b Loss Activated HIF1alpha/pAKT Loop, Leading to Trans-Arterial Chemoembolization Resistance in Hepatocellular Carcinoma. Hepatology. 2020;73(4):1381-1398
30. Yang X, Sun J, Sun H, Wen B, Zhang M, An H. et al. MicroRNA-30a-3p acts as a tumor suppressor in MHCC-97H hepatocellular carcinoma cells by targeting COX-2. Journal of Cancer. 2021;12:3945-57
31. Xie B, Li L, Zhang Z, Zhao L, Cheng J, Zhou C. et al. MicroRNA-1246 by Targeting AXIN2 and GSK-3β Overcomes Drug Resistance and Induces Apoptosis in Chemo-resistant Leukemia Cells. Journal of Cancer. 2021;12:4196-208
32. Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y. et al. Regulation of PTEN transcription by p53. Molecular cell. 2001;8:317-25
33. Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM. PTEN/PTENP1: 'Regulating the regulator of RTK-dependent PI3K/Akt signalling', new targets for cancer therapy. Mol Cancer. 2018;17:37
34. Feng J, Dang Y, Zhang W, Zhao X, Zhang C, Hou Z. et al. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:6868-77
35. He Y, Johnson DT, Yang JS, Wu H, You S, Yoon J. et al. Loss of the tumor suppressor, Tp53, enhances the androgen receptor-mediated oncogenic transformation and tumor development in the mouse prostate. Oncogene. 2019;38:6507-20
36. Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood. 2017;130:699-712
37. Xue Y, San Luis B, Lane DP. Intratumour heterogeneity of p53 expression; causes and consequences. J Pathol. 2019;249:274-85
38. Chen Y, Chen G, Li J, Huang YY, Li Y, Lin J. et al. Association of Tumor Protein p53 and Ataxia-Telangiectasia Mutated Comutation With Response to Immune Checkpoint Inhibitors and Mortality in Patients With Non-Small Cell Lung Cancer. JAMA Netw Open. 2019;2:e1911895
39. Jiao XD, Qin BD, You P, Cai J, Zang YS. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer. 2018;123:70-5
40. La Fleur L, Falk-Sorqvist E, Smeds P, Berglund A, Sundstrom M, Mattsson JS. et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50-8
Author contact
![]() Corresponding author: MengYang Zhao (PhD). Department of Oncology, The People's Hospital of Zhengzhou University, Zhengzhou, China. E-mail: mengyang8715com.
Corresponding author: MengYang Zhao (PhD). Department of Oncology, The People's Hospital of Zhengzhou University, Zhengzhou, China. E-mail: mengyang8715com.

 Global reach, higher impact
Global reach, higher impact