3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(23):7158-7166. doi:10.7150/jca.61567 This issue Cite
Research Paper
GOLM1 Drives Colorectal Cancer Metastasis by Regulating Myeloid-derived Suppressor Cells
Department of Radiation Oncology, Shaanxi Provincial People's Hospital, Xi'an, 710086, China.
Received 2021-4-12; Accepted 2021-9-18; Published 2021-10-21
Abstract
Colorectal cancer (CRC) is the most common digestive neoplasms worldwide, metastasis and recurrence still account for the leading cause for the high mortality rate, but the exact mechanisms remain unclear. More and more evidence has indicated that the deregulation of GOLM1 plays a crucial role in cancer progression. Here, we reported a novel role of GOLM1 in promoting CRC metastasis. In this study, the expression of GOLM1 was detected in human CRC cohort. The function of GOLM1 in CRC metastasis was analyzed by in vivo cecum orthotopic model. We found that the expression of GOLM1 was significantly increased in CRC tissues than adjacent nontumor. Overexpression GOLM1 can promote CRC immune escape and metastasis by recruiting of myeloid-derived suppressor cells (MDSCs) at the same time. PF-04136309, a small molecule and specific inhibitor of CCR2 can largely suppressed GOLM1-mediated CRC metastasis. These results suggest that GOLM1 can promote CRC metastasis and is a prognostic biomarker in human CRC.
Keywords: colorectal cancer, metastasis, GOLM1, myeloid-derived suppressor cells.
Introduction
Colorectal cancer (CRC) is the third most common digestive neoplasms worldwide [1], metastasis and recurrence remains the leading cause for the high mortality rate of CRC patients. Approximately 20% of patients with CRC have metastatic disease at diagnosis [2], although great progress has been made in the treatment of CRC, the overall prognosis of is still poor for patients with metastatic CRC. Hence, it is urgently to deeply understand the underlying mechanisms of metastatic CRC.
Recent work has elucidated that inflammation plays a pivotal role in CRC initiation and metastasis [3]. Human CRC often accomplished by activation of checkpoint signals, such as PD-L1/PD-1, infiltrated of immunosuppressive cells, such as tumor-associated macrophage (TAM) and myeloid-derived suppressor cells (MDSCs) [4, 5]. Mounting data have shown accumulation of MDSCs, which induce local and possibly immunosuppression, is common in tumors and correlated with colorectal cancer progression [4, 6, 7]. However, the ongenic signaling that drives MDSCs recruitment and activation in CRC remains poorly understood.
Golgi membrane protein 1 (GOLM1, also known as GP73 or GOLPH2), a type II transmembrane protein, is widely expressed in normal epithelial cells [8]. Since Golgi apparatus mainly involved in modification and transportation of the proteins, the dysregulation of its associated proteins may lead to markedly altered cellular processes and functions [9]. Accumulating evidence has indicated that the deregulation of GOLM1 gene plays critical roles in cancer progression [10, 11], metastasis [12] and immunosuppression [13]. However, the function and biological function of GOLM1 in human colorectal cancer needs further research.
Here, we found that GOLM1 was overexpression in CRC and associated with poor prognosis in CRC patients. GOLM1 promoted CCL2-induced MDSCs recruitment and facilitated CRC immune escape and progression. Notably, our findings showed that CCR2 inhibitor can prevent MDSCs trafficking in CRC patients with GOLM1 overexpression. These findings reveal the mechanism of GOLM1-mediated CRC metastasis, and are helpful to develop the novel strategies for the treatment of CRC.
Materials and Methods
Real-time PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen), and reverse transcription was performed using the Advantage RT-for-PCR Kit (Takara) according to the manufacturer's instructions. For the real-time PCR analysis, aliquots of double-stranded cDNA were amplified using a SYBR Green PCR Kit (Applied Biosystems). The cycling parameters were as follows: 95°C for 15 s, 55-60°C for 15 s, and 72°C for 15 s for 45 cycles. A melting curve analysis was then performed. The Ct was measured during the exponential amplification phase, and the amplification plots were analyzed using SDS 1.9.1 software (Applied Biosystems). For clinical tissue samples, the fold change of the target gene was determined by the following equation: 2-ΔΔCt (ΔΔCt = ΔCttumor - ΔCtnontumor). This value was normalized to the average fold change in the normal liver tissues, which was defined as 1.0. All reactions were performed in duplicate. The primer sequences for GOLM1 sense 5'- CCGGAGCCTCGAA, AAGAGATT-3', GOLM1 antisense 5'- ATGATCCGTGTCTGGAGGTC -3'.
Immunohistochemistry
This study was approved by the ethics the Committee of Shaanxi Provincial People's Hospital, and informed consent was written and based on the ethical guidelines of the 1975 Declaration of Helsinki. In addition, the privacy rights of human subjects were always observed. CRC specimens and matched adjacent tissues were used to construct a tissue microarray (Shanghai Biochip Co, Ltd. Shanghai, China). The tissue microarray was stained for GOLM1 (Abcam, ab109628), CD11b (Abcam, ab133357) expression. Immunohistochemistry was performed on 4-μm-thick, routinely processed paraffin-embedded sections. Briefly, after baking on a panel at 60 °C for one hour, the tissue sections were deparaffinized with xylene and rehydrated through gradient ethanol immersion. Endogenous peroxidase activity was quenched by 3% (vol/vol) hydrogen peroxide in methanol for 12 min, followed by three 3-min washes with phosphate-buffered saline (PBS). Then the slides were immersed in 0.01mol/L citrate buffer solution (pH 6.0) and placed in a microwave oven for 30 min. After washing in PBS (pH 7.4, 0.01 mol/L), the sections were incubated in a moist chamber at 4 °C overnight with the primary antibody diluted in PBS containing 1% (wt/vol) bovine serum albumin. Negative controls were performed by replacing the primary antibody with preimmunize mouse serum. After three 5 min washes with PBS, the sections were treated with a peroxidase-conjugated second antibody (Santa Cruz) for 30 min at room temperature, followed by additional three 5 min washes with PBS. Reaction product was visualized with diaminobenzidine for 2 min. Images were obtained under a light microscope (Olympus, Japan) equipped with a DP70 digital camera.
Analyses were performed by two independent observers who were blinded to the clinical outcome. The immunostaining intensity was scored on a scale of 0 to 3:0 (negative), 1 (weak), 2 (medium) or 3 (strong). The percentage of positive cells was evaluated on a scale of 0 to 4:0 (negative), 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), or 4 (76%-100%). The final immuno-activity scores were calculated by multiplying the above two scores, resulting an overall score which range from 0~12. Each case was ultimately considered “negative” if the final score ranges from 0~3, and “positive” if the final score ranges from 4~12.
Western Blotting
For Western blotting assay, the lysed cells protein was separated on SDS-PAGE and transferred onto polyvinylidene difluoride membrane. The nonspecific binding was blocked with 10% non-fat milk for one hour. The membranes were incubated with specific antibody overnight at 4°C. Western blotting of β-actin on the same membrane was used as a loading control. Antibody against GOLM1 (Abcam, ab109628) was purchased from Santa Cruz. Antibody against β-actin (A1978) was purchased from sigma. The membranes were then washed with PBS 3 times and incubated with an HRP-conjugated secondary antibody. Proteins were visualized using a ImmobilonTM Western Chemiluminescent HRP substrate (Millipore, USA).
Animal experiment
All animal studies were approved by the Committee on the Use of Live Animals in Teaching and Research, Shaanxi Provincial People's Hospital. Five-weeks-old C57BL/6 male mice were raised in specific pathogen-free conditions in accord with the institutional guidelines for animal care. A metastatic colorectal model in mice was established according to the existing protocol. In brief, mouse MC38 cells (4.0×106) in the 100 μl of phosphate-buffered saline were subcutaneously injected into the cecal wall of C57BL/6 mice under anesthesia (10 for each group). The in vivo tumor formation and metastases were monitored using the bioluminescence. For in vivo signal detection, D-luciferin (Perkin-Elmer) at 100 mg/kg was injected intraperitonially into the nude mice. Bioluminescent images were captured using an IVIS 100 Imaging System (Xenogeny). At the 9 weeks, the mice were sacrificed and the livers and lungs were collected and underwent histological examination. Each experiment was repeated three times and we showed the representative data of three independent experiments.
Flow Cytometric Analysis
Cells were incubated with anti-mouse CD16/CD32 purified antibody (#101302, clone 93, Biolegend) at room temperature to block nonspecific antibodies. Then 1×106 cells were stained with fluorophore-conjugated antibodies for 30 minutes at room temperature, matched isotype antibodies were used as control. Data were analyzed by Flowjio_V10 software (TreeStar, Ashland, OR). Antibodies against CD11b (FITC, #101205), CD45 (PE/Cy7, #103113), Ly-6G/Ly-6C(Gr-1) (PE, #108407) were purchased from biolegend.
Statistical analysis
Statistics were calculated with SPSS software (version 20.0). P values were statistically analyzed by the χ2 test for categorical variables and by Student's test for quantitative data. The recurrence and survival data were analyzed by the Kaplan-Meier method and log-rank test. Cox proportional hazards model was used for univariate and multivariate analyses. Differences were considered statistically significant when p < 0.05.
Results
Elevated expression of GOLM1 in colorectal cancer positively correlates with poor prognosis
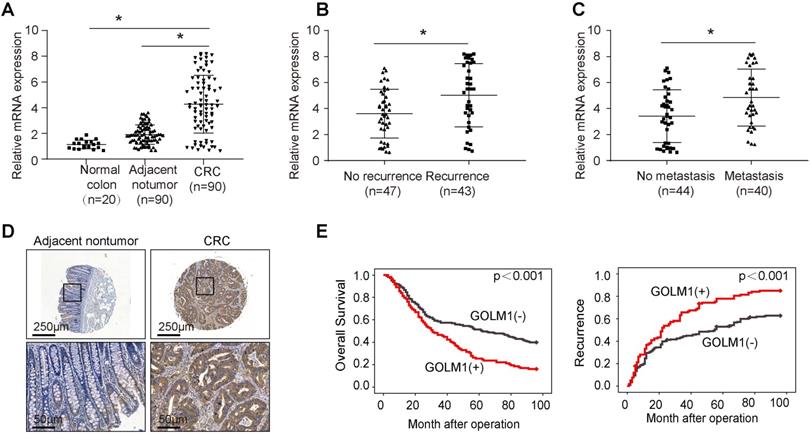
To determine the clinical relevance of GOLM1 expression in CRC, we examined the expression of GOLM1 by RT-PCR in a cohort of 90 paired CRC and adjacent nontumor specimens, and 20 normal colorectal epithelial specimens. We found that the mRNA levels were significantly increased in CRC compared with adjacent nontmor tissues and normal colon tissues (Figure 1A). Furthermore, it is demostrated that GOLM1 protein levels with recurrence or mtatstasis CRC were much higher than patients without recurrence or metastasis (Figure 1B-C). These data showed a close association of GOLM1 with CRC metastasis. To further determine prognostic value of GOLM1 in CRC, we analyzed the GOLM1 protein level in a tissue microrray by IHC study in 341 CRC tissues with complete clinicopathological features and complete follow-up data. We found that GOLM1 is expressed both in adjacent nontumor tissues and CRC species. However, the statistical analysis showed GOLM1 is highly expressed in CRC species than adjacent nontumor tissues (Figure 1D and Supplementary Figure S1). We found that the elevated GOLM1 expression was positively associated with tumor invasion, lymph node metastasis and AJCC stage (Table 1).
Correlation between GOLM1 expression and clinicopathological characteristics of CRC in cohort of human CRC tissues
| Clinicopathological variables | Tumor GOLM1 expression | P Value | ||
|---|---|---|---|---|
| Negative (n=223) | Positive (n=118) | |||
| Age | ≤60 | 69 | 29 | 0.161 |
| >60 | 154 | 89 | ||
| Gender | female | 91 | 61 | 0.054 |
| male | 132 | 57 | ||
| Tumor size | ≤5cm | 91 | 38 | 0.119 |
| >5cm | 132 | 80 | ||
| Tumor differentiation | Well or moderate | 138 | 65 | 0.224 |
| poor | 85 | 53 | ||
| Tumor invasion | T1-T3 | 190 | 88 | 0.016 |
| T4 | 33 | 30 | ||
| Lymph node metastasis | Absent | 147 | 63 | 0.024 |
| Present | 76 | 55 | ||
| Distant metastasis | Absent | 194 | 94 | 0.075 |
| Present | 29 | 24 | ||
| AJCC stage | I-II | 143 | 60 | 0.017 |
| III-IV | 80 | 58 | ||
Survival analysis revealed that patients with elevated GOLM1 expression showed poorer overall survival and higher recurrence rate than patients with low GOLM1 expression (Figure 1E). In Cox proportional hazards regression analyses, GOLM1 expression was an independent predictor for patient's high recurrence rate and poor overall survival (Table 2). Based on the above findings, these indicate that the elevated overexpression is significantly associated with poor prognosis of CRC.
Overexpression of GOLM1 promotes colorectal cancer progression
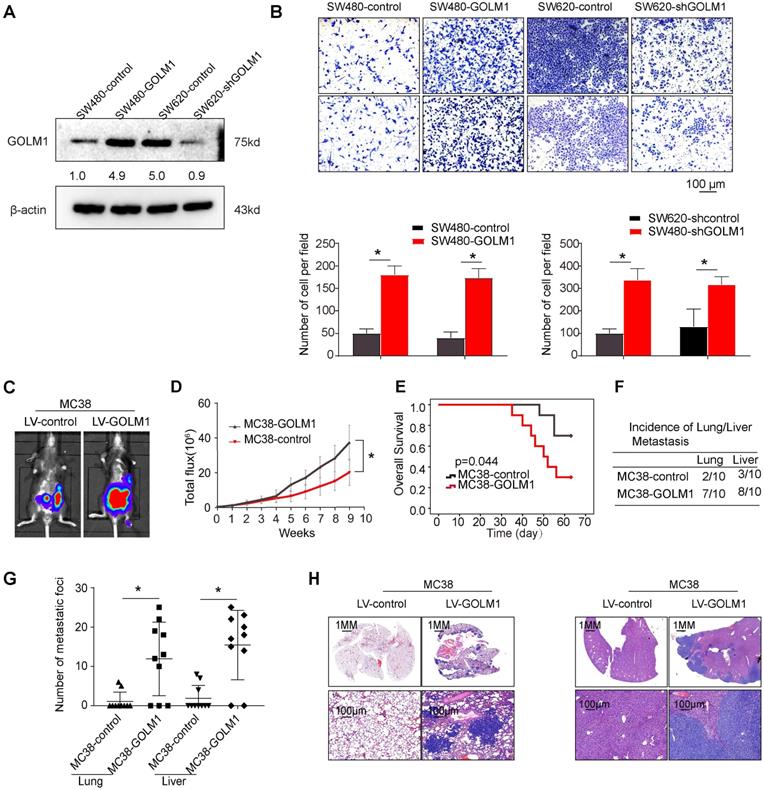
The above data suggest that GOLM1 may play a key role in CRC metastasis. Accordingly, we evaluated the functions of GOLM1 on cell migration in a gain of function experiments both in vitro and in vivo assays. Transwell assay showed that overexpression of GOLM1 can promote CRC invasion and migration, while downregulation of GOLM1 can suppress the invasion and migration abilities of CRC cells (Figure 2A-B). To further investigate the impact of GOLM1 on the metastasis, we established the cecal metastasis model using MC38-GOLM1 cells and its corresponding control cells. In vivo orthotopic implantation of immunocompetent C57BL/6 mice assay showed that GOLM1 upregulation increased the bioluminescence intensity, lung metastasis incidence, while shorten the overall survival of the MC38 group (Figure 2C-H). These above results indicate GOLM1 can promote CRC metastasis both in vitro and in vivo.
GOLM1 promotes CRC progression by recruitment of MDSCs
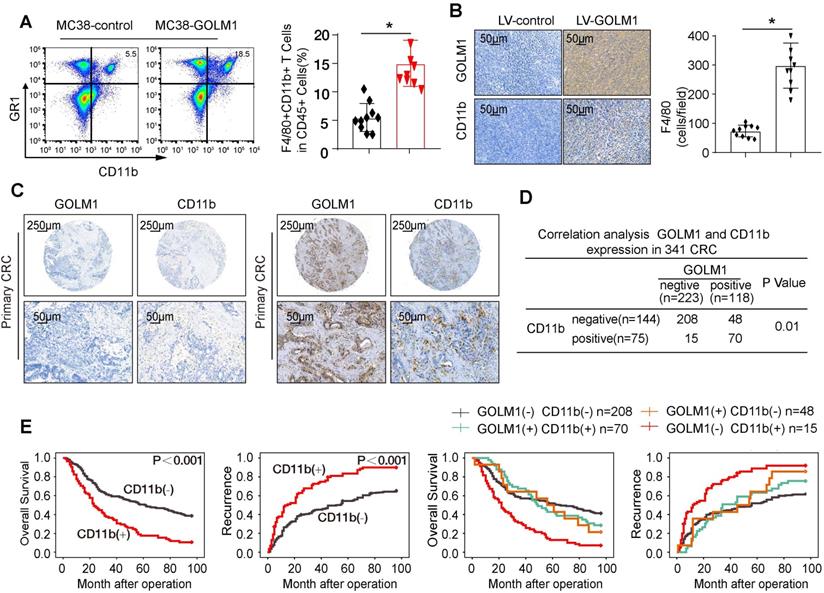
Mounting data have shown accumulation of MDSCs, which induce local and possibly immunosuppression, is correlated with CRC progression [7]. We examined the infiltration of MDSCs by flowcytometric analysis and IHC staining. The results showed that the number of MDSCs markedly elevated in MC38-GOLM1 tumor than MC38-control cells (Figure 3A-B). To verify clinical relevance, we further investigated the expression of CD11b in CRC cohort. The representative images of CD11b were showed in Figure 3C. Correlation assay suggested that GOLM1 positively correlated with CD11b (Figure 3D).
Univariate and multivariate analysis of factors associated with survival and recurrence in two independent cohorts of human CRC.
| Clinical Variables | Time to Recurrence | Overall Survival | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Univariate analysis | ||||
| Age (≤60 vs > 60) | 1.027 (0.771-1.368) | 0.853 | 1.146 (0.877-1.496) | 0.766 |
| Gender (female vs male) | 0.934 (0.724-1.206) | 0.601 | 0.927 (0.716-1.199) | 0.562 |
| Tumor size (≤5 cm vs >5 cm) | 1.073 (0.826-1.395) | 0.596 | 0.597 (0.439-0.821) | 0.318 |
| Tumor differentiation (well/moderate vs poor) | 0.50 (0.387-0.647) | <0.001 | 0.474 (0.366-0.614) | <0.001 |
| Tumor invasion (I-II vs III) | 0.585 (0.432-0.791) | 0.001 | 0.597 (0.439-0.821) | 0.001 |
| Lymph node metastasis (absent vs present) | 0.190 (0.144-0.250) | <0.001 | 0.174 (0.131-0.231) | <0.001 |
| Distant metastasis (absent vs present) | 0.121 (0.085-0.173) | <0.001 | 0.099(0.069-0.1143) | <0.001 |
| AJCC stage (I-II vs III) | 0.173 (0.131-0.229) | <0.001 | 0.159 (0.120-0.210) | <0.001 |
| GOLM1 expression (negative vs positive) | 0.569 (0.439-0.738) | <0.001 | 0.555(0.427-0.7220) | <0.001 |
| Multivariate analysis | ||||
| Tumor differentiation (well/moderate vs poor) | 0.844 (0.632-1.127) | 0.251 | 0.851(0.634-1.140) | 0.279 |
| Tumor invasion (I-II vs III-IV) | 0.748 (0.540-1.035) | 0.080 | 0.789(0.567-1.098) | 0.160 |
| Lymph node metastasis (absent vs present) | 1.175 (0.536-2.576) | 0.686 | 0.937(0.428-2.055) | 0.872 |
| Distant metastasis (absent vs present) | 0.334 (0.225-0.497) | <0.001 | 0.275(0.183-0.412) | <0.001 |
| AJCC stage (I-II vs III) | 0.334 (0.244-0.497) | <0.001 | 0.223 (0.099-0.505) | <0.001 |
| GOLM1 expression (negative vs positive) | 0.194 (0.086-0.438) | 0.002 | 0.641 (0.485-0.846) | 0.002 |
Elevated expression of GOLM1 correlated with poor prognosis in CRC. (A-C) Real time PCR analysis GOLM1 mRNA expression in normal colon tissues(n=20) and paired adjacent nontumor and CRC (n=90), in CRC tissues with (n=43) or without (n=47) recurrence, and in CRC tissues with (n=40) or without (n=44) metastasis. (D) Representative immunohistochemical staining of GOLM1 expression. (E) The association between GOLM1 expression with overall survival and recurrence in CRC cohort.
GOLM1 promotes CRC invasion and metastasis in vivo and in vitro. (A) GOLM1 expression in SW480 and SW620 cells by Western blot. (B) Transwell assays indicated that upregulation of GOLM1 promotes CRC migration and invasion, and depletion of GOLM1 inhibits the migration and invasion potentials of CRC cells. (C-H) Metastasis assays in the immune-competent C57BL/6 mice. Bioluminescent imaging(C), Bioluminescent signals(D), Overall survival(E), The incidence and number of lung and liver colonization(F-G), and HE (H). * P<0.05.
Survival analysis indicated that CRC patients with CD11b expression showed poorer overall survival and elevated recurrence rates than patients without CD11b expression. Furthermore, CRC patients that showed positive coexpression of GOLM1/CD11b had the lowest survival rate and the highest recurrence rate (Figure 3E).
The CCR2 inhibitor PF-04136309 can inhibit GOLM1-mediated CRC metastasis
To study the underlying mechanism of CRC recruit MDSCs, we selected the CRC cell lines with low expression of GOLM1 and constructed SW480-GOLM1 stable cell lines after transfection with lentivirus. By PCR microarray of chemokines and their receptors, we found that GOLM1 overexpression promoted transcriptome changes of a series of chemokines and their receptors such as CCL2, CXCL5, CXCL12, CCL4 (Table 3). Considering the important role of CCL2 in cancer progression, we focused on CCL2 for further study. Real time assay showed that overexpression of GOLM1 markedly upregulated CCL2 expression, whereas downregulation of GOLM1 can reduce the expression levels of CCL2 (Figure 4A).
GOLM1 promotes CRC progression in immunocompetent mice by recruitment of MDSCs. (A) The infiltration of CD11b+GR1+ MDSCs were determined by flow cytometry. (B) The MDSCs infiltration of the indicated groups was evaluated by IHC staining. (C) IHC staining analysis of GOLM1 and CD11b expression in CRC cohort. (D) The expression relationship between GOLM1 and CD11b in CRC cohort. (E) Kaplan-Meier analysis of the association between the expression of CD11b or GOLM1 /CD11b coexpression and recurrence or overall survival in CRC cohort.
Previous study showed that CCL2 recruits immune suppressor cells MDSCs and TAMs to tumor sites by binding to CCR2 receptor. Then, we investigate whether inhibition the CCL2/CCR2 signal can suppress CRC progression. PF-04136309 [14], an antagonist of CCR2, can specifically inhibit the binding of the ligand CCL2 to its receptor CCR2, which may lead to the inhibition of CCR2 activation and signal transduction. We aimed to study whether PF-04136309 treatment affects GOLM1-CCL2 signaling-medicated CRC metastasis. In vivo study showed that PF-04136309 can significantly reduce lung and liver metastasis, metastasis, while prolonged the survival of MC38-GOLM1 group compared to that of the control group.
Discussion
Colorectal cancer is one of the most common digestive system neoplasms with high mortality rate [15]. Although great improvements had been made in screening and treatments, the prognosis is still poor for metastatic CRC. Metastasis still accounts for the main cause of poor clinical outcome of CRC patients [16]. Hence, a deeply study of the underlying mechanisms of metastasis in CRC is crucial to find new treatment strategies for colorectal cancer. Deregulated GOLM1 functioned as an oncogene and correlated with poor prognosis in many types of cancers, such as hepatocellular carcinoma [12, 17], prostate cancer [18], gastric cancer [11], esophagus cancer [19] and so on. In this study, we found that GOLM1 was overexpression in CRC and the elevated GOLM1 correlated with a higher recurrence rate, shorter overall survival and more aggressive tumor phenotype. GOLM1 upregulation increased the bioluminescence intensity, lung metastasis incidence, while shorten the overall survival of the MC38 group in vivo. However, the underlying mechanism of GOLM1 functions as an oncogene in CRC is still unclear. In our previous work, we indeed have studied whether GOLM1 can promote CRC metastasis in both immunocompetent C57BL/6 mice and immune-deficiency nude mice. The SW480-GOLM1 and indicated control cell was injected into the cecal wall of nude mice, while MC38-GOLM1 and indicated control cell was injected into the cecal wall of C57BL/6 mice. These results showed both in nude mice and in C57BL/6 mice, upregulation of GOLM1 can promote CRC metastasis (Figure 2 and Supplementary Figure 2). However, compared with nude mice, GOLM1 promoted CRC metastasis more significantly in C57BL/6 mice. These results indicated that an intact immune system is essential for GOLM1 promote CRC metastasis. However, the underlying mechanism needs further study.
Immune invasion is one of the major hallmarks of cancers [20], often accomplished by expansion of PD-1/PD-L1 axis or the infiltration of immunosuppressive cells such as MDSCs [21-24]. MDSCs represent the universal immunosuppressive population in many pathologic conditions including cancer [24]. Accumulating evidence in recent years has even highlighted the expansion of MDSCs in human CRC as a major barrier to antitumor immunity and escape immune surveillance [24, 25]. More importantly, MDSCs can in turn suppress CD8+ T cell cytotoxicity. Our in vivo study showed that GOLM1 promote CRC metastasis by recruitment of MDSCs. In clinical sample, correlation assay suggested that GOLM1 positively correlated with CD11b. Furthermore, CRC patients with positive coexpression of GOLM1/CD11b had the lowest survival rate and the highest recurrence rate.
Chemokines and Receptors RT2 Profiler PCR Array of SW480-GOLM1 vs SW480 -control
| Gene | Description | SW480-GOLM1 (fold change) |
|---|---|---|
| CCL2 | Chemokine (C-C motif) ligand 2 | 4.95 |
| CXCL5 | Chemokine (C-X-C motif) ligand 5 | 4.31 |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 | 4.15 |
| CCL4 | Chemokine (C-C motif) ligand 4 | 3.14 |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 | 3.45 |
| CCR7 | Chemokine (C-C motif) receptor 7 | 3.12 |
| CCL5 | Chemokine (C-C motif) ligand 5 | 2.85 |
| CCL22 | Chemokine (C-C motif) ligand 22 | 2.85 |
| CCR9 | Chemokine (C-C motif) receptor 9 | 2.84 |
| ACKR1 | Duffy blood group, chemokine receptor | 2.74 |
| CCR4 | Chemokine (C-C motif) receptor 4 | 2.66 |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 2.56 |
| CCL26 | Chemokine (C-C motif) ligand 26 | 2.55 |
| ACKR4 | Chemokine (C-C motif) receptor-like 1 | 2.54 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 2.34 |
| IL16 | Interleukin 16 | 2.32 |
| CCR8 | Chemokine (C-C motif) receptor 8 | 2.31 |
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 2.31 |
| CKLF | Chemokine-like factor | 2.21 |
| CCL20 | Chemokine (C-C motif) ligand 20 | 2.2 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | 2.15 |
| CCL19 | Chemokine (C-C motif) ligand 19 | 2.11 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 2.1 |
| CXCL14 | Chemokine (C-X-C motif) ligand 14 | 1.95 |
| ACKR3 | Chemokine (C-X-C motif) receptor 7 | 1.85 |
| PPBP | Pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 1.85 |
| CCR2 | Chemokine (C-C motif) receptor 2 | 1.83 |
| CMTM2 | CKLF-like MARVEL transmembrane domain containing 2 | 1.83 |
| TLR4 | Toll-like receptor 4 | 1.81 |
| CCR10 | Chemokine (C-C motif) receptor 10 | 1.75 |
| CXCL6 | Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 1.66 |
| CMTM3 | CKLF-like MARVEL transmembrane domain containing 3 | 1.63 |
| CCL24 | Chemokine (C-C motif) ligand 24 | 1.59 |
| CCL25 | Chemokine (C-C motif) ligand 25 | 1.56 |
| CCL27 | Chemokine (C-C motif) ligand 27 | 1.55 |
| CCL14 | Chemokine (C-C motif) ligand 14 | 1.53 |
| CCR5 | Chemokine (C-C motif) receptor 5 | 1.51 |
| CCL11 | Chemokine (C-C motif) ligand 11 | 1.51 |
| CCL15 | Chemokine (C-C motif) ligand 15 | 1.47 |
| CCL13 | Chemokine (C-C motif) ligand 13 | 1.47 |
| CXCR2 | Chemokine (C-X-C motif) receptor 2 | 1.46 |
| TLR2 | Toll-like receptor 2 | 1.45 |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 | 1.44 |
| CXCL3 | Chemokine (C-X-C motif) ligand 3 | 1.44 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 1.44 |
| CCR3 | Chemokine (C-C motif) receptor 3 | 1.44 |
| CCL1 | Chemokine (C-C motif) ligand 1 | 1.41 |
| CXCR1 | Chemokine (C-X-C motif) receptor 1 | 1.33 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 1.23 |
| SLIT2 | Slit homolog 2 (Drosophila) | 1.23 |
| CCRL2 | Chemokine (C-C motif) receptor-like 2 | 1.21 |
| CCL7 | Chemokine (C-C motif) ligand 7 | 1.10 |
| IL1B | Interleukin 1, beta | 1.03 |
| CXCR5 | Chemokine (C-X-C motif) receptor 5 | 0. |
| CCL8 | Chemokine (C-C motif) ligand 8 | -1.17 |
| CCL21 | Chemokine (C-C motif) ligand 21 | -1.22 |
| TYMP | Thymidine phosphorylase | -1.23 |
| CCL23 | Chemokine (C-C motif) ligand 23 | -1.33 |
| CMKLR1 | CHEMOKINE-LIKE RECEPTOR 1 | -1.33 |
| CCL3 | Chemokine (C-C motif) ligand 3 | -1.34 |
| GPR17 | G protein-coupled receptor 17 | -1.44 |
| PF4V1 | Platelet factor 4 variant 1 | -1.66 |
| CXCL16 | Chemokine (C-X-C motif) ligand 16 | -1.84 |
| ACKR2 | Chemokine binding protein 2 | -1.85 |
| TNF | Tumor necrosis factor | -1.89 |
| CCL28 | Chemokine (C-C motif) ligand 28 | -2.11 |
| CCL16 | Chemokine (C-C motif) ligand 16 | -2.14 |
| C5 | Complement component 5 | -2.32 |
| FPR1 | Formyl peptide receptor 1 | -2.33 |
| CMTM1 | CKLF-like MARVEL transmembrane domain containing 1 | -2.34 |
| CCR1 | Chemokine (C-C motif) receptor 1 | -2.39 |
| CCL17 | Chemokine (C-C motif) ligand 17 | -2.44 |
| XCL1 | Chemokine (C motif) ligand 1 | -2.45 |
| C5AR1 | Complement component 5a receptor 1 | -2.45 |
| XCR1 | Chemokine (C motif) receptor 1 | -2.45 |
| CCL18 | Chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | -2.85 |
| CMTM4 | CKLF-like MARVEL transmembrane domain containing 4 | -2.88 |
| IL8 | Interleukin 8 | -3.11 |
| XCL2 | Chemokine (C motif) ligand 2 | -3.11 |
| CCR6 | Chemokine (C-C motif) receptor 6 | -3.12 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | -3.15 |
| HIF1A | Hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | -3.31 |
| CX3CL1 | Chemokine (C-X3-C motif) ligand 1 | -3.55 |
| IL4 | Interleukin 4 | -3.65 |
The CCR2 inhibitor PF-04136309 can inhibit GOLM1-mediated CRC metastasis. (A) GOLM1 and CCL2 expression in the indicated cell by Real-time PCR analysis. (B-G) PF-04136309 (100 mg/kg, subcutaneously, twice, daily, for 9 weeks) reduced the MC38-GOLM1 cells tumor metastasis. Bioluminescence images(B), The incidence of lung and liver colonization (C), Bioluminescence signals(D), Overall survival (E), The number of lung and liver colonization(F), HE staining (G). * P<0.05.
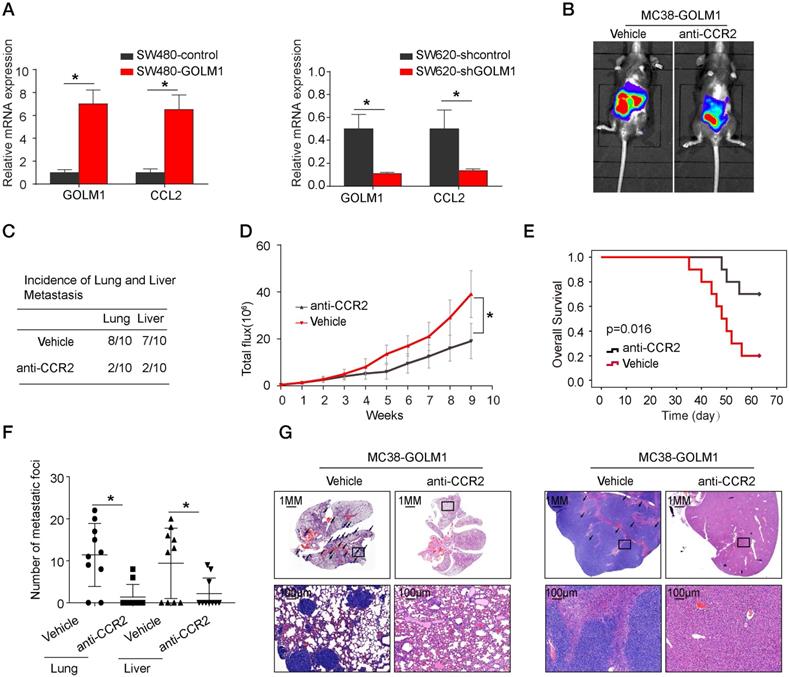
Our results showed that GOLM1 can upregulate the expression of CCL2, which leads to the recruitment of MDSCs. CCL2, highly expressed in many types of cancers, can recruit monocyte and macrophages into tumors to promote tumor cell survival and contribute immune evasion. Previous studies reported that PF-04136309, a specific CCR2 inhibitor, can augment antitumor immunity and improved response to chemotherapy [26]. In this study, we found that PF-04136309 can significantly inhibited MDSCs cells recruitment. In addition, in vivo PF-04136309 can reduce the metastasis rate of CRC, while prolonged the survival time of C57 mice.
In conclusion, we found that CRC was overexpression in CRC and associated with poor prognosis. GOLM1 promoted induced MDSCs recruitment by CCL2/CCR2 pathway to facility CRC immune escape and progression. PF-04136309, a small molecule and specific inhibitor of CCR2, inhibit GOLM1-induced CRC metastasis and MDSCs recruitment.
Abbreviations
CRC: Colorectal cancer; HE: hematoxylin and eosin; IHC: immunohistochemistry; BLI: bioluminescent imaging; GOLM1: Golgi membrane protein 1; MDSCs: myeloid-derived suppressor cells.
Supplementary Material
Supplementary figures.
Acknowledgements
We are appreciating to every author for their assistance with the experiments and constructive comments on the manuscript.
Ethics approval and consent to participate
Ethical approval was given by the Ethics Committee of Shaanxi Provincial People's Hospital.
Funding
This study was supported by Scientific and Technological Talents Support Program Foundation of Shaanxi Provincial People's Hospital (No.2021JY-05).
Author contributions
Yunzhi Dang designed the studies and wrote the paper. Jiao Yu assisted in immunohistochemical staining and animal experiments. Shuhong Zhao provided assistance in collecting tissue samples. Long Jin and Ximing Cao provided assistance in conceiving experiments and analyzing data. Qing Wang provided assistance in performing the experiments and writing the paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RC. et al. Colorectal cancer statistics. Cancer J Clin. 2014;64:104-117
2. Jemal AD, Siegel RM, Ward DE. et al. Cancer statistics. Cancer J Clin. 2008;58:71-96
3. Terzi J, Grivennikov S, Karin E. et al. Inflammation and colon cancer. Gastroenterology. 2010;138:2101-2114
4. Sieminska I, Baran J. Myeloid-derived suppressor cells in colorectal cancer. Front Immunol. 2020;11:1526
5. Eynde VD. et al. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell. 2018;34:1012-1026
6. Serafini P, Borrello V, Bronte V. Myeloid suppressor cells in cancer: Recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53-65
7. Liao WT, Michael J. et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell. 2019;35:559-572
8. Zhou Y, Li LK Hu LB. et al. Golgi phosphoprotein 2 (GOLPH2/GP73/GOLM1) interacts with secretory clusterin. Mol Biol Rep. 2011;38:1457-1462
9. Kladney RD, Bulla GA, Guo L. et al. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53-65
10. Duan J, Li X, Huang S. et al. GOLPH2, a gene downstream of ras signaling, promotes the progression of pancreatic ductal adenocarcinoma. Mol Biol Rep. 2018;17:4187-4194
11. Liu G, Zhang Y, He F. et al. Expression of GOLPH2 is associated with the progression of and poor prognosis in gastric cancer. Oncol Rep. 2014;32:2077-2085
12. Ye QH, Zhu WW, Zhang JB. et al. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;12:444-458
13. Yan J, Zhou B, Guo L. et al. GOLM1 upregulates expression of PD-L1 through EGFR/STAT3 pathway in hepatocellular carcinoma. Am J Cancer Res. 2020;10:3705-3720
14. Xue CB, Wang A, Han Q. et al. Discovery of INCB8761/PF-4136309, a potent, selective, and orally bioavailable CCR2 antagonist. ACS Med Chem Lett. 2011;2:913-918
15. Dekker E, Tanis PJ, Vleugels JLA. et al. Colorectal cancer. The Lancet. 2019;394:1467-1480
16. Shami KE, Oeffiger KC, Erb NL. et al. American cancer society colorectal cancer survivorship care guidelines. Cancer J Clin. 2014;64:225-249
17. Chen MH, Jan YH, Hsin PM. et al. Expression of GOLM1 correlates with prognosis in human hepatocellular carcinoma. Ann Surg Oncol. 2013;20:616-624
18. Yan G, Ru Y, Wu KR. et al. GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate. 2018;78:166-177
19. Wang X, Wu T, Wang P. et al. Circular RNA 103862 promotes proliferation and invasion of laryngeal squamous cell carcinoma cells through the miR-493-5p/GOLM1 axis. Front Oncol. 2020;10:1064
20. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674
21. El-Khoueiry A B, Sangro B, Yau T. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492-2502
22. Rebekka W, Viktor F, Hu X. et al. Myeloid-derived suppressor cells hinder the anti-cancer activity of immune checkpoint inhibitors. Front Immunol. 2018;9:1310
23. Abiko K, Mandai J, Hamanishi Y. et al. PD-L1 on tumor cells is induced in ascites and promotes peritoneal dissemination of ovarian cancer through CTL dysfunction. Clin Cancer Res. 2013;19:1363-1374
24. Veglia F, Perego M, Gabrilovich D. et al. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108-119
25. Hoechst B, Ormandy LA. et al. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology. 2008;135:234-243
26. Nywening TM, Belt BA, Cullinan DR. et al. Targeting both tumour-associated CXCR2+ neutrophils and CCR2+ macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut. 2018 313738
Author contact
![]() Corresponding author: Dr. Yunzhi Dang, Department of Radiation Oncology, Shaanxi Provincial People's Hospital, Xi'an 710086, China; Tel: +86-29-8525 2140; Email: dangyunzhiedu.cn
Corresponding author: Dr. Yunzhi Dang, Department of Radiation Oncology, Shaanxi Provincial People's Hospital, Xi'an 710086, China; Tel: +86-29-8525 2140; Email: dangyunzhiedu.cn

 Global reach, higher impact
Global reach, higher impact